Abstract
The Hrp (type III protein secretion) system is essential for the plant parasitic ability of Pseudomonas syringae and most Gram-negative bacterial plant pathogens. AvrB and AvrPto are two P. syringae proteins that have biological activity when produced via heterologous gene expression inside plant cells or when produced by Hrp+ bacteria. Avr-like proteins, presumably injected by the Hrp system on bacterial contact with plant cells, appear to underlie pathogenic interactions, but none has been observed outside of the bacterial cytoplasm, and identifying novel genes encoding them is tedious and uncertain without a phenotype in culture. Here we describe a cloned Hrp secretion system that functions heterologously in Escherichia coli to secrete AvrB and AvrPto in culture and to promote AvrB and AvrPto biological activity in inoculated plants. The hrp gene cluster, carried on cosmid pCPP2156, was cloned from Erwinia chrysanthemi, a pathogen that differs from P. syringae in being host promiscuous. E. coli DH5α carrying pCPP2156, but not related Hrp-deficient cosmids, elicited a hypersensitive response in Nicotiana clevelandii only when also expressing avrB in trans. The use of pAVRB-FLAG2 and pAVRPTO-FLAG, which produce Avr proteins with a C-terminal FLAG-epitope fusion, enabled immunoblot detection of the secretion of these proteins to E. coli(pCPP2156) culture media. Secretion was Hrp dependent, occurred without leakage of a cytoplasmic marker, and did not occur with E. coli(pHIR11), which encodes a functional P. syringae Hrp system. E. coli(pCPP2156) will promote investigation of Avr protein secretion and systematic prospecting for the effector proteins underlying bacterial plant pathogenicity.
The hrp genes of the common bacterial pathogens of plants, in the genera Erwinia, Pseudomonas, Xanthomonas, and Ralstonia, are essential for the characteristic ability of these bacteria to colonize the intercellular spaces of plant tissues and to trigger plant cell death. hrp genes are clustered and encode a type III protein secretion pathway that can deliver Avr (avirulence) proteins into plant cells (1). avr genes are so named because their experimental transfer from one pathogen to another can convert the recipient from virulence to avirulence on host plants carrying a resistance (R) gene that interacts with the avr gene (2). These gene-for-gene interactions are now thought to have a molecular basis in the interactions of Avr proteins and R gene products within the plant cell, and they trigger the hypersensitive response (HR), a defense-associated programmed death of plant cells at the site of pathogen invasion (1). Some avr genes contribute measurably to bacterial virulence (in hosts lacking a cognate R gene) (3), and it is now thought that avr genes collectively are required for the parasitic and pathogenic abilities of these bacteria (4). This hypothesis is based on the observation that hrp mutants, which cannot transfer Avr proteins, fail to multiply in planta, to elicit the HR in nonhosts, or to produce disease in hosts.
Despite the emerging importance of Avr proteins, we have no direct evidence that they travel the Hrp pathway and no knowledge of their function in virulence. We have identified probably only a subset of those that are produced by typical host-specific pathogens, and we have no evidence that they are produced at all by host-promiscuous pathogens. The evidence that Avr proteins are transferred by the Hrp pathway into plants is most complete, although still indirect, with the P. syringae AvrB and AvrPto proteins. Nonpathogenic Escherichia coli and Pseudomonas fluorescens cells that harbor the functional cluster of P. syringae hrp genes carried on cosmid pHIR11 can elicit an HR that depends on both the type III secretion system and AvrB or AvrPto (5, 6). Both Avr proteins trigger an R gene-dependent HR when transiently expressed inside plant cells (5, 7–9), and the interaction of AvrPto and Pto in the yeast two-hybrid system correlates with biological activity (8, 9). However, neither P. syringae, E. coli(pHIR11), nor P. fluorescens(pHIR11) secrete AvrB or AvrPto in culture, presumably because these proteins travel the type III pathway directly into host cells and only on host cell contact, as with the Yop virulence proteins of Yersinia spp. (5, 10). Other known Avr proteins have been observed only in the bacterial cytoplasm (3, 11–14).
The full arsenal of Avr proteins produced by P. syringae and other host-specific pathogens is unknown, and barring a positive avirulence test, it is not possible to determine whether a novel gene encodes one of these proteins. This problem is because Avr proteins lack any common sequence motifs or biochemical activity, and their individual contributions to virulence are typically too weak for reliable detection in mutant screens. Furthermore, their detection via avirulence tests depends unpredictably on test plants carrying a matching R gene (2, 3). Avr proteins have not been reported for host-promiscuous, necrotrophic pathogens like Erwinia chrysanthemi, which characteristically cause soft rots and secrete copious quantities of plant tissue-degrading pectic enzymes via the type II pathway. Nevertheless, E. chrysanthemi carries hrp genes, and pectic enzyme-deficient mutants elicit a typical Hrp-dependent HR in tobacco and other plants (15).
We report here the isolation of a functional cluster of E. chrysanthemi hrp genes that enables E. coli to deliver P. syringae Avr signals in planta and to secrete P. syringae Avr proteins in culture. This finding opens the way for systematic identification of novel P. syringae proteins that travel the type III pathway and provides evidence for Avr protein production by E. chrysanthemi.
