Abstract
Caveolae are cholesterol/sphingolipid-rich microdomains of the plasma membrane that have been implicated in signal transduction and vesicular trafficking. Caveolins are a family of caveolae-associated integral membrane proteins. Caveolin-1 and -2 show the widest range of expression, whereas caveolin-3 expression is restricted to muscle cell types. It has been previously reported that little or no caveolin mRNA species are detectable in the brain by Northern blot analyses or in neuroblastoma cell lines. However, it remains unknown whether caveolins are expressed within neuronal cells. Here we demonstrate the expression of caveolin-1 and -2 in differentiating PC12 cells and dorsal root ganglion (DRG) neurons by using mono-specific antibody probes. In PC12 cells, caveolin-1 expression is up-regulated on day 4 of nerve growth factor (NGF) treatment, whereas caveolin-2 expression is transiently up-regulated early in the differentiation program and then rapidly down-regulated. Interestingly, caveolin-2 is up-regulated in response to the mechanical injury of differentiated PC12 cells; up-regulation of caveolin-2 under these conditions is strictly dependent on continued treatment with NGF. Robust expression of caveolin-1 and -2 is also observed along the entire cell surface of DRG neurons, including high levels on growth cones. These findings demonstrate that neuronal cells express caveolins.
Caveolae are 50- to 100-nm vesicular organelles that are located at or near the plasma membrane (1–3). It has been proposed that caveolae play a pivotal role in a number of essential cellular functions, including signal transduction, lipid metabolism, cellular growth control, and apoptotic cell death. The principal protein components of caveolae are the caveolin family of proteins, termed caveolin-1, -2, and -3 (1, 2). Caveolin-2 shows the same tissue distribution as caveolin-1, colocalizes with caveolin-1, and forms a hetero-oligomeric complex with caveolin-1 in vivo (4). In contrast, caveolin-3 is a muscle-specific caveolin-related protein that is primarily expressed in striated muscle cell types (cardiac and skeletal) (5–7).
It has been proposed that caveolin family members function as scaffolding proteins (8) to organize and concentrate specific lipids [cholesterol and glyco-sphingolipids (9–11)] and lipid-modified signaling molecules (9, 12–16) within caveolae membranes. Caveolins interact directly with a number of caveolae-associated signaling molecules, such as H-Ras, hetero-trimeric G-proteins, epidermal growth factor receptor, protein kinase C, Src-family tyrosine kinases, and nitric oxide synthase isoforms (12–14, 17, 18). In many of these cases, it has been documented that caveolin-binding can effectively inhibit the enzymatic activity of these signaling molecules in vitro.
Caveolae organelles and caveolin-1 protein are both down-regulated in response to activated oncogenes, such as v-Abl and H-Ras (19, 20). The potential “transformation suppressor” activity of caveolin-1 has recently been evaluated by using an inducible expression system to up-regulate caveolin-1 expression in oncogenically transformed cells. Induction of caveolin-1 expression in v-Abl- and H-Ras (G12V)-transformed NIH 3T3 cells abrogated the anchorage-independent growth of these cells in soft agar and resulted in the de novo formation of caveolae (20). Thus, down-regulation of caveolin-1 expression and caveolae organelles may be critical to maintaining the transformed phenotype.
Based on these and other observations, we and others have proposed the “caveolae signaling hypothesis,” which states that caveolar localization of certain inactive signaling molecules could provide a compartmental basis for their regulated activation and explain cross-talk between different signaling pathways (1, 21–23). Thus, we have suggested that caveolin may function as a negative regulator of many different classes of signaling molecules through the recognition of specific caveolin-binding motifs (3, 24).
Are these findings relevant to neuronally based signal transduction? Caveolin mRNAs and proteins have been shown to be virtually undetectable in brain tissue by Northern and Western blot analyses by several independent investigators (4–7, 25–28), which initially suggested that neurons do not express caveolin proteins. In addition, it has been shown that caveolin-1 is not expressed within neuroblastoma cell lines (29). However, this could be secondary to their transformed phenotype, as both caveolin-1 and caveolae are down-regulated in response to cell transformation (19, 20, 30, 31).
Here we demonstrate the expression of caveolin-1 and -2 in differentiating PC12 cells and dorsal root ganglion (DRG) neurons by using mono-specific antibody probes. In support of our current findings, it has been previously shown that caveolae-like domains can be purified from neuronal cell plasma membranes and that they contain receptor tyrosine kinases [including insulin and the neurotrophin receptors, Trk-B and p75 nerve growth factor (NGF) receptors], as well as other signaling molecules, and the scrapie prion protein (32, 33). However, these investigators were unable to detect the presence of caveolin proteins (32, 33). Also, it has been shown that the p75 NGF receptor is associated with caveolae membranes when heterologously expressed in NIH 3T3 cells and is observed to coimmunoprecipitate with caveolin-1 (34).
MATERIALS AND METHODS
Materials.
Antibodies and their sources were as follows: caveolin-1 (pAb N-20; rabbit antipeptide antibody directed against caveolin-1 residues 2–21; Santa Cruz, Biotechnology); caveolin-2 [mAb 65; Transduction Laboratories, Lexington, KY (4)]; GAP-43 (mAb; clone GAP-7B10; Sigma); GAP-43 [pAb (35)]; and neurofilament protein [mAb 3A10; gift of Susan Morton in the Jessel laboratory, Columbia Univ. (36)]. Human brain Northern blots were purchased from Clontech and were probed as suggested by the manufacturer by using the cDNAs to caveolins 1 and 2.
