SUMMARY
In metazoa, pre-mRNA 3′-end formation occurs via two pathways: cleavage/polyadenylation for the majority of RNA polymerase II transcripts and U7-snRNP-dependent cleavage for replication-dependent histone pre-mRNAs. An RNA element derived from a replication-dependent histone gene affects multiple steps of pre-mRNA processing. Here, we demonstrate that a portion of this RNA element, present in the majority of histone mRNAs, stimulates U7-snRNP-dependent cleavage. Surprisingly, this element binds U2 snRNP although it is derived from an intronless mRNA. Specifically, SF3b, a U2 and U12 snRNP component, contacts the RNA element both in vitro and in vivo in conjunction with hPrp43, a DEAH-box helicase. Tethering either U2 or U12 snRNP to histone pre-mRNA substrates stimulates U7-snRNP-dependent cleavage in vitro and in vivo. Finally, we show that U2 snRNP associates with histone pre-mRNAs in vivo. We conclude that U2 snRNP plays a non-splicing role in histone mRNA maturation.
Keywords: 3′-end Processing, snRNP, Histone, SF3b, hPrp43
INTRODUCTION
Replication-dependent histone pre-mRNAs, synthesized during S phase of the cell cycle, are unique RNA polymerase (Pol) II transcripts; they are intronless and cleaved at the 3′ end in a U7-snRNP-dependent manner (Marzluff, 2005). In contrast, most RNA Pol II transcripts are 3′-end processed in a coupled cleavage/polyadenylation (CPA) reaction by several factors, including two multimeric complexes – cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF); the endonuclease is likely CPSF73 (Mandel et al., 2006; Ryan et al., 2004). Recently, CPA factors were implicated in U7-snRNP-dependent cleavage; CPSF73 cross-links to the scissile phosphate of a U7-snRNP substrate (Dominski et al., 2005), and symplekin is a heat-labile factor necessary for U7-snRNP-dependent cleavage (Kolev and Steitz, 2005). Symplekin associates with CPSF and CstF (Hofmann et al., 2002; Takagaki and Manley, 2000) and is involved in cytoplasmic pre-mRNA polyadenylation during embryogenesis (Barnard et al., 2004).
Pre-mRNA processing reactions occur co-transcriptionally (Neugebauer, 2002; Proudfoot et al., 2002), and links between splicing and CPA have been described. SR protein splicing factors, such as SRm160 and U2AF65 (U2 snRNP auxiliary factor), stimulate CPA in vitro and in vivo (McCracken et al., 2002; Millevoi et al., 2002). Moreover, U1A (a U1 snRNP protein) stimulates CPA (Lutz and Alwine, 1994), and an isolated 3′ splice site stimulates CPA (Cooke and Alwine, 1996) either via U2AF65 (Millevoi et al., 2002) or the U2 snRNP interaction with the intronic branchpoint (Kyburz et al., 2006). Kyburz et al. have further shown that CPA stimulation by U2 snRNP involves an interaction between SF3b (a heteroheptameric protein subcomplex of U2 holo-snRNP) and CPSF.
A 101-nt RNA element from a replication-dependent mouse H2a gene was previously shown to affect multiple steps of pre-mRNA processing, including CPA (Huang et al., 1999). A 22-nt portion of this RNA element binds the shuttling SR proteins, SRp20 and 9G8 (Huang and Steitz, 2001), which are known splicing enhancers (Cavaloc et al., 1994; Zahler et al., 1992). Since this 101-nt RNA element is derived from an intronless pre-mRNA processed in a U7-snRNP-dependent manner, we hypothesized that protein binding partners might stimulate U7-snRNP-dependent cleavage. Identification of these proteins surprisingly revealed that U2 snRNP is responsible for this stimulation in vitro. Also, in vivo, U2 snRNP binds intronless replication-dependent histone pre-mRNAs and stimulates U7-snRNP-dependent 3′-end formation.
RESULTS
A 7-nt RNA Motif Is Conserved in Human Histone mRNAs
Five replication-dependent histone proteins (H1, H2a, H2b, H3, and H4) exist in metazoan cells. There are 72 known human histone genes and transcribed pseudogenes located primarily in three genomic clusters (Marzluff et al., 2002). The 22-nt RNA element used in these studies (Figure 1A; Huang and Steitz, 2001) was derived from the 101-nt element within the coding region of a mouse H2a variant (Huang et al., 1999). Within this 22-nt sequence is a 7-nt motif (C/GAAGAAG) that is conserved in 59 of the 72 histone mRNAs and transcribed pseudogenes. Where present, this motif is usually contained in the open-reading frame, with the AAGAAG portion encoding two adjacent lysines and the variable first nucleotide (C or G) representing the third position of the previous codon. Since histone H4 does not contain paired lysines in its amino acid sequence, this 7-nt motif is more degenerate in mRNAs encoding H4 isoforms; but 4 out of 13 genes contain the 7-nt motif in the 5′ untranslated region.
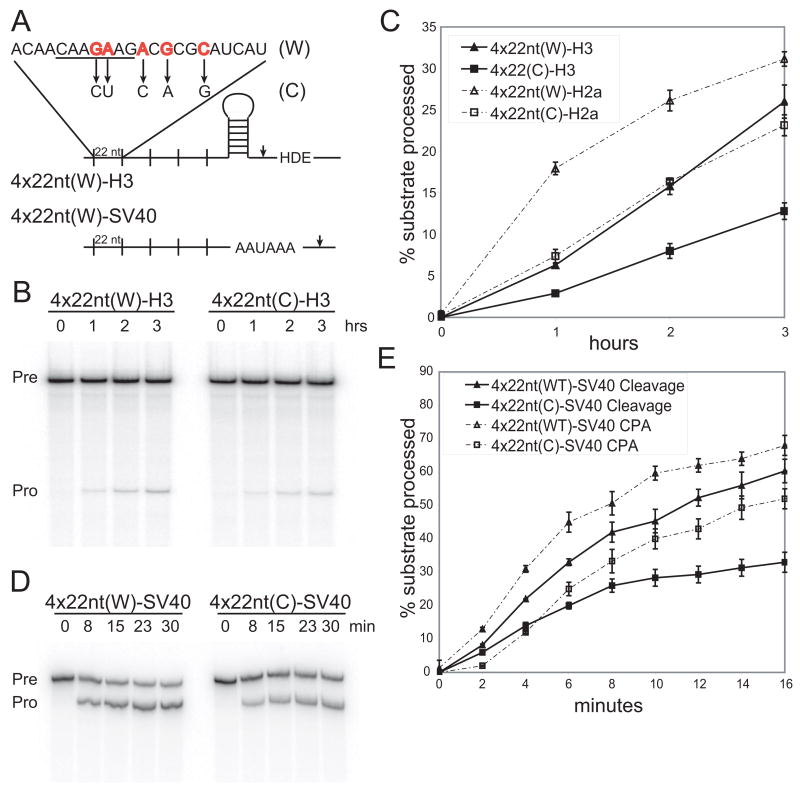
Figure 1. An RNA Element Derived from a Replication-Dependent Histone mRNA Stimulates 3′-end Formation.