MATERIALS AND METHODS
Bacterial Strains, Culture Conditions, and DNA Manipulation Techniques.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were routinely grown in LM medium (16) at 37°C for isolation of plasmids and at 30°C for protein secretion assays. The following concentrations of antibiotics were used in selective media: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; gentamycin (Gm), 10 μg/ml; kanamycin (Km), 50 μg/ml; nalidixic acid (Nx), 20 μg/ml; spectinomycin (Sp), 50 μg/ml; and tetracycline (Tc), 10 μg/ml. Standard procedures were followed for DNA manipulations (17).
Table 1.
Bacterial strains and plasmids used in this work
| Designation | Relevant characteristics and use | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | SupE44 ΔlacU169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1, Nxr | Life Technologies |
| XLOLR | Δ(mcrA) 183 Δ(mcrBC-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 relA1 lac [F′ proAB laqlqZΔM15 Tn10(Tcr)] | Stratagene |
| SM10λpir | SM10 lysogenized with λ-pir for mobilizing pUT∷mini-Tn5Cm, Kmr | 41 |
| MC4100 | F′ araD139 Δ(argF-lacZYA)U169 rpsL150 relA1 flb-5301 ptsF25 deoC1 | 42 |
| Plasmids | ||
| pFLAG-CTC | For construction of C-terminal fusion to FLAG peptide, Apr | Kodak |
| pUT∷mini- Tn5Cm | Mini-Tn5 transposon with Cmr on suicide plasmid pGP704 derivative for transposon mutagenesis, Apr | 43 |
| pML123 | Broad host range expression vector for cloning avrPto-FLAG, Gmr | 44 |
| pPtE6 | An avrPto clone in pDSK519, Kmr | 28 |
| pHIR11 | pLAFR3 carrying Pss61 hrp/hrc cluster, Tcr | 45 |
| pCPP2156 | pCPP19 carrying E. chrysanthemi hrp cluster, Spr | This work |
| pCPP2157 | pCPP19 carrying E. chrysanthemi hrp cluster, Spr | 19 |
| pCPP2329 | pFLAG-CTC carrying avrPto, Apr | This work |
| pAVRB-FLAG2 | pML123 carrying avrB-FLAG, Gmr | 5 |
| pAVRB1 | pDSK519 carrying avrB, Kmr | 27 |
| pAVRPTO-FLAG | pML123 carrying avrPto-FLAG, Gmr | This work |
| pCPP2368 | pCPP2156∷Tn5Cm that has HR− phenotype, Spr, Cmr | This work |
| pCPP2416 | pCPP2156∷Tn5Cm that has HR+ phenotype, Spr, Cmr | This work |
| pCPP2318 | pCPP30 carrying blaM lacking signal peptide sequences, Tcr | 25 |
Ap, ampicillin; GM, gentamycin; Km, kanamycin; Nx, nalidixic acid.
Mini-Tn5Cm Mutagenesis of the hrp Gene Cluster in pCPP2156.
Mini-Tn5Cm mutagenesis of E. coli DH5α(pCPP2156) was initiated by conjugation with E. coli SM10λpir (pUT∷mini-Tn5Cm). Because pUT cannot replicate in E. coli DH5α, Cmr transconjugants have mini-Tn5Cm transposed to the chromosome or pCPP2156. To obtain a pool of pCPP2156∷mini-Tn5Cm plasmids, all Cmr colonies were triparentally mated with E. coli XLOLR (Tcr). The cosmids from E. coli XLOLR transconjugants, selected on LM agar containing Tc, Sp, and Cm, were isolated, and their restriction fragment patterns were compared with each other and pCPP2156. All 46 cosmids initially examined contained random insertions of mini-Tn5Cm in pCPP2156. Based on restriction mapping and DNA sequencing from both ends of mini-Tn5Cm with the primers 5′-AGATCTGATCAAGAGACAG-3′ and 5′-CCGTGTGTATAAGAGTCAG-3′, two different pCPP2156∷mini-Tn5Cm derivatives were chosen. In one of them, Tn5Cm was inserted in the intergenic region between hrpJ and hrcV in the hrpJ operon. This cosmid was named pCPP2368. The other cosmid contained mini-Tn5Cm outside of the hrp cluster and was named pCPP2416. Both cosmids were electroporated into E. coli DH5α.
Plant Bioassays.
Tobacco (Nicotiana tabacum L. cv. Xanthi) and N. clevelandii were grown under greenhouse conditions and then maintained in the lab at room temperature with daylight and supplemental metal halide illumination for HR assays. Tomato (Lycopersicum esculentum Mill. cv. Rio Grande) plants were grown from seeds in pots with Cornell Mix (Cornell University) in the lab at room temperature. E. coli DH5α cells grown overnight on LM plates were washed twice with 5 mM Mes (morpholinoethanesulfonic acid, pH 6.5) by centrifugation and then resuspended in an appropriate volume of the same buffer to an OD600 of 0.8 (experiments involving avrPto used E. coli MC4100 and a 3-fold higher level of inoculum). Previously described procedures were used for the infiltration of bacterial cells into tobacco, tomato, and N. clevelandii leaves (15).
Preparation of AvrB Antibodies.
