Abstract
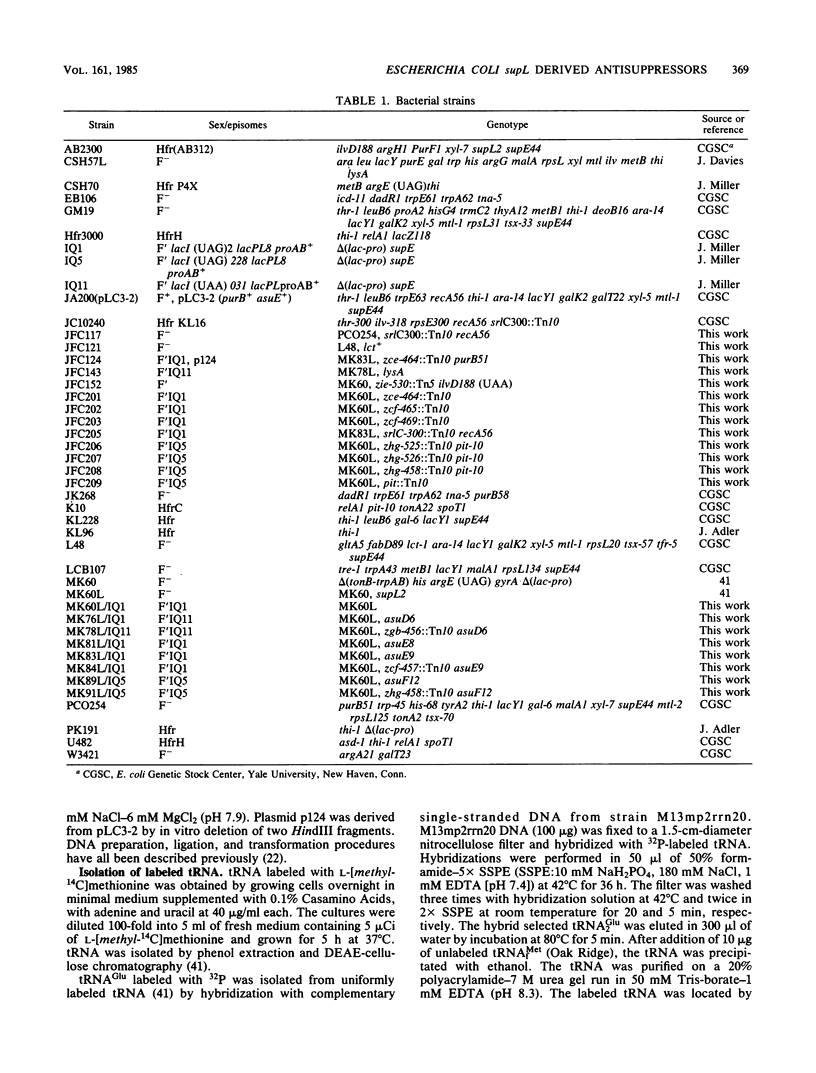
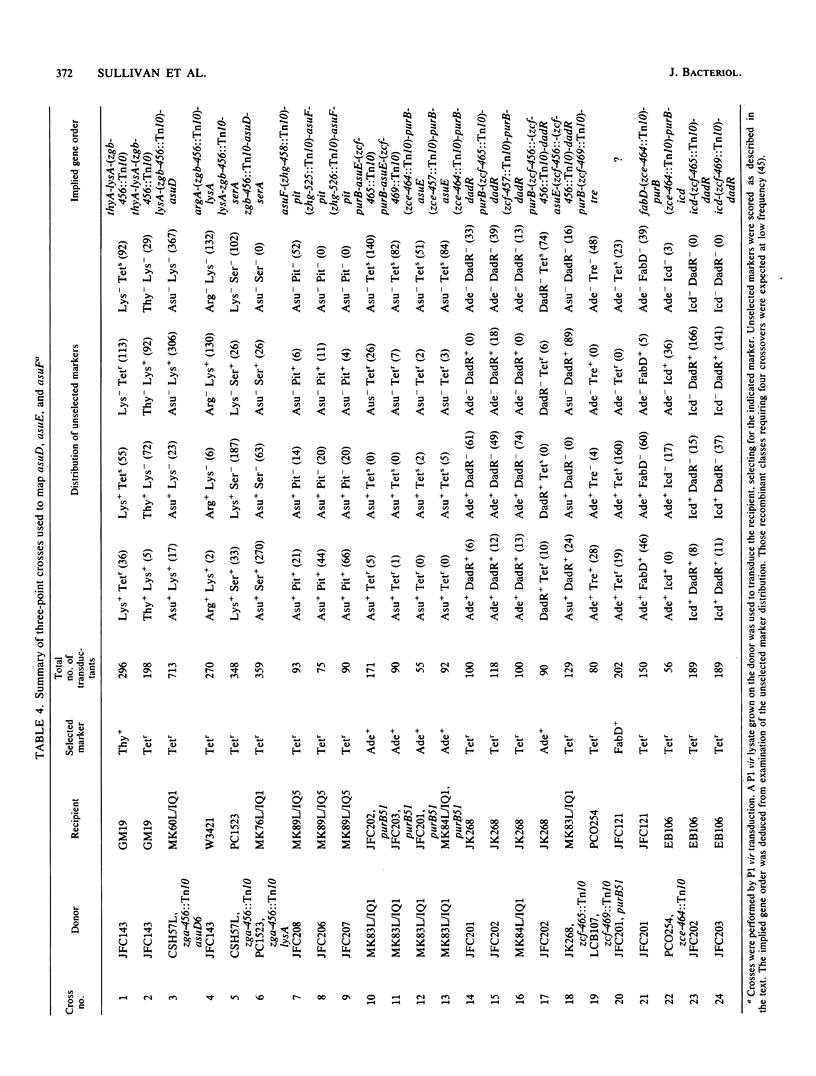
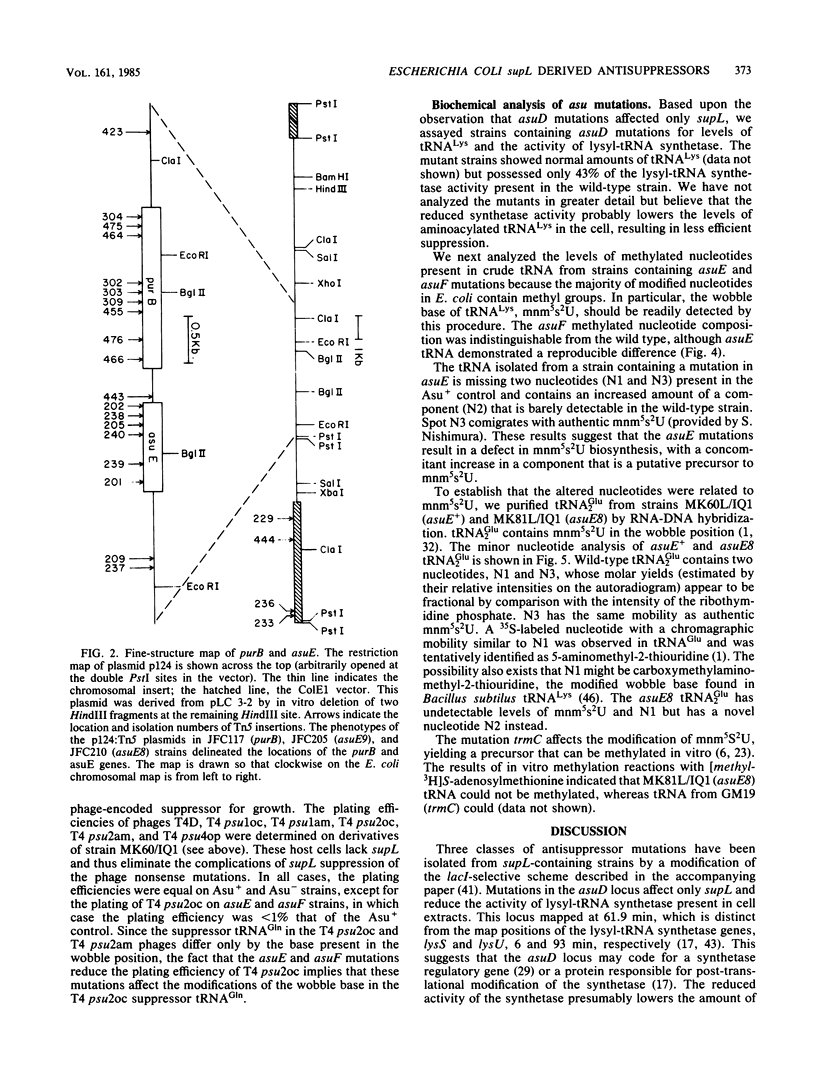
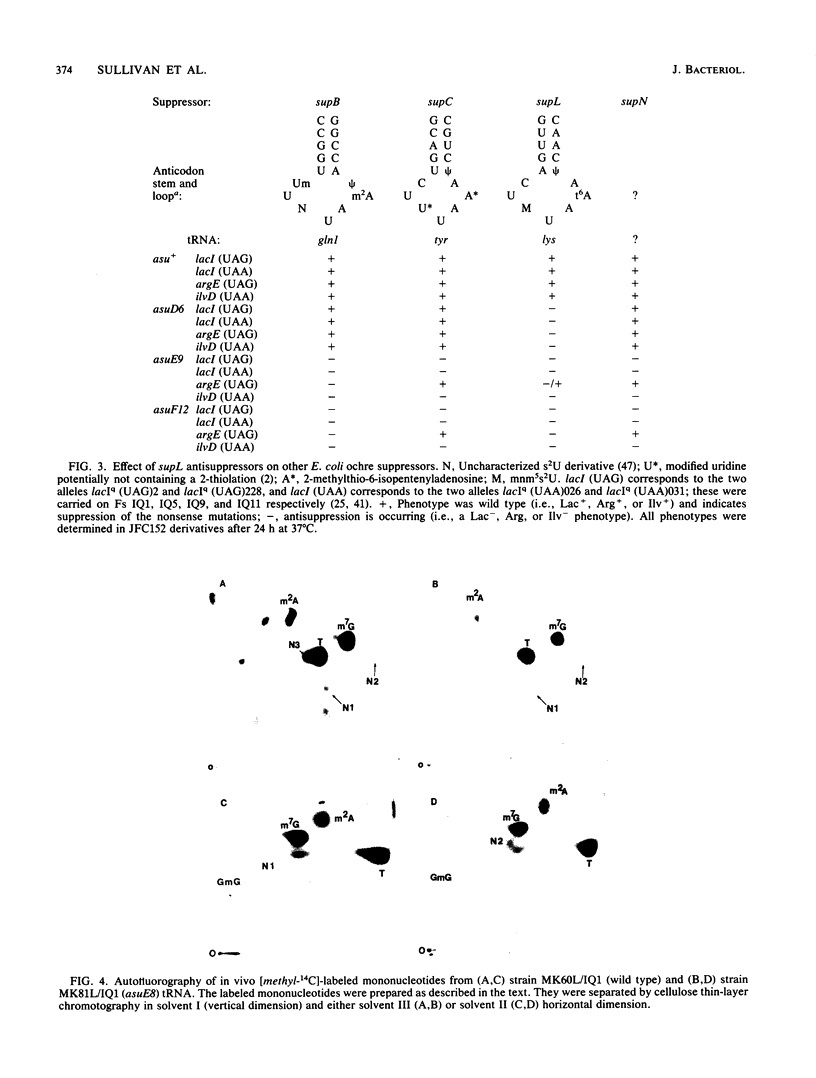
Mutations in three Escherichia coli K-12 genes were isolated that reduce the efficiency of the lysine-inserting nonsense suppressor supL. These antisuppressor mutations asuD, asuE, and asuF map at 61.9, 25.3, and 76.3 min, respectively, on the E. coli chromosome. Biochemical and genetic analysis of the mutant strains revealed the reason for the antisuppressor phenotype for two of these genes. The activity of lysyl-tRNA synthetase was reduced in strains with asuD mutations. The modification of 5-methylaminomethyl-2-thiouridine, the wobble base of tRNALys, was impaired in asuE mutant strains, presumably at the 2-thiolation step.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agris P. F., Söll D., Seno T. Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry. 1973 Oct 23;12(22):4331–4337. doi: 10.1021/bi00746a005. [DOI] [PubMed] [Google Scholar]
- Altman S. A modified uridine in the anticodon of E. coli tRNA I Tyr su + oc. Nucleic Acids Res. 1976 Feb;3(2):441–448. doi: 10.1093/nar/3.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Weiss A., Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980 May;142(2):439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Chakraburtty K., Steinschneider A., Case R. V., Mehler A. H. Primary structure of tRNA-Lys of E. coli B. Nucleic Acids Res. 1975 Nov;2(11):2069–2075. doi: 10.1093/nar/2.11.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Colby D. S., Schedl P., Guthrie C. A functional requirement for modification of the wobble nucleotide in tha anticodon of a T4 suppressor tRNA. Cell. 1976 Nov;9(3):449–463. doi: 10.1016/0092-8674(76)90090-8. [DOI] [PubMed] [Google Scholar]