NGF-Induced Differentiation of PC12 Cells.
PC12 cells were grown in RPMI 1640 medium with 5% fetal bovine serum and 10% horse serum. PC 12 cells were differentiated for 1–4 days by growing cells in low serum medium (RPMI 1640 medium with 1% horse serum) with NGF (100 ng/ml).
Injury of Differentiated PC12 Cells.
Neurite regeneration was carried out with NGF-differentiated PC12 cells. After 4 days in differentiation medium, cells were rinsed five times with medium (without NGF), while the cells were still attached to the substrate. Then the cells were dislodged from the dish by trypsinization and trituration through a Pasteur pipette and washed three times with medium (without NGF) by centrifugation at 600 × g for 5 min at room temperature. Cells were then plated at low density (8 × 105 cells/100-mm dish) in either complete medium (without NGF) or in low serum medium with NGF (100 ng/ml) and cultured for 2 days (37).
Isolation and Culture of DRG Neurons.
DRG neurons were isolated and cultured essentially as described (38). Briefly, DRGs (sensory neurons) were removed from embryonic day-15 rats under aseptic conditions. Ganglia were dissociated before plating onto a rat tail collagen substrate. Cultures of DRG neurons were fed three times per week with minimal essential medium supplemented with 10% fetal bovine serum, 0.4% glucose, and 50 ng/ml NGF. During the first 2 weeks of culture, cells were treated with 5-fluoro-deoxyuridine and uridine (at 10 μM each) on alternate feedings to eliminate mitotically active nonneuronal cells (38, 39). For Western blot analysis of caveolin expression, neurites from undissociated ganglia, cycled as above, were isolated after mechanically excising and discarding the neuronal somas.
Preparation of Cell Extracts and Immunoblotting.
PC12 cells were extracted for 30–45 min on ice with a buffer containing 20 mM Tris (pH 7.5), 0.15 M NaCl, 1% Triton X-100, and 60 mM octyl-glucoside. After extraction, the remaining insoluble material was removed by centrifugation in the microfuge (14,000 × g). For DRG neurons, extracts were divided into Triton-soluble and Triton-insoluble fractions essentially as we have previously described for Madin–Darby canine kidney cells (40, 41). Samples were separated by SDS/PAGE (15% acrylamide) and transferred to nitrocellulose. After transfer, nitrocellulose sheets were stained with Ponceau S to visualize protein bands and subjected to immunoblotting.
Immunostaining of PC12 Cells.
PC12 cells were washed three times with PBS and fixed for 30 min at room temperature with 2% paraformaldehyde in PBS. Fixed cells were rinsed with PBS and permeabilized with 0.1% Triton X-100/0.2% BSA for 10 min. Cells were then treated with 25 mM NH4Cl in PBS for 10 min at room temperature to quench free aldehyde groups. The cells were rinsed with PBS and incubated with primary antibodies for 1 h at room temperature: either anti-caveolin-1 IgG (pAb; directed against caveolin-1 residues 2–21), or anti-caveolin-2 IgG (pAb, directed against the extreme C terminus of caveolin-2), and anti-GAP-43 (mAb; clone GAP-7B10; Sigma) diluted into PBS with 0.1% Triton X-100/0.2% BSA. After three washes with PBS (10 min each), cells were incubated with the secondary antibodies for 1 h at room temperature: lissamine rhodamine B sulfonyl chloride-conjugated goat anti-rabbit antibody (5 μg/ml), and fluorescein isothiocyanate-conjugated goat anti-mouse antibody (5 μg/ml). Cells were washed three times with PBS (10 min for each wash). Slides were mounted with Slow-Fade antifade reagents (Molecular Probes) and examined by confocal microscopy.
Immunostaining of DRG Neurons.
For immunofluorescent double-label analysis, DRG neurons were fixed with 4% para-formaldehyde for 15 min and permeabilized with 0.2% Triton X-100 for 20 min at room temperature, rinsed in PBS, and blocked for 1 h in L-15 containing 10% heat-inactivated horse serum. Fixed and permeabilized cells were incubated overnight with an antineurofilament (mAb 3A10) and anti-caveolin-1 (pAb N-20) antibodies. After washing three times, coverslips were incubated for 1 h at room temperature with biotinylated anti-rabbit IgG (1:100 dilution; The Jackson Laboratories), followed by strepavidin-fluorescein (1:300 dilution) and Texas red-conjugated anti-mouse (1:100 dilution; The Jackson Laboratories). Finally, cultures were rinsed in PBS and mounted with Citifluor medium (Ted Pella, Redding, CA) containing Hoechst dye. Digital photographs were taken with a charged-couple device (CCD) camera and analyzed with metamorph software (Universal Imaging, Media, PA). A similar procedure was performed for double labeling of DRG cultures with a GAP-43 [pAb (35)] and caveolin-2 (mAb 65) antibodies. In this case, for secondary preabsorbed antibodies we used biotinylated anti-mouse (The Jackson Laboratories) and rhodamine-conjugated anti-rabbit IgG (Chemicon).