(A) The U7-snRNP (derived from mouse histone H3 mRNA) and CPA (SV40 late) substrates are schematized with sites of cleavage indicated by arrowheads. The 22-nt RNA element (W) and a control element (C), with changed nucleotides indicated by arrows, have been described (Huang and Steitz, 2001). The 7-nt conserved motif is underlined.
(B and C) Time courses of U7-snRNP-dependent cleavage of two U7-snRNP substrates (H3 and H2a) containing four copies of the 22-nt W element or the C element were performed in HeLa nuclear extract (Kolev and Steitz, 2005). The H3 substrate (Pre→Pro) is shown in B; processing of both substrates was quantitated (error bars represent standard deviations; panel C) as the ratio of the 5′ product to total RNA. We verified that the substrates are cleaved in a U7-snRNP-dependent manner (data not shown; Bond et al., 1991; Huang et al., 2003).
(D and E) Processing time courses of the SV40 late polyadenylation substrate (Pre) with four copies of either the 22-nt W element or the C element were performed in HeLa nuclear extract (Ryan et al., 2004); note the shorter time required for comparable extent of cleavage in E versus C). In D, cordycepin was added to inhibit polyadenylation of the cleavage product (Pro). E shows quantitations, as in panel C, for both the uncoupled and coupled CPA reactions.
The 22-nt RNA Element Stimulates U7-snRNP-dependent Cleavage and CPA In Vitro
Previously, the 22-nt RNA element was shown to enhance CPA in vivo (Huang and Steitz, 2001). Four copies of either the 22-nt RNA element (W) or a control element (C) bearing five point mutations, which does not enhance CPA in vivo, were inserted into two U7-snRNP substrates derived from mRNAs encoding mouse H2a and H3 variants (Figure 1A; Bond et al., 1991; Huang et al., 2003). Under standard in vitro U7-snRNP dependent cleavage conditions, both substrates in HeLa nuclear extract revealed a twofold stimulation by the 22-nt RNA element versus the control element (Figures 1B and 1C). This is comparable to the effect on CPA (Figures 1D and 1E). Four copies of the 22-nt RNA element placed 5′ to the SV40 late polyadenylation signal (Figure 1A; Wilusz and Shenk, 1988) stimulated the cleavage reaction [poly-A tail elongation was inhibited by cordycepin (3′-deoxy-ATP)] compared to a construct containing four copies of the control RNA sequence, which has the same processing efficiency as that of the SV40 signal alone (data not shown). Four copies of the 22-nt RNA element also stimulated coupled CPA two-fold (Figure 1E), and this stimulation is consistent with that by the complete 101-nt H2a element previously reported (Huang et al., 1999).
The RNA Element Cross-links Specifically to a 150 kDa Protein in HeLa Nuclear Extract and Xenopus laevis Germinal Vesicles
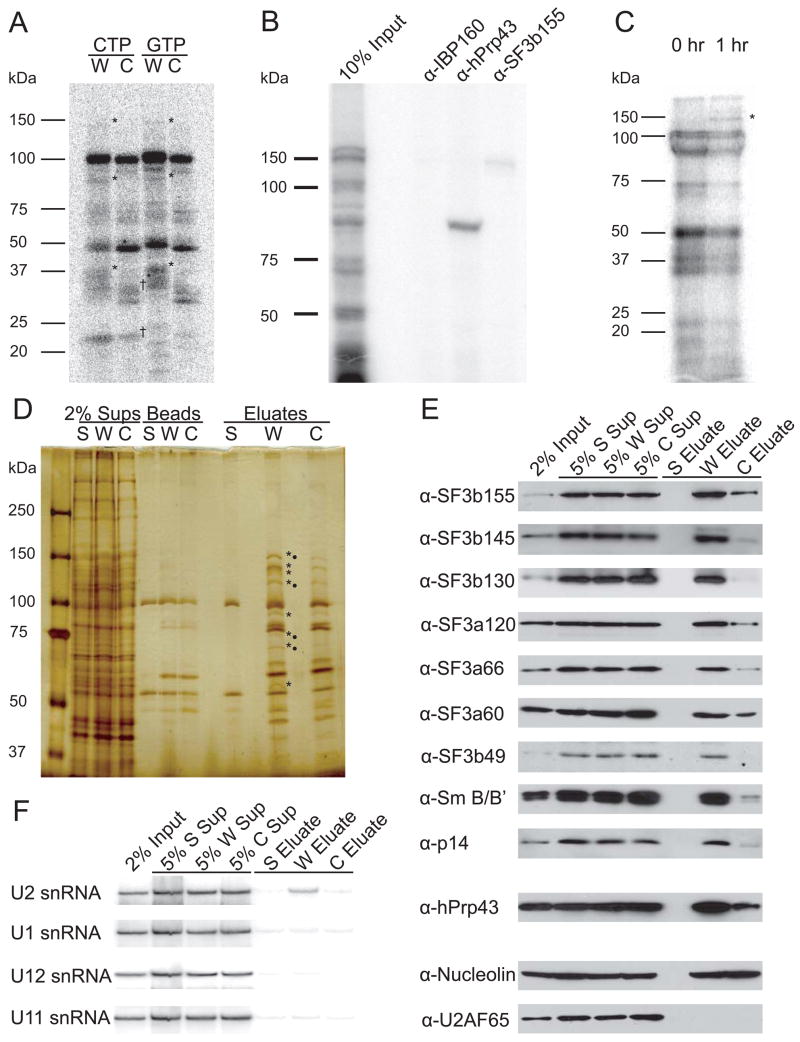
UV cross-linking analysis of a radiolabeled construct containing four copies of the 22-nt RNA element but devoid of 3′-end formation signals was performed in HeLa nuclear extract under CPA conditions. Specific cross-links to ~150, 93, and 37 kDa proteins were observed (*) for the 22-nt RNA element (W) relative to the mutant element (C), whether the RNAs were labeled with [α-32P]-CTP or [α-32P]-GTP (Figure 2A). Similar cross-links were observed under conditions used for U7-snRNP-dependent cleavage, but with higher background (data not shown). The shuttling SR proteins, SRp20 and 9G8, which were previously reported to specifically cross-link to the 22-nt RNA element (Huang and Steitz, 2001), are best seen with C-label (†; Figure 2A).
Figure 2. U2 snRNP Binds the 22-nt RNA Element In Vitro and In Vivo.
(A)RNAs containing four copies of the 22-nt (W) or control (C) element were UV cross-linked under CPA conditions (Ryan et al., 2004). RNAs were synthesized in the presence of 32P-labeled CTP or GTP as indicated, and cross-linked proteins were separated by 5–15% gradient SDS-PAGE. The 22-nt RNA element (W) cross-linked specifically to three proteins (*) compared to the control RNA (C). SRp20 and 9G8, previously characterized as cross-linking to the RNA element (Huang and Steitz, 2001), are detected for both RNAs labeled at C-residues (†).