- Comer M. M., Foss K., McClain W. H. A mutation of the wobble nucleotide of a bacteriophage T4 transfer RNA. J Mol Biol. 1975 Dec 5;99(2):283–293. doi: 10.1016/s0022-2836(75)80146-x. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Greener A., Hill C. W. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980 Oct;144(1):312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Harada F., Nishimura S. Possible anticodon sequences of tRNA His , tRNA Asm , and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry. 1972 Jan 18;11(2):301–308. doi: 10.1021/bi00752a024. [DOI] [PubMed] [Google Scholar]
- Hirshfield I. N., Bloch P. L., Van Bogelen R. A., Neidhardt F. C. Multiple forms of lysyl-transfer ribonucleic acid synthetase in Escherichia coli. J Bacteriol. 1981 Apr;146(1):345–351. doi: 10.1128/jb.146.1.345-351.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Liu C., Yeh F. M. Two modes of metabolic regulation of lysyl-transfer ribonucleic acid synthetase in Escherichia coli K-12. J Bacteriol. 1977 Aug;131(2):589–597. doi: 10.1128/jb.131.2.589-597.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kao S. H., McClain W. H. U-G-A suppressor of bacteriophage T4 associated with arginine transfer RNA. J Biol Chem. 1977 Nov 25;252(22):8254–8257. [PubMed] [Google Scholar]