Caveolin-2 Antipeptide Antibodies.
An antibody directed against a caveolin-2-derived peptide was prepared by using standard methodology, as described (42). Briefly, the peptide, VGRCFSSVSLQLSQD (the extreme 15 C-terminal residues of human caveolin-2) was coupled to keyhole limpet hemocyanin and injected into rabbits. Pre-immune and immune serum were collected and used to immunoprecipitate caveolin-2 from cellular extracts.
Immunoprecipitation.
Immunoprecipitations were carried out by using protein-A Sepharose CL-4B (Pharmacia) as described (40), with minor modifications. Briefly, mouse muscle tissue was homogenized in a buffer containing 10 mM Tris (pH 8.0), 0.15 M NaCl, 5 mM EDTA, 1% Triton X-100, and 60 mM octyl-glucoside with a Polytron tissue grinder. After clarification by low-speed centrifugation (14,000 × g), the supernatant was subjected to immunoprecipitation with anti-caveolin-2 antipeptide antibody directed against the extreme 15 C-terminal residues of human caveolin-2. After extensive washing, samples were separated by SDS/PAGE (15% acrylamide) and transferred to nitrocellulose. Blots were then probed with IgGs directed against caveolin-2 (mAb 65).
Reverse Transcription–PCR.
Total RNA was extracted from one 10-cm dish of cells by using RNeasy Mini Kit (Qiagen, Chatsworth, CA). Next, 1 μg of total RNA was used for first-strand cDNA synthesis by using the Advantage RT-for-PCR Kit (Clontech). We used the following two primer pairs for PCR amplification: caveolin-1-F, 5′-CTACAAGCCCAACAACAAGGC-3′; caveolin-1-R, 5′-AGGAAGCTCTTGATGCACGGT-3′ caveolin-2-F, 5′-GCTCAACTCTCATCTCAAGCT-3′; caveolin-2-R, 5′-TCTGTCACACTCTTCCATATT-3′.
The the expected size of the amplification product was 340 bp for caveolin-1 and 260 bp for caveolin-2. The correct DNA sequence was verified by DNA sequencing.
RESULTS AND DISCUSSION
Expression and Localization of Caveolins 1 and 2 in Differentiating PC12 Cells.
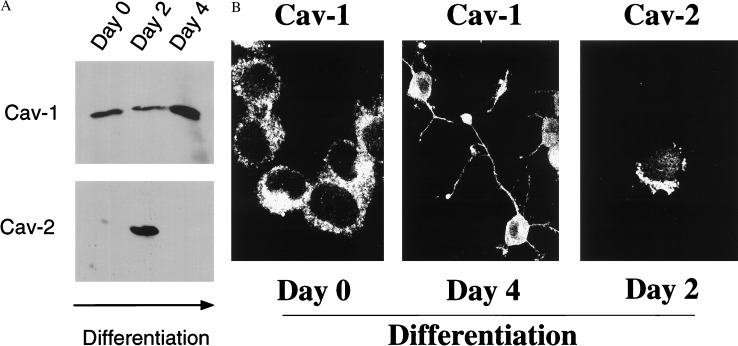
The PC12 cell line, derived from a rat pheochromocytoma, has become a preferred model to study neuronal development. In high mitogen medium, these cells adopt a round morphology and proliferate. In contrast, in low mitogen medium supplemented with NGF, these cells enter G0 and extend long neurites and acquire many of the characteristics of sympathetic neurons. Fig. 1A shows that both caveolins 1 and 2 are coexpressed in PC12 cells. However, their patterns of expression during differentiation are dramatically different. Caveolin-1 is expressed well in undifferentiated PC12 cells and is up-regulated by ≈2- to 3-fold by day 4 of NGF treatment. In contrast, caveolin-2 is not expressed in undifferentiated PC12 cells, is dramatically up-regulated by day 2 of NGF-induced differentiation, and is again down-regulated by day 4 of differentiation. Immunolocalization of caveolins 1 and 2 is shown in Fig. 1B. In undifferentiated PC12 cells, caveolin-1 immunostaining revealed characteristic punctate labeling seen previously in fibroblasts, such as NIH 3T3 cells (20, 21, 43). By day 4 of differentiation, caveolin-1 was localized primarily to the soma or cell body, although some staining of projections was observed. At day 2 of differentiation, caveolin-2 also revealed a characteristic punctate pattern, as shown previously in fibroblasts and adipocytes (4, 28).
Figure 1.
Regulated expression of caveolins 1 and 2 during NGF-induced differentiation of PC12 cells. (A) Western Blot analysis. Lysates were prepared from undifferentiated (day 0) and differentiating (days 2 and 4) PC12 cells and subjected to immunoblot analysis with caveolin-1 (pAb N-20) and caveolin-2 (mAb 65) specific antibody probes. Note that caveolin-1 is induced ≈2-fold between days 2 and 4. In contrast, caveolin-2 is transiently up-regulated on day 2 and down-regulated again by day 4. Equal amounts of protein were loaded in each lane. (B) Immunolocalization. Caveolin-1 (pAb N-20) and caveolin-2 (mAb 65) specific antibody probes were used to localize caveolin-1 on days 0 and 4 (Left and Center) and caveolin-2 on day 2 (Right) of differentiation. Note that both caveolins 1 and 2 are most highly expressed on the soma or cell body and less apparent within cellular projections.