(B) A CTP-labeled RNA containing four copies of the 22-nt element (W) was incubated and UV-irradiated as in A; immunoprecipitations were performed with the indicated antibodies followed by 5–15% gradient SDS-PAGE.
(C) CTP-labeled RNA containing the complete H2a 101-nt RNA element 5′ to the H3-derived U7-snRNP substrate was injected into GVs and UV-irradiated after 0 or 1 hr; cross-linked proteins were separated by 12% SDS-PAGE. Proteins visible at zero time most likely represent a pre-processing heterogeneous RNP, as described for in vitro CPA (Skolnik-David et al., 1987) and serve as a control. A specific 150 kDa cross-linked protein was observed (*), but could not be identified by α-SF3b155 immunoprecipitation because the signal was too weak.
(D) Affinity purification was performed using the biotinylated 22-nt RNA element (W, in four copies), the mutated control (C, in four copies), or streptavidin beads (S) alone. Supernatants (Sups), proteins retained on the beads after RNase elution (Beads), and the eluted proteins (Eluates) were separated by 5–15% gradient SDS-PAGE. Proteins enriched in the purification with W relative to C are indicated (*), as are proteins identified by mass spectrometric analysis (•).
(E) Western blots were performed on affinity-purified samples to confirm and extend the mass spectrometry results. U2B″ and U2A′ were also enriched in W versus C (data not shown).
(F) Northern blotting identified U2 snRNA in the eluate from the W affinity purification.
Because U2 snRNP couples splicing and CPA (Kyburz et al., 2006), we reasoned that two of the cross-linked proteins might be SF3b155 (155 kDa) and hPrp43 (a 93 kDa protein previously observed in crude U2 snRNP preparations; Will et al., 2002). Both should exhibit RNA-binding activity since SF3b155 cross-links to introns during splicing (Wang et al., 1998), and hPrp43 is a predicted DEAH-box helicase (Martin et al., 2002). Immunoprecipitation confirmed cross-linked protein identities (Figure 2B).
Finally, protein-RNA cross-links were examined in vivo using isolated Xenopus laevis germinal vesicles (GVs; oocyte nuclei). A radiolabeled U7-snRNP substrate containing the complete H2a 101-nt RNA element 5′ of a histone stem-loop was microinjected, and the GVs were UV-irradiated. At a time when processed product appeared (1 hr, data not shown), a cross-link to a protein of similar size to the Xenopus homolog of SF3b155 (Schmidt-Zachmann et al., 1998) specifically appeared (Figure 2C). We conclude that SF3b155 and hPrp43 bind the 22-nt RNA element in vitro and that SF3b155 may bind the H2a 101-nt RNA element in GVs.
Affinity Purification from Nuclear Extract Reveals that U2 snRNP Binds the 22-nt RNA Element
For affinity purification, an RNA consisting either of four copies of the 22-nt element or the control element (Figure 1A) was synthesized with biotinylated CTP and incubated in CPA extract under conditions similar to in vitro cross-linking but without the cross-linking step. After selection on immobilized streptavidin, RNA-bound proteins were eluted by RNase treatment and separated by SDS-PAGE. Several proteins (* in Figure 2D) were enriched in the eluate from the 22-nt RNA (W) versus the control (C) element purification. Tryptic digestion followed by mass spectrometric analysis yielded peptide matches for four U2 snRNP proteins (•): SF3b155 and all three proteins of SF3a (60, 66, and 120 kDa; Brosi et al., 1993). The only identified protein enriched in the 22-nt RNA element purification (W) that was not a U2 snRNP component was hPrp43 (93 kDa).
The identities of all eight enriched proteins were confirmed using Western blots (Figure 2E), which were also probed for additional proteins present in the U2 holo-snRNP (Behrens et al., 1993; Will et al., 2001). Enrichment in the eluate from the 22-nt RNA element (W) purification relative to the control (C) was observed for Sm B/B′ protein and the following proteins of the SF3b heteroheptamer: SF3b155, SF3b145, SF3b130, SF3b49, and p14. Probing with α-U2AF65 antibody did not detect this protein. Nucleolin, equally present in both RNA element purifications (W and C), served as a loading control.
When the affinity purification was repeated with the RNase elution step omitted, an enrichment of U2 snRNA with the 22-nt RNA element (W) was detected by Northern blotting (Figure 2F). No other examined snRNAs were detectably enriched, including U12 snRNA. We conclude that the RNA element binds intact U2 holo-snRNP under in vitro CPA conditions.
Isolated SF3b Cross-links to the RNA Element, Requiring the Conserved 7-nt Motif
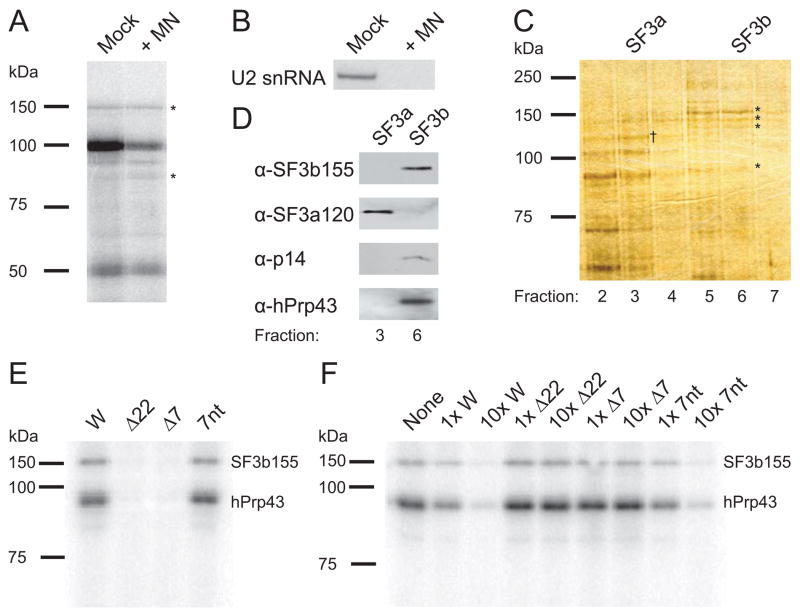
U2 snRNA binds the intronic branchpoint during splicing by base-pairing, but the 22-nt RNA element is not significantly complementary to the branchpoint recognition region or any other U2 snRNA sequence. This suggested that U2 snRNA might be unnecessary for SF3b to contact the 22-nt RNA element. Indeed, nuclear extract treated with micrococcal nuclease (MN) to destroy intact U2 snRNA still generated UV cross-links between an RNA consisting of four copies of the 22-nt RNA element and both SF3b155 and hPrp43 (Figure 3A). Successful destruction of U2 snRNA was monitored by Northern blotting (Figure 3B).