- LaRossa R., Vögell G., Low K. B., Söll D. Regulation of biosynthesis of aminoacyl-tRNA synthetases and of tRNA in Escherichia coli. II. Isolation of regulatory mutants affecting leucyl-tRNA synthetase levels. J Mol Biol. 1977 Dec 25;117(4):1033–1048. doi: 10.1016/s0022-2836(77)80011-9. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R., Söll D., Kwong T. C. Isolation and partial characterization of three Escherichia coli mutants with altered transfer ribonucleic acid methylases. J Bacteriol. 1975 Apr;122(1):257–265. doi: 10.1128/jb.122.1.257-265.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Guthrie C., Barrell B. G. The psu1+ amber suppressor gene of bacteriophage T4: identification of its amino acid and transfer RNA. J Mol Biol. 1973 Dec 5;81(2):157–171. doi: 10.1016/0022-2836(73)90186-1. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Albertini A. M. Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983 Feb 15;164(1):59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Morgan S. D., Söll D. Regulation of the biosynthesis of aminoacid: tRNA ligases and of tRNA. Prog Nucleic Acid Res Mol Biol. 1978;21:181–207. doi: 10.1016/s0079-6603(08)60270-6. [DOI] [PubMed] [Google Scholar]
- Morgan S., Körner A., Low K. B., Söll D. Regulation of biosynthesis of aminoacyl-tRNA synthetases and of tRNA in Escherichia coli. I. Isolation and characterization of a mutant with elevated levels of tRNAGln 1. J Mol Biol. 1977 Dec 25;117(4):1013–1031. doi: 10.1016/s0022-2836(77)80010-7. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oashi Z., Saneyoshi M., Harada F., Hara H., Nishimura S. Presumed anticodon structure of glutamic acid tRNA from E. coli: a possible location of a 2-thiouridine derivative in the first position of the anticodon. Biochem Biophys Res Commun. 1970 Aug 24;40(4):866–872. doi: 10.1016/0006-291x(70)90983-6. [DOI] [PubMed] [Google Scholar]