Caveolin-2 and GAP-43 Are Both Transiently Up-Regulated During PC12 Cell Differentiation: Effects of Cellular Injury.
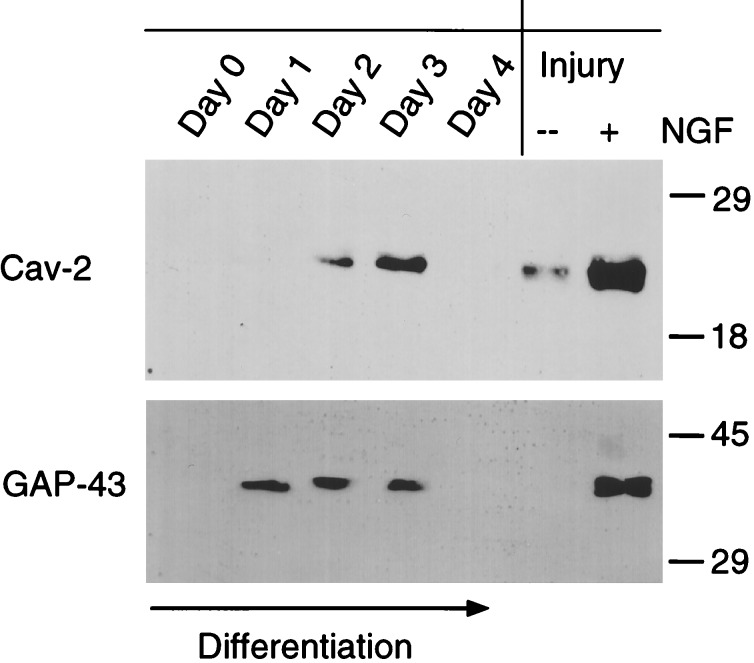
As caveolin-2 protein expression was transiently up-regulated during NGF-induced differentiation of PC12 cells, we analyzed the time-course of its expression more extensively (Fig. 2 Left). Caveolin-2 protein expression was abruptly up-regulated on day 2 of differentiation and reached maximal levels on day 3. This pattern of protein expression was strikingly similar to another marker of neuronal differentiation, GAP-43, whose expression is primarily localized to neuronal growth cones.
Figure 2.
Both caveolin-2 and GAP-43 are transiently up-regulated during PC12 cell differentiation; effects of cellular injury. (Left) Lysates were prepared from undifferentiated (day 0) and differentiating (days 1, 2, 3, and 4) PC12 cells and subjected to immunoblot analysis with anti-caveolin-2 (mAb 65) and GAP-43 (mAb; clone GAP-7B10) specific mAb probes. Note that both caveolin-2 and GAP-43 are transiently up-regulated between days 1 and 3 of differentiation and then both are down-regulated on day 4. (Right) After 4 days of differentiation, PC12 cells were trypsinized, subjected to physical injury by repeating pipeting, and replated in the absence (−) or presence (+) of NGF and allowed to recover for 2 days. Lysates were then prepared and subjected to immunoblot analysis with anti-caveolin-2 (mAb 65) and GAP-43 (mAb; clone GAP-7B10). Note that both caveolin-2 and GAP-43 are up-regulated in response to cellular injury and recovery in NGF. Equal amounts of protein were loaded in each lane.
As GAP-43 is known to be up-regulated in response to neuronal injury, we used PC12 cells as a model system to study whether caveolin-2 behaves in a similar fashion. For this purpose, we used an established protocol developed for differentiated PC12 cells to simulate neuronal injury. Briefly, after 4 days in differentiation medium, PC12 cells were rinsed five times with medium (without NGF), while the cells were still attached to the substrate. Differentiated PC12 cells were then mechanically dislodged from the dish by trituration through a Pasteur pipette and washed three times with medium (without NGF). Then the cells were re-plated at low density in either complete medium (without NGF) or low serum medium with NGF (100 ng/ml) and allowed to recover for 2 days. Fig. 2 Right shows that both caveolin-2 and GAP-43 responded similarly to injury. Caveolin-2 and GAP-43 were most highly expressed in injured differentiated PC12 cells that were allowed to recover in the presence of NGF. Thus, both cell injury and NGF can transiently up-regulate the expression of caveolin-2.
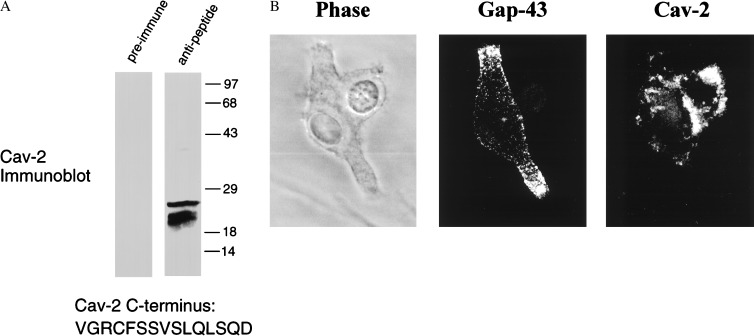
As both caveolin-2 and GAP-43 showed similar patterns of induction and response to injury, we next examined whether caveolin-2 and GAP-43 are colocalized at the cellular level. For this purpose, we generated a rabbit antipeptide antibody directed against the extreme C terminus of caveolin-2. Fig. 3A shows the characterization of this antibody probe and that this antibody specifically recognizes caveolin-2. Fig. 3B shows double labeling of PC12 cells with a mouse mAb directed against GAP-43 and a rabbit polyclonal antipeptide antibody generated against the unique C terminus of caveolin-2. Note that caveolin-2 immunostaining appears primarily as punctate areas localized close to the soma or cell body. In contrast, GAP-43 is most highly concentrated in cellular outgrowths that most likely represent growth cones. Thus, despite similar patterns of regulated expression, caveolin-2 and GAP-43 do not share the same subcellular distribution in differentiating PC12 cells.