Figure 3. Isolated SF3b/hPrp43 Requires the 7-nt RNA Motif for Binding.
(A) Nuclear extract digested with micrococcal nuclease (MN) or mock treated was incubated with [α-32P]-CTP-labeled RNA containing four copies of the 22-nt RNA element and UV-irradiated. The profile of cross-linked proteins separated by 8% SDS-PAGE indicated comparable cross-linking of the 150 and 93 kDa proteins (*).
(B) A Northern blot was performed to confirm destruction of U2 snRNA by MN-treatment.
(C) Glycerol gradient fractions 2–7 (top-bottom) from the SF3b/hPrp43 purification were analyzed by 8% SDS-PAGE and silver stained. SF3b155, SF3b145, SF3b130, and hPrp43 are visible in fractions 5–7 (*), whereas SF3a components peak in fraction 3 (SF3a120 is indicated, †). A similarly sized band to hPrp43 is seen in fractions 2–4, but must be a different protein (see D below).
(D) Fractions containing SF3b (fraction 6) and SF3a (fraction 3) were compared by Western blotting for SF3b155, SF3a120, p14, and hPrp43. Fraction 3 does not contain detectable levels of hPrp43.
(E) Purified SF3b/hPrp43 was tested for cross-linking in vitro to the H2a 101-nt element (W) and to mutant RNAs with either the 22-nt RNA element (Δ22) or conserved 7-nt motif (Δ7) deleted or a 37-nt RNA containing the 7-nt motif flanked by different sequences (7nt; see Supplemental Data). Three independent experiments showed >10-fold stronger cross-linking to W than to either mutant.
(F) Purified SF3b/hPrp43 was cross-linked in vitro to the radiolabeled H2a 101-nt RNA element in the presence of the indicated unlabeled competitor RNAs at low (1x) and high (10x) concentrations. Comparable results were obtained in three independent experiments. The band at ~80 kDa is variable, probably a breakdown product.
A modified protocol (Will et al., 2001) was used to purify SF3b. After the glycerol gradient step, fractions were analyzed by SDS-PAGE (Figure 3C). Asterisks indicate three SF3b proteins, as well as a fourth protein co-sedimenting with the SF3b complex. Western blots confirmed the identity of SF3b155 and the integrity of the SF3b complex by detecting the presence of p14; moreover, hPrp43 (93 kDa) co-sediments with the SF3b complex (Figure 3D). These proteins were not observed in a slower sedimenting fraction containing crude SF3a (fraction 3).
Finally, sequence specificity of SF3b/hPrp43 binding to the complete H2a 101-nt RNA element (W) was tested by UV cross-linking analysis. Mutants with either the 22-nt RNA element (Δ22) or the conserved 7-nt motif (Δ7) deleted or the 7-nt motif flanked by different 15-nt RNA sequences (7nt) were synthesized in the presence of [α-32P]-ATP and 6-thioguanosine triphosphate to improve cross-linking efficiency. Addition to purified SF3b/hPrp43 followed by UV cross-linking revealed that both SF3b155 and hPrp43 (93 kDa) cross-linked more weakly to the deletion constructs than to the complete 101-nt RNA element (W; Figure 3E) whereas cross-linking to the 7nt RNA is equally intense. Moreover, competition experiments using excess unlabeled RNAs demonstrated that SF3b155 and hPrp43 associate only when the construct contains the 7-nt RNA motif (Figure 3F). We conclude that the 7-nt RNA motif is necessary and sufficient for SF3b/hPrp43 interaction with the H2a 101-nt RNA element in the absence of U2 snRNA.
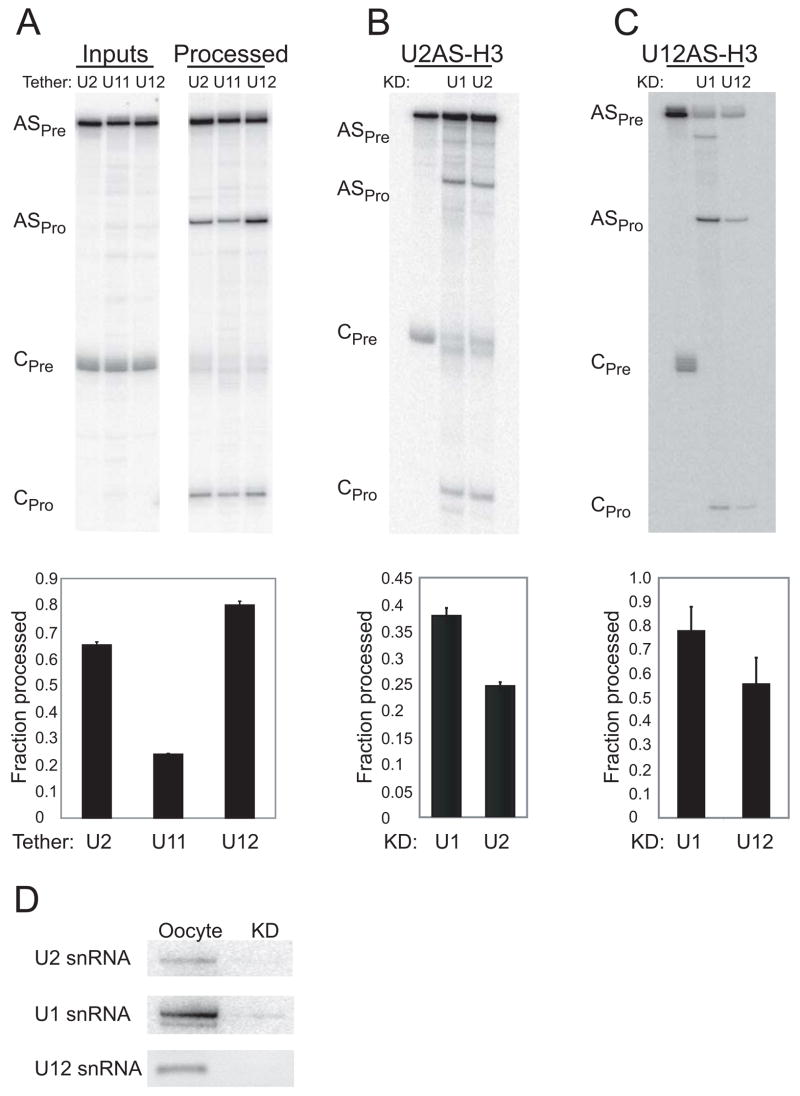
Tethering Either U2 or U12 snRNP to an RNA Enhances U7-snRNP-dependent Cleavage In Vitro and In Vivo
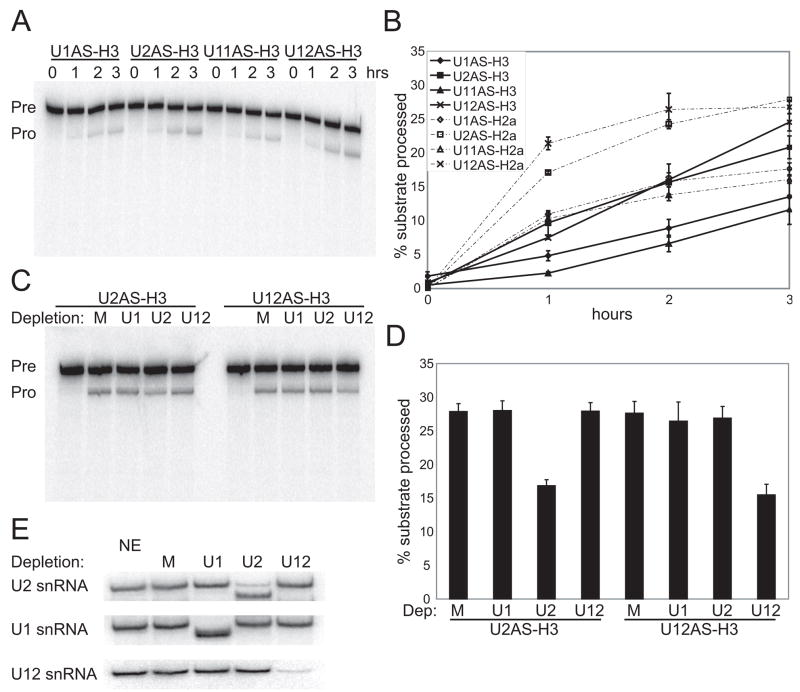
To confirm that U2 snRNP stimulates histone mRNA 3′-end formation, we asked whether four copies of the 22-nt RNA element could be replaced by a sequence that base-pairs with U2 snRNA. Single, 10-nt RNA sequences complementary to the 5′ end of U1 or of U11 snRNA or to the branchpoint recognition region of U2 or of U12 snRNA were substituted for the four 22-nt elements in the U7-snRNP substrates described in Figure 1A. U12 snRNP-tethering constructs were also examined since U12 snRNP likewise contains the SF3b complex (Will et al., 1999), and SF3b components interact with CPSF (Kyburz et al., 2006).
