Abstract
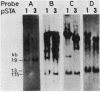
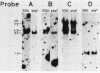
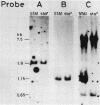
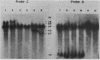
DNA coding for extracellular glucoamylase genes STA1 and STA3 was isolated from DNA libraries of two Saccharomyces diastaticus strains, each carrying STA1 or STA3. Cells transformed with a plasmid carrying either the STA1 or STA3 gene secreted glucoamylases having the same enzymatic and immunological properties and the same electrophoretic mobilities in acrylamide gel electrophoresis as those of authentic glucoamylases. Southern blot analysis of genomic DNA from S. diastaticus and a glucoamylase-non-secreting yeast, Saccharomyces cerevisiae, revealed that the STA1 and STA3 loci of S. diastaticus showed a high degree of homology, and that both yeast species (S. diastaticus and S. cerevisiae) contained DNA segments highly homologous to those of the extracellular glucoamylase genes. Restriction maps of the homologous DNA segments suggested that the extracellular glucoamylase genes of S. diastaticus may have arisen from recombination among the resident DNA segments in S. cerevisiae.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna W. J., Magee P. T. Glycogenolytic enzymes in sporulating yeast. J Bacteriol. 1978 Jun;134(3):844–853. doi: 10.1128/jb.134.3.844-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Brown R. D. Sequences of 5S ribosomal RNA from Xenopus mulleri and the evolution of 5S gene-coding sequences. Cell. 1976 Aug;8(4):485–493. doi: 10.1016/0092-8674(76)90216-6. [DOI] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Nienhuis A. W., Stamatoyannopoulos G. Hemoglobin switching. Cell. 1978 Sep;15(1):307–315. doi: 10.1016/0092-8674(78)90106-x. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Imai Y., Yamashita I., Fukui S. In vivo ligation of linear DNA molecules to circular forms in the yeast Saccharomyces cerevisiae. J Bacteriol. 1983 Aug;155(2):747–754. doi: 10.1128/jb.155.2.747-754.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Hu S. Z., Wang T. S., Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase alpha. J Biol Chem. 1982 Jul 25;257(14):8386–8390. [PubMed] [Google Scholar]
- Yamashita I., Suzuki K., Fukui S. Nucleotide sequence of the extracellular glucoamylase gene STA1 in the yeast Saccharomyces diastaticus. J Bacteriol. 1985 Feb;161(2):567–573. doi: 10.1128/jb.161.2.567-573.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]