Figure 3.
Dual immunolabeling of GAP-43 and caveolin-2 in differentiating PC12 cells. (A) Characterization of an antipeptide antibody directed against the extreme C terminus of caveolin-2. An antipeptide antibody was raised against the peptide VGRCFSSVSLQLSQD that is derived from the extreme C terminus of human caveolin-2 (pAb C-15). Pre-immune and immune serum were incubated in the presence of protein A-Sepharose to purify the IgGs and used to immunoprecipitate a muscle cell lysate that is known to contain caveolin-2. Immunoprecipitates were subjected to Western blot analysis with an anti-caveolin-2 (mAb 65) mAb probe. Note that this caveolin-2 antipeptide antibody (pAb C-15) can be used to immunoprecipitate caveolin-2 from a cellular extract. (B) Dual immunolabeling of GAP-43 and caveolin-2. Caveolin-2 (pAb C-15) and GAP-43 (mAb; clone GAP-7B10) specific antibody probes were used to localize these proteins on day 3 of differentiation. Note that caveolin-2 and GAP-43 do not colocalize. GAP-43 is most highly expressed within the developing cellular projections, whereas caveolin-2 is localized more closely to the soma or cell body and absent from the projections. (Left) Phase image. (Center) GAP-43 immunostaining. (Right) Caveolin-2 immunostaining.
Expression and Localization of Caveolins 1 and 2 in DRG Neurons.
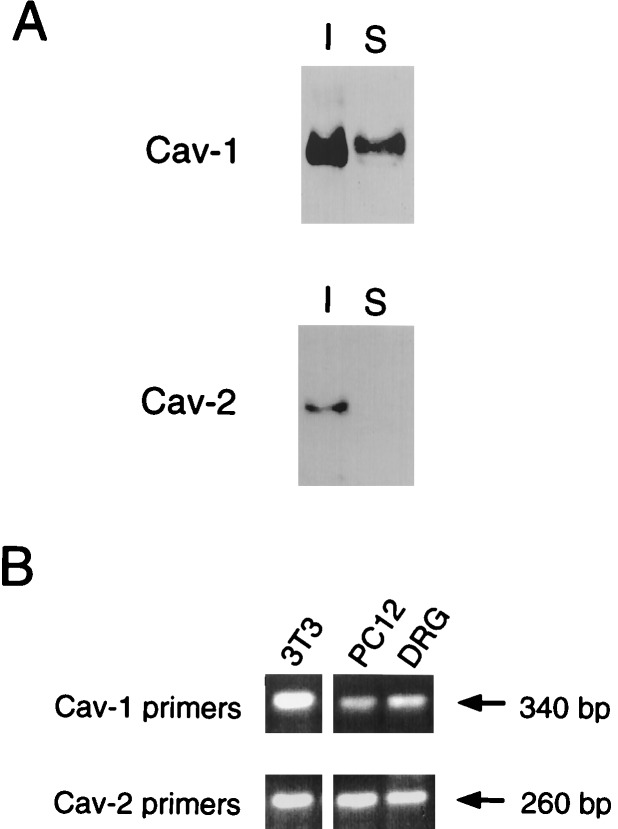
As PC12 cells only represent a model for the differentiation of peripheral neurons, we next examined whether caveolins are indeed expressed within neurons. Primary cultures of DRG neurons were examined by Western blot analysis and immunofluorescence microscopy. Fig. 4A shows that both caveolins 1 and 2 are coexpressed in lysates prepared from neurites isolated from cultures of DRG neurons. Both caveolin-1 and caveolin-2 behaved as Triton-insoluble proteins, as we and others have shown previously in other cell types (21, 40, 44). Triton-insolubility is a characteristic feature of the caveolin proteins (3). In support of these findings, expression of caveolins 1 and 2 in PC12 cells and DRG neurons was also observed by RT-PCR (Fig. 4B). The correct DNA sequences were verified by sequencing.
Figure 4.
Expression of caveolins 1 and 2 in DRG neurons. (A) Immunoblot analysis. Lysates were prepared from cultures of rat DRG neurons and divided into Triton-soluble (S) and Triton-insoluble (I) fractions. Caveolin-1 (pAb N-20) and caveolin-2 (mAb 65) specific antibody probes were then used to probe these cellular extracts by immunoblotting. Note that both caveolins 1 and 2 are expressed and behave as Triton-insoluble components. (B) RT-PCR analysis. Total RNA isolated from PC12 cells and DRG neurons was subjected to RT-PCR analysis by using primers directed against the known sequences of caveolins 1 and 2; NIH 3T3 cells served as a postitive control.