In nuclear extract, tethering either U2 or U12 snRNP to the RNA substrate stimulated U7-snRNP-dependent cleavage (Figures 4A and 4B) comparably to the effect observed with four copies of the 22-nt RNA element (Figures 1B and 1C). Two-fold stimulation by tethering U2 or U12 relative to U1 or U11 snRNP was seen for both U7-snRNP substrates. Enhanced processing of the U2 antisense (AS) element-containing construct was abrogated by DNA-directed RNase H cleavage of U2 snRNA present in the extract (Figures 4C and 4D), as was processing of the U12 AS-containing construct upon destruction of U12 snRNA. Knockdown efficiency was determined by Northern blotting (Figure 4E), showing >90% depletion of U1, U2, and U12 snRNAs.
Figure 4. Tethering Either U2 or U12 snRNP Stimulates U7-snRNP-Dependent Cleavage In Vitro.
A single copy of a 10-nt sequence complementary to the 5′-end of U1 or of U11 snRNA (U1AS, U11AS) or the branchpoint recognition region of U2 or of U12 snRNA (U2AS, U12AS) was substituted for 4x22nt in the H3 and H2a U7-snRNP substrates schematized in Figure 1A.
(A and B) A processing time course of the antisense (AS) element-containing H3-derived substrates in HeLa nuclear extract is shown in A. Quantitations (error bars represent standard deviations) for both the H2a and H3 substrates are in B.
(C and D) In vitro processing of the U2 or U12 tethering construct was monitored in extracts with U1, U2, or U12 snRNAs depleted by DNA-directed RNase H treatment. M is a mock-treated sample. Representative processing reactions are shown in C; the fraction processed at the 3-hr timepoint (error bars represent standard deviations) is graphed in D.
(E) Northern blotting determined the knockdown efficiencies for the DNA-directed RNase H-treated extracts used in C, compared to untreated (NE) and mock-treated (M) extracts. 3′ fragments of U1 and U2 snRNAs generated by RNase H are visualized, but the 3′ U12 snRNA fragment ran off the gel (data not shown). Knockdown efficiencies were U1 snRNA (98%), U2 snRNA (90%), and U12 snRNA (94%).
To demonstrate the in vivo activity of U2 or U12 snRNP in stimulating U7-snRNP-dependent cleavage, we used isolated Xenopus GVs. The oocyte has the advantage that various snRNAs can be knocked down prior to assaying U7-snRNP-dependent cleavage. In contrast, knockdown of SF3a proteins in HeLa cells causes cell-cycle arrest (Tanackovic and Krämer, 2005), and histone pre-mRNA processing occurs during S phase of the cell cycle (Zheng et al., 2003). We used the U2 or U12 AS-containing H3-derived substrates (U2AS-H3 and U12AS-H3). When normalized to a control U7-snRNP substrate (CPre and CPro), stimulation of U7-snRNP-dependent cleavage was observed by tethering either U2 or U12 snRNP in comparison to the substrate containing a sequence complementary to the 5′ end of U11 snRNA (Figure 5A). Because the U1 AS-containing H3-derived substrate experienced rapid degradation in multiple experiments in germinal vesicles (in contrast to nuclear extract, data not shown), we were unable to use this construct as a control. Knockdown of the corresponding snRNA using DNA-directed RNase H cleavage (Hamm et al., 1989; Yu et al., 1998) resulted in no stimulation for the U2 AS-containing substrate (Figure 5B) or the U12 AS-containing substrate (Figure 5C). After this treatment, no detectable U2 or U12 snRNA remained (Figure 5D); U1 snRNA provided a control. We conclude that the 22-nt RNA element can be replaced by tethering either U2 or U12 snRNP to a U7-snRNP substrate in vitro or in vivo.
Figure 5. Tethering either U2 or U12 snRNP Stimulates U7-snRNP-Dependent Cleavage In Vivo.
(A) The U2 or U12 snRNA antisense element-containing transcripts (ASPre; from Figure 4) were microinjected into Xenopus GVs, and U7-snRNP-dependent cleavage (ASPro) was monitored after 1 hr. AS processing was normalized (below) to a different, co-microinjected control H2a-derived U7-snRNP substrate (CPre and CPro; efficiency set to 1). U2 and U12 tethering constructs displayed enhanced processing compared to a U11 antisense element-containing construct (from Figure 4) where error bars represent standard deviations (p<0.001).
(B and C) Processing of the U2AS-H3 and U12AS-H3 constructs were monitored in GVs where either U1, U2, or U12 snRNA was knocked down by DNA-directed RNase H activity. Results are quantitated as in panel A (p<0.001 for U2AS and p<0.005 for U12AS).