AvrB-FLAG was purified from E. coli DH5α(pFLAG-CTC∷AvrB) by affinity chromatography as described (5), followed by precipitation of aliquots containing 1 mg of partially purified protein with trichloroacetic acid (20% final concentration), resuspension in SDS polyacrylamide gel loading buffer (17), and electrophoresis on 1.5 mm × 11 cm × 10 cm 12% polyacrylamide preparative gels. The AvrB band was excised from each gel after brief staining with a solution of 0.2% Coomassie R350 (Pharmacia Biotech) dissolved in water. Subsequent extraction of AvrB from the gel matrix and generation of polyclonal rabbit anti-AvrB antisera were performed by the Immunological Resource Center at the University of Illinois, Urbana. Before usage, the antisera was delipified with sodium dextran sulfate (average molecular weight of 500,000) to a final concentration of 0.25% and CaCl2 to a final concentration of 1.0% followed by incubation at 4°C for 8–12 h (18). This mixture was clarified by centrifugation at 12,000 × g at 4°C for 10 min. Proteins were precipitated by the addition of ammonium sulfate at 50% saturation, followed by incubation at 4°C for 8–12 h, and then collected by centrifugation at 12,000 × g at 4°C for 10 min and resuspended in their original volume with PBS.
Construction of pAVRPTO-FLAG.
The avrPto gene was isolated by PCR with Pfu DNA polymerase (Stratagene) and pPtE6 as the template. The upper primer was 5′-GAGCGAGCATATGGGAAATATATGTGTCGGC-3′ with an NdeI site, and the lower primer was 5′-ATTGTAGTCGACTTGCCAGTTACGGTACGGG-3′ with a SalI site. The reaction products from 30 PCR cycles were resolved by electrophoresis through 0.7% agarose, and the avrPto DNA was isolated by using an Eluquick kit (Schleicher & Schuell), followed by digestion with NdeI and SalI. This DNA was cloned into pFLAG-CTC, previously digested with NdeI and SalI and named pCPP2329. The avrPto-FLAG DNA was isolated from pCPP2329 by digestion with SspI and cloned into pML123, which had been previously digested with BamHI and blunted with Klenow polymerase, producing pAVRPTO-FLAG. As with avrB-FLAG2 (5), avrPto-FLAG is expressed by both the tac promoter (from pFLAG-CTC) and the pML123 nptII promoter, thus permitting constitutive expression in LM medium and in planta.
Preparation of Protein Samples from Supernatant and Cell Fractions.
Bacteria grown overnight on LM plates at 37°C were washed twice by centrifugation and resuspended in LM broth. Each bacterial suspension was diluted to OD600 = 0.2 in 40 ml of LM broth containing appropriate antibiotics and cultured at 30°C in a rotary shaking incubator at 220 rpm until the OD600 reached 0.8. Centrifugations for the separation of bacterial cultures into cell-bound and supernatant fractions were performed with an SS-34 rotor (DuPont) at 4°C.
Forty milliliters of culture was initially centrifuged at 4,300 × g for 15 min. For the supernatant fraction, the upper 20 ml of supernatant was carefully transferred to a new centrifuge tube and further centrifuged at 17,200 × g for 40 min, followed by transfer of the upper 10 ml of supernatant to a new tube. Six milliliters of 25% trichloroacetic acid was added to the supernatant fraction, which then was kept on ice for 3–4 h followed by centrifugation at 17,200 × g for 40 min. The pellet subsequently was washed with 20 ml of ice-cold acetone and then resuspended in 200 μl or 100 μl of 1× SDS sample buffer (New England Biolabs). For the cell fraction, the pellet from the initial centrifugation was resuspended in 4 ml of LM broth. One hundred microliters of bacterial cell suspension was mixed with 50 μl of 3× SDS sample buffer. Each protein sample was held in a boiling water bath for 5 min before electrophoresis, and then 15 μl of each sample was loaded onto the gel.
Immunoblot Analysis.
Protein samples were separated by electrophoresis through a 10% SDS/PAGE. Proteins in the gel then were electrotransferred to Immobilon-P membrane (Millipore) with a Semi-Phor system (Hoefer). AvrB-FLAG and AvrPto-FLAG were detected with the Western-Light Plus kit (Tropix, Bedford, MA) by using anti-FLAG M2 antibodies (Kodak Scientific Imaging Systems) and anti-mouse IgG alkaline phosphatase conjugate (Sigma) as primary and secondary antibodies, respectively. AvrB and β-lactamase were detected with the same system except by using anti-AvrB antibodies or anti-β-lactamase antibodies (5 prime → 3 prime) and anti-rabbit IgG alkaline phosphatase conjugate (Sigma) as primary and secondary antibodies, respectively.
Primers, DNA Sequencing, and Data Analysis.
Oligonucleotide synthesis and DNA sequencing were performed at the Cornell Biotechnology Center. DNA sequence data were managed and analyzed with the DNAStar program (DNAStar, Madison, WI).
RESULTS
Cosmid pCPP2156, but Not pCPP2157, Carries an Intact E. chrysanthemi hrp Gene Cluster.