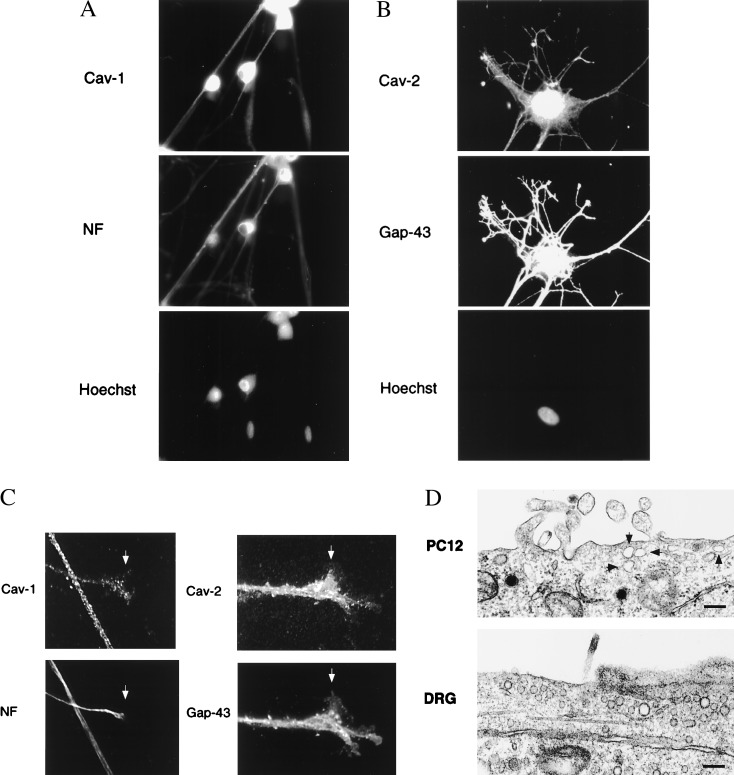
To rule out the possibility that caveolin expression reflected contamination by nonneuronal cell types that occasionally persist in primary cultures of DRG neurons, we performed double-labeling experiments with antibodies directed against caveolins 1 and 2 and specific neuronal markers [neurofilament proteins (NF) or GAP-43]. Fig. 5 A and B shows that both caveolins 1 and 2 are expressed specifically within DRG neurons (NF or GAP-43-positive cells). Expression was observed over the entire neuronal cell surface, including substantial levels in the growth cones (Fig. 5C). Caveolae and caveolae-like structures were also observed by transmission electron microscopy in both PC12 cells and DRG neurons (Fig. 5D). These results demonstrate that neuronal cells express caveolins and contain caveolae.
Figure 5.
Localization of caveolins 1 and 2 in cultured DRG neurons. (A and B) Caveolin-1 [pAb N-20 (A)] and caveolin-2 [mAb 65 (B)] specific antibody probes were used to localize caveolins 1 and 2 in cultures of DRG neurons. Neurons were counterstained with antineurofilament protein (NF) or anti-GAP-43 antibodies to identify neuronal cells. Nuclei were visualized by Hoechst staining. (C). Expression of caveolins 1 and 2 in the growth cones of DRG neurons. Arrowheads point at growth cones showing double-labeling with anti-caveolin-1 and antineurofilament antibodies; and double-labeling with anti-caveolin-2 and GAP-43 antibodies. (D). Caveolae-like structures in PC12 cells and DRG neurons. Caveolae were identified by their size (50–100 nm in diameter) and location at or near the plasma membrane. (Bar = 200 nm.)
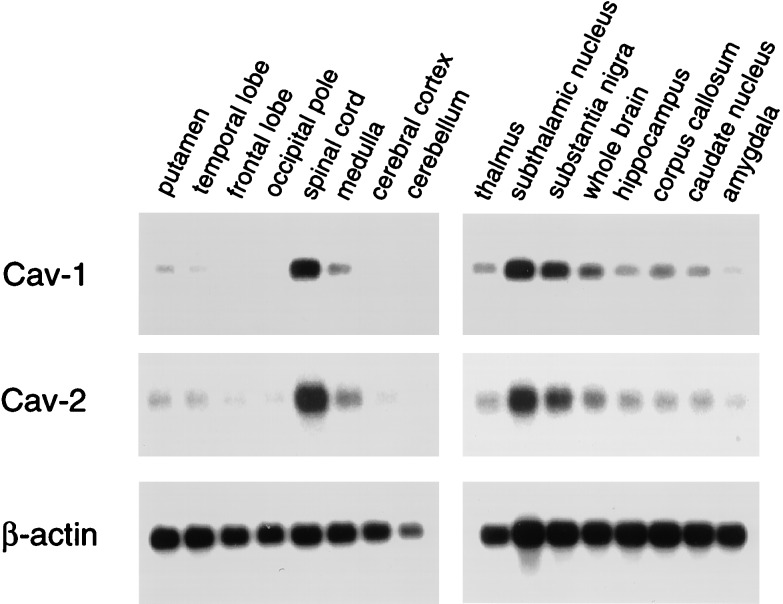
In accordance with these findings, Northern blot analysis reveals that caveolins 1 and 2 are coexpressed within the brain (Fig. 6). mRNA species corresponding to caveolins 1 and 2 showed a similar pattern of expression and were highest in the spinal cord, subthalamic nucleus, and substantia nigra, relative to whole brain. The distribution of β-actin mRNA served as a control for equal loading.