(D) Knockdown (KD) efficiency was determined relative to untreated GVs by Northern blotting for U1 and U2 snRNAs and RT-PCR for U12 snRNA. The Xenopus laevis homolog of U11 snRNA has not yet been identified, so it could not be knocked down.
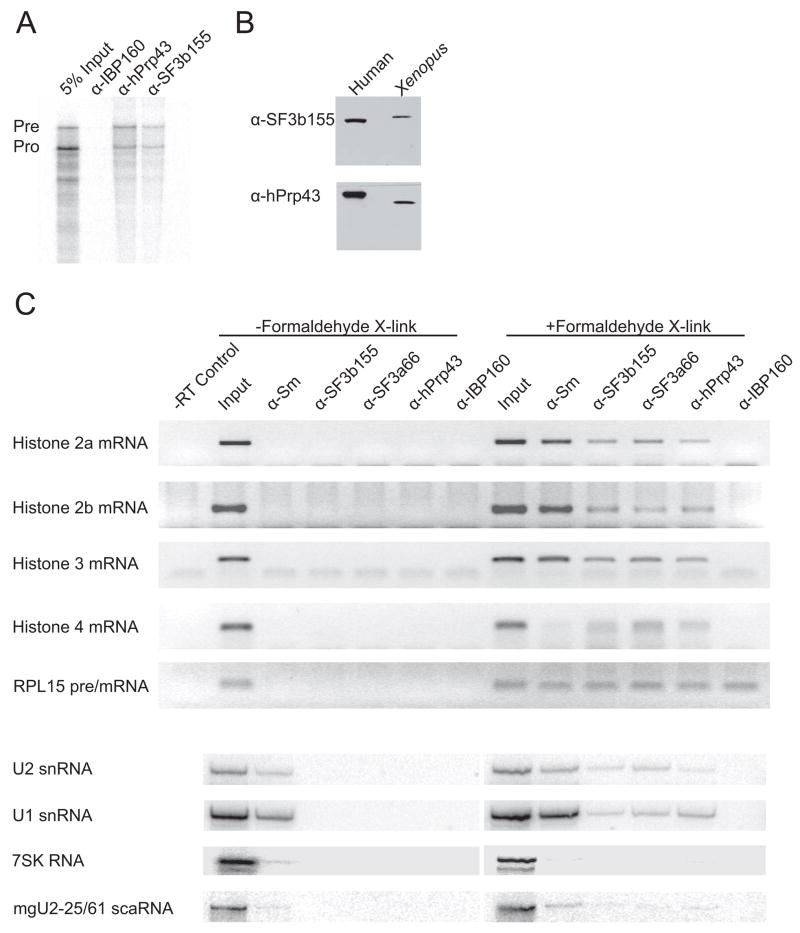
U2 snRNP Associates with Histone Pre-mRNAs In Vivo
Our observations predicted that an association between the U2 snRNP and histone pre-mRNAs should be detected in vivo. We injected the cross-linking substrate from Figure 2C into GVs and incubated the GVs to yield both unprocessed and processed RNA. The GVs were then UV-irradiated, lysed, and incubated under denaturing conditions with α-hPrp43, α-SF3b155, or control (α-IBP160, an intronic binding protein; Hirose et al., 2006) antibodies; separating co-immunoprecipitated RNAs on a denaturing gel showed an enrichment of unprocessed RNAs in both the α-hPrp43 and α-SF3b155 immunoprecipitates, but not in the control, α-IBP160, immunoprecipitate (Figure 6A). The small amount of co-immunoprecipitated RNA was consistent with the low cross-linking efficiency of UV-irradiation. Since these human antibodies have not been shown to recognize the Xenopus homologs of hPrp43 and SF3b155, Western blots were performed to verify that the antibodies bind the homologous proteins specifically (Figure 6B). We conclude that the Xenopus homologs of hPrp43 and SF3b155 both bind unprocessed histone pre-mRNA preferentially to processed mRNA.
Figure 6. Intact U2 snRNP Associates with Histone Pre-mRNAs In Vivo.
(A) A labeled U7-snRNP substrate containing the H2a 101-nt RNA element was microinjected into Xenopus GVs and incubated for 1 hr. After UV-irradiation, extracts were prepared and immunoprecipitated with α-hPrp43 or α-SF3b155 antibodies; α-IBP160 antibody served as a control. Pre-mRNA (Pre) enrichment of ~2-fold over processed product (Pro) was observed in the α-hPrp43 and α-SF3b155 precipitations in two separate experiments.
(B) Western blots with the indicated antibodies were performed on GV extract and HeLa nuclear extract, separated by SDS-PAGE.
(C) Immunoprecipitations using various α-U2 snRNP antibodies and α-IBP160 as a control were performed on extracts prepared with (+) or without (−) prior formaldehyde fixation of the HeLa cells. Histone mRNAs and RPL15 mRNA were detected via RT-PCR. The snRNAs, 7SK, and mgU2-25/61 scaRNA were detected by Northern blotting.
To verify that U2 holo-snRNP and hPrp43 associate with endogenous mammalian histone mRNAs, we performed RNA immunoprecipitation (RIP) experiments on nuclear extract prepared from in vivo formaldehyde cross-linked HeLa cells (prevents RNP rearrangement; Mili and Steitz, 2004). Among the antibodies directed against U2 snRNP proteins (α-Sm, α-SF3b155, and α-SF3a66), the α-SF3a antibody was included since this heterotrimer is found exclusively in U2 holo-snRNP (Will et al., 1999). The α-Sm antibody served as a positive control since it immunoprecipitates U7 snRNP. RT-PCR analysis showed enrichment of four histone mRNAs in the α-hPrp43 and all three α-U2 snRNP immunoprecipitates from formaldehyde cross-linked cells, but not in the control antibody (α-IBP160) precipitate (Figure 6C). A histone H4 mRNA, which lacks the exact 7-nt motif required for SF3b/hPrp43 binding in vitro, also co-immunoprecipitates. Primers directed against the exonic regions of an intron-containing pre-mRNA encoding ribosomal protein L15 (RPL15) demonstrated co-immunoprecipitation of this pre-mRNA with the control antibody directed against an intron-binding protein (α-IBP160), as well as with the various α-U2 snRNP and α-hPrp43 antibodies.
Other controls showed expected results. U2 snRNA was co-immunoprecipitated, as detected by Northern blotting (Figure 6C), with all antibodies directed against U2 snRNP components as was U1 snRNA when the cells were treated with formaldehyde (due to interactions in the spliceosome). Neither U2 nor U1 snRNA was detected in immunoprecipitates derived from cells that were not formaldehyde treated; only the antibody against Sm proteins, which do not readily dissociate from snRNAs under these conditions, co-immunoprecipitated snRNAs from untreated cells. Other nuclear RNAs, 7SK (involved in transcription elongation) and mgU2-25/61 (an independently RNA Pol II-transcribed U2 snRNA methylation guide), were co-immunoprecipitated at background levels with the α-U2 snRNP and α-hPrp43 antibodies even from cross-linked cells [the exception is mgU2-25/61 which co-immunoprecipitates with α-Sm antibody (Tycowski et al., 2004) because it associates with U2 snRNA to guide modification before SF3a and SF3b associate; Donmez et al., 2004; Yu et al., 1998].