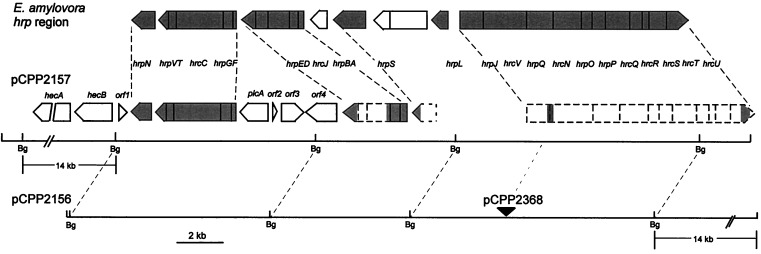
Several cosmids carrying E. chrysanthemi EC16 hrp genes previously were isolated on the basis of their ability to hybridize with an E. amylovora DNA fragment carrying the hrpJ operon (15). One of these cosmids, pCPP2157, is shown in Fig. 1 and subsequently was found to also carry hrpN and the complete hrpC operon (19, 20). Although pCPP2157 appeared to carry both borders of the E. chrysanthemi hrp cluster, E. coli(pCPP2157) failed to elicit an HR in tobacco leaves. DNA sequencing of the right end of the pCPP2157 insert revealed that hrcU was missing the last 180 nucleotides, as suggested by comparison with the E. amylovora hrcU gene (21)(data not shown). hrcU is one of nine former hrp genes that encode core components of the type III secretion system, are broadly conserved in plant and animal pathogenic bacteria, and have been renamed as hrc (HR and conserved) genes (22). Because of the hrcU truncation, additional cosmids hybridizing with probes carrying hrpN and hrcU were analyzed. pCPP2156 was one of those. Partial DNA sequence analysis and physical map comparisons with the E. amylovora hrp genes suggested that pCPP2156 carried the entire E. chrysanthemi hrp gene cluster, including at least one intercalated region not obviously related to Hrp function and 14 kb of additional DNA beyond hrcU (Fig. 1). However, pCPP2156 failed to elicit an HR in tobacco (data not shown).
Figure 1.
Physical maps of pCPP2156 and pCPP2157, which contain the E. chrysanthemi hrp region, and comparison of the hrp regions of E. chrysanthemi and E. amylovora (19–21, 46–48). Arrow-shaped boxes denote putative transcriptional units. Shaded areas in E. amylovora hnp region denote hrp/hrc genes. Dashed boxes denote transcriptional units predicted on the basis of the homology and spacing of partially sequenced regions (shaded areas) in comparison with the corresponding E. amylovora hrp genes. The filled triangle indicates the location of mini-Tn5Cm in pCPP2368.
E. coli(pCPP2156) Enables Elicitation of an AvrB-Dependent HR in N. clevelandii.
We had determined previously that pHIR11, which carries the intact P. syringae pv syringae 61 hrp cluster, enables E. coli to elicit an HR in tobacco because it also carries hrmA, an avr-like gene whose transient expression in tobacco cells is lethal (23, 24). This finding suggested the possibility that pCPP2156 failed to elicit an HR in tobacco because it did not carry an appropriate avr gene. To test this possibility, we transformed pAVRB-FLAG2 into E. coli DH5α cells carrying either pCPP2156 or pCPP2157. pAVRB-FLAG2 expresses the P. syringae pv glycinea avrB gene such that the product has an 8-aa FLAG epitope C-terminal fusion (5). Transformants were infiltrated at a concentration of 5 × 108 cfu/ml into N. clevelandii, a plant that reacts hypersensitively to Hrp+ bacteria carrying avrB (R.W. Innes, personal communication). A typical HR developed within 24 h in panels inoculated with bacteria carrying both avrB and pCPP2156, but there was no response in panels inoculated with bacteria lacking avrB or carrying an incomplete hrp cluster (Fig. 2), or inoculated with bacteria carrying only pAVRB-FLAG2 (data not shown).
Figure 2.
Elicitation of Hrp-dependent HR in leaves of N. clevelandii by E. coli DH5α carrying E. chrysanthemi hrp clusters that are either intact (pCPP2156, pCPP2416) or defective (pCPP2157, pCPP2368). N. clevelandii leaves were infiltrated with bacteria at a concentration of 5 × 108 cfu/ml. Leaves were photographed 48 h after infiltration. Tissue collapse occurred within 24 h. The area below each number on the leaf was infiltrated with E. coli DH5α carrying the following constructs: 1, pCPP2156; 2, pCPP2156 and pAVRB-FLAG2; 3, pCPP2157; 4, pCPP2157 and pAVRB-FLAG2; 5, pCPP2416 and pAVRB-FLAG2; and 6, pCPP2368 and pAVRB-FLAG2.
The failure of E. coli(pCPP2157, pAVRB-FLAG2) to elicit an HR in N. clevelandii suggested that this ability was Hrp dependent. However, an explanation based on differences in the DNA flanking the hrp gene clusters in pCPP2156 and pCPP2157 remained a formal possibility. To resolve this problem, we mutated pCPP2156 with mini-Tn5Cm and isolated two derivatives. Restriction mapping and DNA sequence analysis revealed that pCPP2416 and pCPP2368 carried insertions in the 14-kb region beyond hrcU and in the intergenic region between hrpJ and hrcV, respectively (Fig. 1). The mutation in pCPP2368 would be expected to block transcription of hrcV and downstream genes in the putative hrpJ operon, and a polar mutation in the hrpJ operon of P. syringae pv syringae 61 has been shown to result in accumulation of the HrpZ harpin within the bacterial cytoplasm (25). E. coli cells carrying pCPP2416 and pCPP2368 were transformed with pAVRB-FLAG2 and tested for their ability to elicit the HR in N. clevelandii. An HR was observed only with E. coli(pCPP2416, pAVRB-FLAG2) (Fig. 2).