Figure 6.
Distribution of caveolins 1 and 2 in the brain. mRNA species corresponding to caveolins 1 and 2 showed a similar pattern of expression and were highest in the spinal cord, subthalamic nucleus, and substantia nigra, relative to whole brain. The distribution of β-actin mRNA is shown as a control for equal loading.
As caveolae and caveolins have been implicated in a wide variety of signal transduction processes in nonneuronal cell types, differentiated PC12 cells and primary cultures of DRG neurons will serve as important model systems for establishing the functional significance of caveolins proteins in neuronal signal transduction. The high level of expression in growth cones is of interest in this regard and may suggest a role in neurite outgrowth. Although caveolins 1 and 2 are expressed in differentiated PC12 cells and DRG neurons, these findings do not rule out the existence of neuron-specific caveolins. In this regard, these two model cell systems will be invaluable in the search for other caveolin family members that may be expressed in the nervous system.
Caveolin-1 is selectively down-regulated in NIH 3T3 fibroblasts in response to cell transformation by activated oncogenes (19, 20). This result may explain why we and other groups failed to detect caveolin-1 protein expression in transformed neuroblastoma cell lines, such as the SKN-SH (N-Ras activated) and N2a cell lines (4, 29). Conversely, caveolins 1 and 2 are dramatically induced ≈10- to 20-fold during the process of adipocyte differentiation; similarly, caveolin-3 is absent in precursor myoblasts and abundant in differentiated myotubes (4–7, 27, 28). Thus, induction of caveolins 1 and 2 during NGF-induced differentiation of PC12 cells is consistent with the idea that caveolins are generally induced during differentiation processes and that caveolin expression is highest in terminally differentiated nondividing cells.
Acknowledgments
This work was supported by a National Institutes of Health FIRST Award (GM-50443 to M.P.L. and MH-56036 to T.O.), and grants from the Charles E. Culpeper Foundation (to M.P.L.), G. Harold and Leila Y. Mathers Charitable Foundation (to M.P.L. and P.E.S.), and the Sidney Kimmel Cancer Research Foundation (to M.P.L.). P.E.S. was supported by a grant from Pfizer Corp., a pilot grant from the Albert Einstein College of Medicine Diabetes Research and Training Center, and by a research grant from the American Diabetes Association. T.O. was supported by an NIH FIRST Award (MH-56036).
ABBREVIATIONS
- DRG
dorsal root ganglion
- NGF
nerve growth factor
- RT-PCR
reverse transcription–PCR
References
- 1.Lisanti M P, Scherer P, Tang Z-L, Sargiacomo M. Trends Cell Biol. 1994;4:231–235. doi: 10.1016/0962-8924(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 2.Couet J, Li S, Okamoto T, Scherer P S, Lisanti M P. Trends Cardiovasc Med. 1997;7:103–110. doi: 10.1016/S1050-1738(97)00001-7. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto T, Schlegel A, Scherer P E, Lisanti M P. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 4.Scherer P E, Lewis R Y, Volonté D, Engelman J A, Galbiati F, Couet J, Kohtz D S, van Donselaar E, Peters P, Lisanti M P. J Biol Chem. 1997;272:29337–29346. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- 5.Song K S, Scherer P E, Tang Z-L, Okamoto T, Li S, Chafel M, Chu C, Kohtz D S, Lisanti M P. J Biol Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- 6.Tang Z-L, Scherer P E, Okamoto T, Song K, Chu C, Kohtz D S, Nishimoto I, Lodish H F, Lisanti M P. J Biol Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- 7.Way M, Parton R. FEBS Lett. 1995;376:108–112. doi: 10.1016/0014-5793(95)01256-7. [DOI] [PubMed] [Google Scholar]
- 8.Sargiacomo M, Scherer P E, Tang Z-L, Kubler E, Song K S, Sanders M C, Lisanti M P. Proc Natl Acad Sci USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Song K S, Lisanti M P. J Biol Chem. 1996;271:568–573. [PubMed] [Google Scholar]
- 10.Murata M, Peranen J, Schreiner R, Weiland F, Kurzchalia T, Simons K. Proc Natl Acad Sci USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fra A M, Masserini M, Palestini P, Sonnino S, Simons K. FEBS Lett. 1995;375:11–14. doi: 10.1016/0014-5793(95)95228-o. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Okamoto T, Chun M, Sargiacomo M, Casanova J E, Hansen S H, Nishimoto I, Lisanti M P. J Biol Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- 13.Song K S, Li S, Okamoto T, Quilliam L, Sargiacomo M, Lisanti M P. J Biol Chem. 1996;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Couet J, Lisanti M P. J Biol Chem. 1996;271:29182–29190. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaul P W, Smart E J, Robinson L J, German Z, Yuhanna I S, Ying Y, Anderson R G W, Michel T. J Biol Chem. 1996;271:6518–6522. doi: 10.1074/jbc.271.11.6518. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Cardena G, Oh P, Liu J, Schnitzer J E, Sessa W C. Proc Natl Acad Sci USA. 1996;93:6448–6453. doi: 10.1073/pnas.93.13.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couet J, Sargiacomo M, Lisanti M P. J Biol Chem. 1997;272:30429–30438. doi: 10.1074/jbc.272.48.30429. [DOI] [PubMed] [Google Scholar]