Although the data in Figure 6C do not discriminate between histone pre-mRNAs and mRNAs, coupled with the Xenopus GV experiments (Figures 6A and 6B), the results suggest a U2 snRNP-(pre)mRNA association that continues after 3′-end formation. Co-immunoprecipitation with an α-SF3a protein antibody strongly argues that intact U2 snRNP binds histone pre-mRNAs in vivo, but cannot rule out that U12 snRNP or SF3b alone may bind.
DISCUSSION
Our observations demonstrate that an RNA element from mouse histone H2a mRNA stimulates U7-snRNP-dependent cleavage via interaction with U2 snRNP protein components and the DEAH-box helicase hPrp43 (Figures 1B and 1C). We show that this 22-nt element also stimulates CPA in vitro (Figures 1D and 1E). Direct contacts between SF3b155 and hPrp43 and the 22-nt RNA element are observed in vitro (Figures 2A and 2B), resulting in the binding of U2 holo-snRNP (Figures 2D, 2E, and 2F). Purified SF3b/hPrp43 interacts with a conserved (present in the majority of histone genes) 7-nt motif contained in the 22-nt RNA element in the absence of U2 snRNA and other U2 snRNP proteins (Figure 3), arguing for underlying protein-RNA interactions. Yet, tethering either U2 or U12 snRNP to U7-snRNP substrates via RNA-RNA interactions likewise promotes 3′-end formation in vitro and in vivo (Figures 4 and 5). Finally, we have documented U2 snRNP association with replication-dependent histone pre-mRNAs inside HeLa cells and Xenopus GVs (Figures 2C and 6), supporting the in vivo relevance of our results.
The RNA Element Represents Splicing-Independent Binding Specificity for U2 snRNP
Within U2 snRNP, the SF3b complex contacts the intron during splicing. Of this heteroheptameric complex, p14 specifically cross-links to the branchpoint adenosine (Will et al., 2001), while two members – SF3b155 and SF3b49 – cross-link to nonconserved sequences flanking the branchpoint (Gozani et al., 1996; Wang et al., 1998). Accordingly, SELEX experiments using recombinant SF3b155 did not reveal a consensus RNA binding sequence (Cass and Berglund, 2006). Therefore, our finding that SF3b, complexed with hPrp43, interacts with a specific sequence that is not the CURAYY branchpoint consensus (Patel and Steitz, 2003) represents a new binding specificity, but does not indicate which protein provides this specificity. We deduce that U2 snRNP binds a larger set of pre-mRNAs than only those containing introns.
The conserved 7-nt motif within the 22-nt RNA element (C/GAAGAAG) encodes two adjacent lysines in the amino acid sequence of H1, H2a, H2b, and H3 isoforms. Histone mRNAs codons are biased toward G/C residues at the degenerate third position (DeBry and Marzluff, 1994), and the AAG lysine codon frequency is 78% in human histone mRNAs (our observation) which predicts the 7-nt RNA motif would occur 61% of the time when adjacent lysines are encoded. While a small number of replication-dependent histone genes do not contain this conserved 7-nt motif (mostly H4 isoforms, one of which was co-immunoprecipitated with α-U2 snRNP antibodies; Figure 6C), these mRNAs contain pyrimidine-rich sequences that may bind U2AF to recruit U2 snRNP. In fact, the Drosophila homolog of U2AF65, dU2AF50, binds a large number of intronless genes (Blanchette et al., 2004), suggesting that U2 snRNP may be recruited to intronless genes in that organism.
Stimulation of 3′-end Formation by U2 snRNP Is Conserved between CPA and U7-snRNP-dependent Cleavage
The links between splicing and pre-mRNA 3′-end formation are well documented. The presence of a 3′ splice site stimulates pre-mRNA CPA, and U2 snRNP couples splicing and CPA (Kyburz et al., 2006). Kyburz et al. concluded that contacts between SF3b (a complex shared by U2 and U12 snRNPs; Will et al., 1999) and CPSF (a complex shared by CPA and U7-snRNP-dependent cleavage) mediate this interaction since both SF3b130 and SF3b49 interact in vitro with CPSF100. Our work indicates that this mechanism is conserved for replication-dependent histone mRNAs since either U2 or U12 snRNP, which both contain SF3b, can stimulate U7-snRNP-dependent cleavage (Figures 4 and 5). However, the mode of snRNP recruitment is different – via RNA-RNA interactions at the branchpoint for CPA (Kyburz et al., 2006) versus protein-RNA interactions for histone mRNA 3′-end formation (Figures 3 and 7).
Figure 7. Model for Interactions between U2 snRNP and Histone Pre-mRNAs to Stimulate U7-snRNP-dependent Cleavage.
The 7-nt conserved motif, histone 3′ stem-loop, and histone downstream element (HDE) that base-pairs with U7 snRNA are schematized. Stimulation of cleavage is proposed to occur when SF3b130 and SF3b49 mediate contact between U2 snRNP and CPSF, which contains the putative endonuclease for U7-snRNP-dependent cleavage, CPSF73 (Dominski et al., 2005; Kolev and Steitz, 2005). In contrast to the stimulation of CPA, where U2 snRNA-branchpoint interactions are critical (Kyburz et al., 2006), here the stimulatory RNA element is recognized by protein (but can be substituted by RNA-RNA) interactions.
Other proteins, mainly those containing RS domains, also stimulate CPA. The link between U2AF65 and CPA occurs via the U2AF65 RS domain interacting with the CF Im 68 RS domain (Dettwiler et al., 2004; Millevoi et al., 2006). Other SR proteins may similarly stimulate CPA, but not U7-snRNP-dependent cleavage since CF Im has not been identified as present in the heat-labile complex required for histone pre-mRNA processing (Kolev and Steitz, 2005). Thus, binding certain shuttling SR proteins (9G8 and SRp20) may be an additional function of the 22-nt RNA element that facilitates histone mRNA export to the cytoplasm (Huang and Steitz, 2001). At present, it is also unclear whether the 22-nt RNA element contacts SF3b, hPrp43, and SR proteins in a single RNP.
hPrp43 Collaborates with SF3b to Bind the Conserved RNA Element
S. cerevisiae Prp43p has been implicated in several nuclear functions. It is involved in lariat release during splicing (Arenas and Abelson, 1997); it interacts with Ntr1p and Ntr2p to facilitate the release and recycling of U2 snRNP after splicing (Tsai et al., 2005); and it contributes to ribosomal RNA biogenesis in the nucleolus (Lebaron et al., 2005). The human homolog of Prp43p (hPrp43 or DDX15) co-purifies with U2 snRNP (Will et al., 2002), suggesting that its role in intron release and recycling is conserved in higher eukaryotes. In our studies, SF3b and hPrp43 likewise co-purify and bind the conserved 7-nt motif specifically. It is unclear whether an SF3b component or hPrp43 makes initial contacts with the RNA and whether binding to the RNA element requires complex assembly. It is tempting to speculate that hPrp43 may release SF3b from bound RNA, a step that would be required in vivo since bound U2 snRNP would likely prevent efficient histone mRNA export to the cytoplasm.