E. coli(pCPP2156) Secretes AvrB in Culture in a Hrp-Dependent Manner While Retaining β-Lactamase.
The secretion of Avr proteins by P. syringae is presumed to depend on host cell contact because Avr proteins remain cytoplasmic in culture even when the Hrp system is actively secreting harpins (1). The seemingly less specialized interaction of E. chrysanthemi with its hosts suggested the possibility that Avr secretion may be regulated less tightly. To test this, we exploited the FLAG epitope-tagged AvrB encoded by pAVRB-FLAG2 to determine whether E. coli(pCPP2156) could secrete AvrB in culture. Bacterial cultures in late logarithmic-phase growth were separated into supernatant and cell-bound fractions by centrifugation, and then proteins in both fractions were resolved by SDS/PAGE. AvrB-FLAG was visualized by immunoblotting with anti-FLAG mAbs and chemiluminescent detection. AvrB-FLAG was found in the supernatant of E. coli(pCPP2156)(Fig. 3). Although much of the AvrB remained in the cell-bound fraction, secretion was Hrp dependent and specific in that no AvrB-FLAG was found in the supernatant of E. coli(pCPP2368)(Fig. 3), and Coomassie staining revealed equally low levels of protein in the supernatant fractions of all of the bacteria tested (data not shown).
Figure 3.
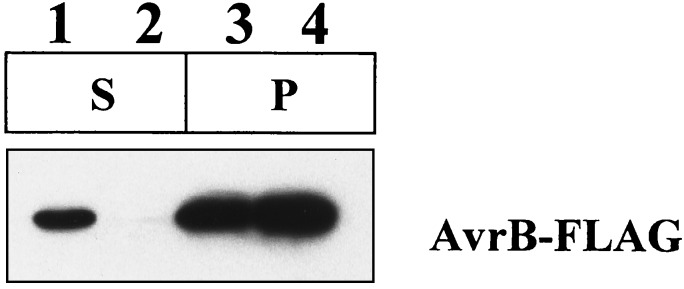
Differential secretion of AvrB-FLAG by E. coli DH5α carrying either a wild-type (pCPP2156) or mutant (pCPP2368) E. chrysanthemi hrp cluster. The supernatant fraction (S) was concentrated 7.5 times more than the cell pellet fraction (P). Lane 1, E. coli(pCPP2156, pAVRB-FLAG2); lane 2, E. coli(pCPP2368, pAVRB-FLAG2); lane 3, E. coli(pCPP2156, pAVRB-FLAG2); and lane 4, E. coli(pCPP2368, pAVRB-FLAG2).
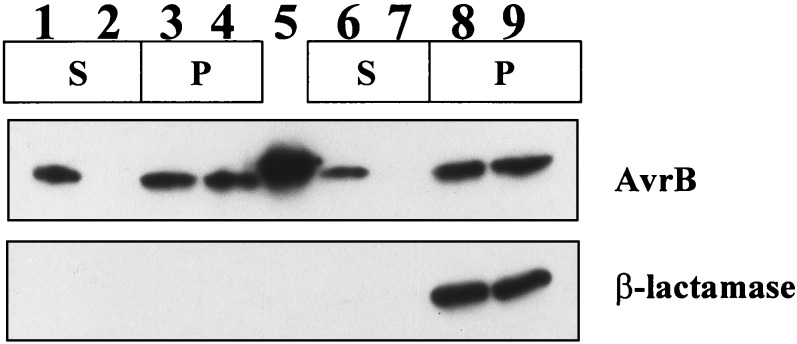
To confirm that the presence of AvrB-FLAG in the E. coli(pCPP2156) medium resulted from specific secretion and not cell lysis, and that secretion was not because of the FLAG epitope, we simultaneously monitored the localization of AvrB and mature β-lactamase (Fig. 4). E. coli cells carrying pCPP2156 or pCPP2368 first were transformed with pAVRB1, which expresses AvrB from the lac promoter (27), and pCPP2318, which encodes a mature β-lactamase that lacks its N-terminal signal peptide and can be used as a cytoplasmic marker (25). The distribution of AvrB and β-lactamase in the same supernatant and cell-bound fraction samples was monitored by immunoblotting with appropriate antibodies. The E. coli(pCPP2156) supernatant sample contained AvrB but no β-lactamase (Fig. 4), indicating that AvrB secretion occurred without the FLAG epitope and without cell lysis.
Figure 4.