- 18.Oka N, Yamamoto M, Schwencke C, Kawabe J, Ebina T, Couet J, Lisanti M P, Ishikawa Y. J Biol Chem. 1997;272:33416–33421. doi: 10.1074/jbc.272.52.33416. [DOI] [PubMed] [Google Scholar]
- 19.Koleske A J, Baltimore D, Lisanti M P. Proc Natl Acad Sci USA. 1995;92:1381–1385. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman J A, Wycoff C C, Yasuhara S, Song K S, Okamoto T, Lisanti M P. J Biol Chem. 1997;272:16374–16381. doi: 10.1074/jbc.272.26.16374. [DOI] [PubMed] [Google Scholar]
- 21.Sargiacomo M, Sudol M, Tang Z-L, Lisanti M P. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisanti M P, Scherer P E, Vidugiriene J, Tang Z-L, Hermanoski- Vosatka A, Tu Y-H, Cook R F, Sargiacomo M. J Cell Biol. 1994;126:111–126. doi: 10.1083/jcb.126.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson R G W. Proc Natl Acad Sci USA. 1993;90:10909–10913. doi: 10.1073/pnas.90.23.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couet J, Li S, Okamoto T, Ikezu T, Lisanti M P. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 25.Glenney J R. FEBS Lett. 1992;314:45–48. doi: 10.1016/0014-5793(92)81458-x. [DOI] [PubMed] [Google Scholar]
- 26.Glenney J R, Soppet D. Proc Natl Acad Sci USA. 1992;89:10517–10521. doi: 10.1073/pnas.89.21.10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherer P E, Lisanti M P, Baldini G, Sargiacomo M, Corley-Mastick C, Lodish H F. J Cell Biol. 1994;127:1233–1243. doi: 10.1083/jcb.127.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scherer P E, Okamoto T, Chun M, Nishimoto I, Lodish H F, Lisanti M P. Proc Natl Acad Sci USA. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorodinsky A, Harris D A. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman J A, Lee R J, Karnezis A, Bearss D J, Webster M, Siegel P, Muller W J, Windle J J, Pestell R G, Lisanti M P. J Biol Chem. 1998;273:20448–20455. doi: 10.1074/jbc.273.32.20448. [DOI] [PubMed] [Google Scholar]
- 31.Lee S W, Reimer C L, Oh P, Campbel L D B, Schnitzer J E. Oncogene. 1998;16:1391–1397. doi: 10.1038/sj.onc.1201661. [DOI] [PubMed] [Google Scholar]
- 32.Wu C, Butz S, Ying Y, Anderson R G W. J Biol Chem. 1997;272:3554–3559. doi: 10.1074/jbc.272.6.3554. [DOI] [PubMed] [Google Scholar]
- 33.Vey M, Pilkuhn S, Holger W, Nixon R, DeArmond S J, Smart E J, Anderson R G W, Taraboulos A, Prusiner S B. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilderback T R, Grigsby R J, Dobrowsky R T. J Biol Chem. 1997;272:10922–10927. doi: 10.1074/jbc.272.16.10922. [DOI] [PubMed] [Google Scholar]
- 35.Williams E J, Doherty P, Turner G, Reid R A, Hemperley J J, Walsh F S. J Cell Biol. 1992;119:885–892. doi: 10.1083/jcb.119.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenfeld D P, Hynes M A, Skoler K M, Furley A J, Jessel T M. Neuron. 1994;12:675–690. doi: 10.1016/0896-6273(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 37.Jaiswal R K, Weissinger E, Kolch W, Landreth G E. J Biol Chem. 1996;271:23626–23629. doi: 10.1074/jbc.271.39.23626. [DOI] [PubMed] [Google Scholar]
- 38.Rosen C L, Lisanti M P, Salzer J L. J Cell Biol. 1992;117:617–627. doi: 10.1083/jcb.117.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Einheber S, Zanazzi G, Ching W, Scherer S, Milner T A, Peles E, Salzer J L. J Cell Biol. 1997;139:1495–1506. doi: 10.1083/jcb.139.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisanti M P, Tang Z-L, Sargiacomo M. J Cell Biol. 1993;123:595–604. doi: 10.1083/jcb.123.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song K S, Tang Z-L, Li S, Lisanti M P. J Biol Chem. 1997;272:4398–4403. doi: 10.1074/jbc.272.7.4398. [DOI] [PubMed] [Google Scholar]
- 42.Bickel P E, Scherer P E, Schnitzer J E, Oh P, Lisanti M P, Lodish H F. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 43.Scherer P E, Tang Z-L, Chun M C, Sargiacomo M, Lodish H F, Lisanti M P. J Biol Chem. 1995;270:16395–16401. doi: 10.1074/jbc.270.27.16395. [DOI] [PubMed] [Google Scholar]
- 44.Schnitzer J E, Oh P, Jacobson B S, Dvorak A M. Proc Natl Acad Sci USA. 1995;92:1759–1763. doi: 10.1073/pnas.92.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]