- Randerath E., Yu C. T., Randerath K. Base analysis of ribopolynucleotides by chemical tritium labeling: a methodological study with model nucleosides and purified tRNA species. Anal Biochem. 1972 Jul;48(1):172–198. doi: 10.1016/0003-2697(72)90181-9. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Comer M. M., McClain W. H. Nucleotide alterations in the bacteriophage T4 glutamine transfer RNA that affect ochre suppressor activity. J Mol Biol. 1974 Dec 25;90(4):677–689. doi: 10.1016/0022-2836(74)90532-4. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Takeishi K., Ukita T. Specificity of yeast glutamic acid transfer RNA for codon recognition. Biochim Biophys Acta. 1969 Jun 17;182(2):411–426. doi: 10.1016/0005-2787(69)90192-0. [DOI] [PubMed] [Google Scholar]
- Semple K. S., Silbert D. F. Mapping of the fabD locus for fatty acid biosynthesis in Escherichia coli. J Bacteriol. 1975 Mar;121(3):1036–1046. doi: 10.1128/jb.121.3.1036-1046.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G. C., Ghosh H. P. Role of modified nucleosides in tRNA: effect of modification of the 2-thiouridine derivative located at the 5'-end of the anticodon of yeast transfer RNA Lys2. Nucleic Acids Res. 1976 Mar;3(3):523–535. doi: 10.1093/nar/3.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. A., Bock R. M. Isolation and characterization of antisuppressor mutations in Escherichia coli. J Bacteriol. 1985 Jan;161(1):377–384. doi: 10.1128/jb.161.1.377-384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Anzaldo F. J., Bastarrachea F. Genetic characterization of streptomycin-resistant and -dependent mutants of Escherichia coli K12. Mol Gen Genet. 1974 Apr 9;130(1):47–64. doi: 10.1007/BF00270518. [DOI] [PubMed] [Google Scholar]