Differential secretion of AvrB by E. coli DH5α carrying either a wild-type (pCPP2156) or mutant (pCPP2368) E. chrysanthemi hrp cluster. pCPP2318 encodes mature BlaM, used as a cytoplasmic marker. The supernatant fraction (S) was concentrated 7.5 times more than the cell pellet fraction (P) for lanes 1 and 2 and 15× more for lanes 6 and 7. Lane 1, E. coli(pCPP2156, pAVRB1); lane 2, E. coli(pCPP2368, pAVRB1); lane 3, E. coli(pCPP2156, pAVRB1); lane 4, E. coli(pCPP2368, pAVRB1); lane 5, purified AvrB; lane 6, E. coli(pCPP2156, pAVRB1, pCPP2318); lane 7, E. coli(pCPP2368, pAVRB1, pCPP2318); lane 8, E. coli(pCPP2156, pAVRB1, pCPP2318); and lane 9, E. coli(pCPP2368, pAVRB1, pCPP2318).
E. coli(pCPP2156) Secretes AvrPto in Culture in a Hrp-Dependent Manner.
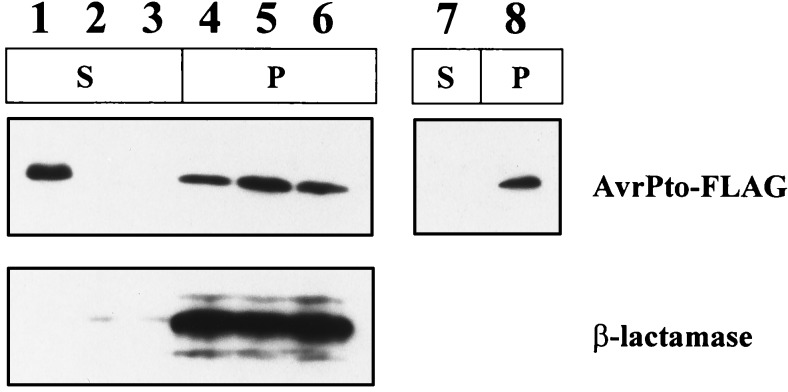
The evidence for Avr action inside plant cells after Hrp-dependent transfer is strongest with AvrPto, whose structural gene originally was isolated from P. syringae pv tomato (1, 8, 9, 28). Consequently, we chose AvrPto to test whether the ability of E. coli(pCPP2156) to deliver P. syringae Avr signals in planta and to secrete Avr proteins in culture would extend beyond AvrB. We first constructed pAVRPTO-FLAG, which encodes AvrPto with a C-terminal FLAG epitope fusion. E. coli cells carrying pAVRPTO-FLAG and pCPP2156 (but not pAVRPTO-FLAG alone, pCPP2156 alone, or pAVRPTO-FLAG with pCPP2368) elicited an HR in tomato cultivar Rio Grande carrying the Pto resistance gene (data not shown). The secretion of AvrPto was determined with the same methods used for AvrB-FLAG and AvrB. AvrPto-FLAG was secreted by E. coli(pCPP2156) but not by E. coli(pCPP2157) or E. coli(pCPP2368)(Fig. 5). Thus, secretion of AvrPto-FLAG was Hrp dependent, and it also occurred without leakage of β-lactamase. In contrast, no AvrPto-FLAG was found in the supernatant of E. coli(pHIR11), which expresses the intact P. syringae Hrp system (Fig. 5).
Figure 5.
Differential secretion of AvrPto-FLAG by E. coli DH5α carrying either intact (pCPP2156) or defective (pCPP2157, pCPP2368) E. chrysanthemi hrp clusters and E. coli MC4100 carrying pHIR11. The supernatant fraction (S) was concentrated 7.5 times more than the cell pellet fraction (P). Lane 1, E. coli(pCPP2156, pCPP2318, pAVRPTO-FLAG); lane 2, E. coli(pCPP2157, pCPP2318, pAVRPTO-FLAG); lane 3, E. coli(pCPP2368, pCPP2318, pAVRPTO-FLAG); lane 4, E. coli(pCPP2156, pCPP2318, pAVRPTO-FLAG); lane 5, E. coli(pCPP2157, pCPP2318, pAVRPTO-FLAG); lane 6, E. coli(pCPP2368, pCPP2318, pAVRPTO-FLAG); lane 7, E. coli(pHIR11, pAVRPTO-FLAG); and lane 8, E. coli(pHIR11, pAVRPTO-FLAG).
DISCUSSION
The isolation of a cluster of E. chrysanthemi hrp genes that directs E. coli to secrete P. syringae Avr proteins in culture and deliver Avr signals in planta has several implications for the pathogenic biology of E. chrysanthemi and P. syringae. As representative necrotrophic and biotrophic parasites, respectively, these two bacteria mark the extremes in the pathogenic personalities of the common Gram-negative phytopathogenic bacteria. Nevertheless, we now can propose that they elicit the HR and initiate parasitic attack in fundamentally similar ways, that they may be able to interchange their avr genes without loss of function, and that their cloned hrp clusters can be used to biochemically investigate Avr protein secretion and to systematically prospect for the proteins injected into plants by many plant pathogenic bacteria.