- Taya Y., Nishimura S. Biosynthesis of 5-methylaminomethyl-2-thiouridylate. I. Isolation of a new tRNA-methylase specific for 5-methylaminomethyl-2-thiouridylate. Biochem Biophys Res Commun. 1973 Apr 16;51(4):1062–1068. doi: 10.1016/0006-291x(73)90035-1. [DOI] [PubMed] [Google Scholar]
- VanBogelen R. A., Vaughn V., Neidhardt F. C. Gene for heat-inducible lysyl-tRNA synthetase (lysU) maps near cadA in Escherichia coli. J Bacteriol. 1983 Feb;153(2):1066–1068. doi: 10.1128/jb.153.2.1066-1068.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsky G. R., Bennett R. L., Malamy M. H. Inorganic phosphate transport in Escherichia coli: involvement of two genes which play a role in alkaline phosphatase regulation. J Bacteriol. 1973 Feb;113(2):529–539. doi: 10.1128/jb.113.2.529-539.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Murao K., Ishikura H. 5-(carboxymethylaminomethyl)-2-thiouridine, a new modified nucleoside found at the first letter position of the anticodon. Nucleic Acids Res. 1981 Apr 24;9(8):1933–1939. doi: 10.1093/nar/9.8.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M., Folk W. R. The nucleotide sequences of the two glutamine transfer ribonucleic acids from Escherichia coli. J Biol Chem. 1975 May 10;250(9):3243–3253. [PubMed] [Google Scholar]
- de Lares L. B., Ratouchniak J., Casse F. Chromosomal location of gene governing the trehalose utilization in Escherichia coli K12. Mol Gen Genet. 1977 Mar 28;152(1):105–108. doi: 10.1007/BF00264946. [DOI] [PubMed] [Google Scholar]