E. chrysanthemi and P. syringae appear to elicit the HR by the same mechanism in that their cloned hrp clusters depend on an appropriate avr gene for elicitation of the HR when heterologously expressed in nonpathogens. Thus, cosmid pHIR11 (P. syringae hrp cluster) directs HR elicitation in tobacco because it carries the avr-like hrmA gene. Cosmid pCPP2156 (E. chrysanthemi hrp cluster) fails to elicit the HR in tobacco, N. clevelandii, or tomato because it does not carry an avr gene that is recognized by these plants, but when provided with avrB or avrPto it appropriately directs elicitation of the HR in N. clevelandii and tomato cultivar Rio Grande. This finding has two implications regarding HR elicitation by E. chrysanthemi. First, the harpin encoded by pCPP2156, like that encoded by pHIR11, is apparently insufficient for bacterial HR elicitation (although both harpins can elicit programmed cell death when delivered exogenously)(19, 29). Second, E. chrysanthemi must carry avr genes somewhere outside the region cloned in pCPP2156 because it is able to elicit a Hrp-dependent HR in tobacco without provision of a heterologous avr gene (15).
E. chrysanthemi hrp mutants also are reduced in their ability to elicit infection at low levels of inoculum. Because it now appears that the primary function of the Hrp system is to deliver Avr-like proteins to host cells, identifying these proteins and determining their function will be key to understanding how E. chrysanthemi initiates infection. Recent observations with E. amylovora indicate that homologous avr-like genes are present in Erwinia spp. and P. syringae (30, 31). Specifically, dspE, which is required for the pathogenicity of E. amylovora, is a homolog of avrE, a gene that contributes quantitatively to the virulence of P. syringae pv tomato strain PT23 on tomato and has an Avr phenotype in P. syringae pv glycinea when tested on a variety of soybean cultivars (32, 33). The ability of avrE to restore the pathogenicity of an E. amylovora dspE mutant provides direct evidence that a P. syringae avr gene can function biologically in an Erwinia background. (31). Furthermore, DspE-specific antibodies and appropriate hrp mutants have been used to establish that E. amylovora secretes DspE in a Hrp-dependent manner in culture (34). However, it is not known whether AvrE can be secreted in culture by E. amylovora (or P. syringae) or whether DspE and AvrE function inside plant cells.
The regulation of the E. chrysanthemi Hrp system appears more relaxed in two ways in comparison with host-specific pathogens like P. syringae. First, the E. chrysanthemi hrp genes are not repressed by complex media (which enhances the utility of the system for secretion studies) (35). Second, the E. chrysanthemi Hrp system does not appear to be gated in culture with respect to the secretion of Avr proteins. Although the P. syringae hrp cluster carried on pHIR11 enables delivery of AvrB and AvrPto signals (presumably the Avr proteins themselves) to plant cells, it does not direct secretion of these proteins in culture (Fig. 5) (5). Because E. chrysanthemi and P. syringae possess similar Hrp systems (both in group I) (1, 20), comparisons and genetic exchanges between them are likely to be useful for elucidating the mechanisms controlling Avr protein secretion in P. syringae.
There is no direct evidence yet for the Hrp-mediated transfer of any Avr protein into plant cells, although the indirect evidence for this is particularly compelling with AvrB and AvrPto, as discussed above. Our observation that these two proteins can travel the Hrp pathway to the bacterial milieu now provides direct confirmation of the first step in the translocation process. More importantly, the targeting signals controlling secretion and other aspects of the secretion process now can be explored in vitro. In this regard, the differing traffic specificities of the type II and type III protein secretion systems of E. chrysanthemi are noteworthy, especially because both systems function heterologously in E. coli. The cloned cluster of out (type II secretion) genes from E. chrysanthemi EC16 directs the secretion of pectate lyase isozymes expressed from E. chrysanthemi pel genes but not from E. carotovora pel genes (36). This species-specific secretion occurs despite the fact that the Out systems and some of the Pels of these two species are homologous (37). The construction of hybrid Pels has shown that the targeting information controlling species-specific secretion resides in the tertiary structure of these proteins (38). In contrast, the E. chrysanthemi Hrp (type III) system lacks even genus specificity for its traffic, and the secreted proteins may be devoid of targeting information. This notion is based on the possibility that targeting information resides in the mRNA encoding the N termini of these proteins, as has been demonstrated recently for the YopE and YopN proteins secreted by the Yersinia type III pathway (39). Use of the cloned E. chrysanthemi Hrp secretion system should make testing this hypothesis and the identification of targeting signals straightforward.
E. coli heterologously expressing the E. chrysanthemi Hrp system also can be used to systematically prospect for genes from E. chrysanthemi, P. syringae, and possibly other bacteria that encode Avr-like effector proteins. However, two factors may limit universal application of this system. First, some Avr-like proteins may require a dedicated chaperone, as has been observed with Yersinia Yops (although characteristic gene arrangements and structural properties of the chaperones may help identify them) (40). Second, we do not know whether E. coli(pCPP2156) will secrete Avr-like proteins derived from pathogens like Ralstonia solanacearum and Xanthomonas spp., which possess group II Hrp systems (1).
Several questions arise when the actions of putative Avr-like effector proteins are considered in the context of the host-promiscuous and necrotrophic parasitism of E. chrysanthemi. Will they include suppressors that “disarm” defenses associated with the HR? Will they include homologs of known Avr proteins and reveal evidence of horizontal transfer with P. syringae and other host-specific pathogens? Will heterologously expressed P. syringae avr genes confer avirulence phenotypes and restrict the host range of E. chrysanthemi? In summary, pCPP2156 is likely to promote new investigations into mechanisms of Hrp secretion and the functions of effector proteins in diverse plant pathogens.
Acknowledgments
We thank Noel T. Keen for pAVRB1, Kent Loeffler for photography, and Amy O. Charkowski for helpful discussions. We also thank Jihyun F. Kim for providing the oligonucleotides from which both ends of pCPP2157 were sequenced. This work was supported by Grants MCB-9631530 from the National Science Foundation and 97-35303-4488 from the National Research Initiative Competitive Grants Program/U.S. Department of Agriculture.
ABBREVIATIONS
- avr
avirulence
- HR
hypersensitive response
- Cm
chloramphenical
- Sp
spectinomycin
- Tc
tetracycline
References
- 1.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keen N T. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 3.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 4.Alfano J R, Collmer A. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Mol Plant–Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 7.Leister R T, Ausubel F M, Katagiri F. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2062. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 9.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis G R, Wolf-Watz H. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 11.Knoop V, Staskawicz B, Bonas U. J Bacteriol. 1991;173:7142–7150. doi: 10.1128/jb.173.22.7142-7150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown I, Mansfield J, Irlam I, Conrads-Strauch J, Bonas U. Mol Plant–Microbe Interact. 1993;6:376–386. [Google Scholar]
- 13.Young S A, White F F, Hopkins C M, Leach J E. Mol Plant–Microbe Interact. 1994;7:799–804. doi: 10.1094/mpmi-7-0799. [DOI] [PubMed] [Google Scholar]
- 14.Puri N, Jenner C, Bennet M, Stewart R, Mansfield J, Lyons N, Taylor J. Mol Plant–Microbe Interact. 1997;10:247–256. doi: 10.1094/MPMI.1997.10.2.247. [DOI] [PubMed] [Google Scholar]
- 15.Bauer D W, Bogdanove A J, Beer S V, Collmer A. Mol Plant–Microbe Interact. 1994;7:573–581. doi: 10.1094/mpmi-7-0573. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Walton K W, Scott P J. J Clin Pathol. 1964;17:627–643. doi: 10.1136/jcp.17.6.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer D W, Wei Z-M, Beer S V, Collmer A. Mol Plant–Microbe Interact. 1995;8:484–491. doi: 10.1094/mpmi-8-0484. [DOI] [PubMed] [Google Scholar]
- 20.Kim J F, Ham J H, Bauer D W, Collmer A, Beer S V. Mol Plant–Microbe Interact. 1998;11:563–567. doi: 10.1094/MPMI.1998.11.6.563. [DOI] [PubMed] [Google Scholar]
- 21.Bogdanove A J, Wei Z-M, Zhao L, Beer S V. J Bacteriol. 1996;178:1720–1730. doi: 10.1128/jb.178.6.1720-1730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 23.Alfano J R, Bauer D W, Milos T M, Collmer A. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 24.Alfano J R, Kim H-S, Delaney T P, Collmer A. Mol Plant–Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 25.Charkowski A O, Huang H-C, Collmer A. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staskawicz B, Dahlbeck D, Keen N, Napoli C. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamaki S, Dahlbeck D, Staskawicz B, Keen N T. J Bacteriol. 1988;170:4846–4854. doi: 10.1128/jb.170.10.4846-4854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronald P C, Salmeron J M, Carland F M, Staskawicz B J. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He S Y, Huang H-C, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 30.Gaudriault S, Malandrin L, Paulin J-P, Barny M-A. Mol Microbiol. 1997;26:1057–1069. doi: 10.1046/j.1365-2958.1997.6442015.x. [DOI] [PubMed] [Google Scholar]
- 31.Bogdanove A J, Kim J F, Wei Z, Kolchinsky P, Charkowski A O, Conlin A K, Collmer A, Beer S V. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorang J M, Shen H, Kobayashi D, Cooksey D, Keen N T. Mol Plant–Microbe Interact. 1994;7:508–515. [Google Scholar]
- 33.Lorang J M, Keen N T. Mol Plant–Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 34.Bogdanove A J, Bauer D W, Beer S V. J Bacteriol. 1998;180:2244–2247. doi: 10.1128/jb.180.8.2244-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collmer A, Bauer D W, Alfano J R, Preston G, Loniello A O, Huang H-C, He S Y. In: Advances in Molecular Genetics of Plant-Microbe Interactions. Daniels M J, editor. Vol. 3. Dordrecht, The Netherlands: Kluwer; 1994. pp. 49–56. [Google Scholar]
- 36.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindeberg M, Salmond G P C, Collmer A. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 38.Lindeberg M L, Boyd C M, Keen N T, Collmer A. J Bacteriol. 1998;180:1431–1437. doi: 10.1128/jb.180.6.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson D M, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 40.Wattiau P, Woestyn S, Cornelis G R. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 41.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 43.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labes M, Puhler A, Simon R. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 45.Huang H-C, Schuurink R, Denny T P, Atkinson M M, Baker C J, Yucel I, Hutcheson S W, Collmer A. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Z-M, Laby R J, Zumoff C H, Bauer D W, He S Y, Collmer A, Beer S V. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 47.Wei Z-M, Beer S V. J Bacteriol. 1993;175:7958–7967. doi: 10.1128/jb.175.24.7958-7967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J F, Wei Z-M, Beer S V. J Bacteriol. 1997;179:1690–1697. doi: 10.1128/jb.179.5.1690-1697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]