U2 snRNP Binds Intronless Histone Pre-mRNAs In Vivo
The 22-nt RNA element interacts directly with SF3b, a heteroheptameric complex found in both U2 and U12 snRNPs. Our inability to detect U12 snRNP binding to the 22-nt RNA element (Figure 2) does not necessarily mean that this interaction cannot occur in vivo. In fact, when tethered, U12 snRNP stimulates U7 snRNP-dependent cleavage in vivo (Figure 5). However, the co-immunoprecipitation of comparable levels of histone mRNAs by both α-SF3b and α-SF3a antibodies (Figure 6; SF3a is exclusive to U2 snRNP; Will et al., 1999) argues that U2 holo-snRNP is the major source of histone mRNA contacts with SF3b in vivo.
There is evidence that a fraction of U2 snRNP is present at nuclear locations where histone pre-mRNAs are transcribed and processed. A significant portion of U7 snRNP co-localizes with Cajal bodies, as do the replication-dependent histone gene clusters in both Xenopus oocytes (Wu and Gall, 1993) and mammalian cells (Frey and Matera, 1995). During biogenesis, the nascent U2 snRNP transits through Cajal bodies after cytoplasmic cap-trimethylation and association of the Sm core (Nesic et al., 2004). In the Cajal body, both SF3a and SF3b are assembled onto the 11S U2 core snRNP to complete assembly of the 17S U2 holo-snRNP (Krämer et al., 2005). Additionally, at the G1 to S transition, SF3b155 is a substrate for cyclin E, which is present at histone transcription sites (Seghezzi et al., 1998; White et al., 2007). Thus, U2 snRNP is likely present in Cajal bodies during S phase (Boudonck et al., 1999), when replication-dependent histone pre-mRNAs are synthesized (Zheng et al., 2003), providing the opportunity for it to enhance U7-snRNP-dependent maturation of histone mRNAs.
EXPERIMENTAL PROCEDURES
Supplemental Data contain descriptions of constructs, nuclear extract preparation, in vitro U7-snRNP-dependent cleavage and CPA reaction conditions, MN-treatment, pCp-labeling, snRNA knockdown, and the RIP protocol.
UV Cross-linking Analysis
5 fmol of radiolabeled substrate RNAs in 1 μL of TE buffer (10 mM Tris pH 7.5 and 1 mM EDTA), 7 μL UV cross-linking buffer (0.8 mM MgCl2, 250 mM KCl, 4 μg/mL yeast total RNA, 0.8 mg/mL heparin, 15 mM HEPES pH 7.9, 2.5% glycerol, and 0.5 mM DTT), and 1 μL of nuclear extract or purified SF3b/hPrp43 were incubated (final volume of 10 μL) for 5 min at 30°C and UV-irradiated (wavelength 254 nm, distance 5 cm, 2400 mJ) at 25°C in a Stratalinker (Stratagene). 1 μL of RNase A and 2 μL of HeLa nuclear extract (which prevents non-specific interactions between the cross-linked proteins and glass during electrophoresis) were added to the samples and incubated at 37°C for 15 min. SDS-PAGE loading dye was added, and samples were analyzed either by 5–15% gradient SDS-PAGE for nuclear extract analysis or by 12% SDS-PAGE for the SF3b/hPrp43 analysis. 6-thioguanosine triphosphate (gift from Scott Strobel) was incorporated into the H2a 101-nt RNA element to improve cross-linking efficiency of purified SF3b/hPrp43 (UV irradiation at 365 nm).
Affinity Purification with the RNA Element
2.5 pmol of biotinylated 4x22-nt substrate RNA was in vitro transcribed with a CTP:biotin-14-CTP (Invitrogen) ratio to incorporate eight biotin molecules per RNA. 300 μL CPA extract was pre-cleared using 50 μL (bed volume) of streptavidin-agarose (Pierce) at 4°C for 1 hr. Substrate RNAs were resuspended after precipitation with ethanol in 210 μL of UV cross-linking buffer, 90 μL of pre-cleared extract was added, and the solution was mixed and incubated at 30°C for 5 min. The mixture was added to 50 μL (bed volume) of streptavidin-agarose and nutated for 4 hrs at 4°C. Some supernatant was saved; beads were washed with NET-2 buffer (50 mM Tris pH 7.5, 150 mM NaCl, and 0.05% NP-40) and resuspended in 50 μL of NET-2 buffer supplemented with 1 mM MgCl2. 1 μL of RNase One (Promega) was added, and samples were incubated 15 min at 37°C. Eluates were saved, and proteins were separated by SDS-PAGE and silver stained. Mass spectrometry was performed by Eugene Davidov at Yale University.
RNA was prepared by omitting RNase digestion. Beads were resuspended in G50 buffer (20 mM Tris pH 7.5, 2 mM EDTA, 300 mM NaOAc, and 0.25% SDS), treated with proteinase K, and extracted with PCA (phenol:chloroform:isoamyl alcohol, 25:24:1) followed by precipitation with ethanol. Samples were separated by denaturing PAGE, and Northern blots were probed for snRNAs.
Antibodies
Monoclonal anti-Sm (Y12) antibody has been described (Lerner et al., 1981). Anti-SF3b145 (Seghezzi et al., 1998) and anti-SF3b130 (Das et al., 1999) were provided by Robin Reed (Harvard Medical School). Anti-SF3b155 and anti-p14 (Will et al., 2001), anti-hPrp43 (Fouraux et al., 2002) and anti-IBP160 (Hirose et al., 2006) were all provided by Reinhard Lührmann (Max Planck Institute, Göttingen). Anti-SF3a120 (Krämer et al., 1994) and anti-SF3a66 (Brosi et al., 1993) were from Angela Krämer (University of Geneva). Anti-U2AF65 was provided by Juan Valcarcel (CBMSO, Madrid; Gama-Carvalho et al., 1997). Anti-SF3b49 (Abcam) and anti-SF3a60 (Novus Biologicals) were purchased.
SF3b/hPrp43 Purification
The protocol, modified from Will et al. (2001), used standard Dignam nuclear extract prepared from 3 billion HeLa cells (final volume of 9 mL). Y12 monoclonal antibody (anti-Sm) was immobilized on protein-A sepharose (100 μL bed volume), and nuclear extract was incubated with the antibody for 4 hrs at 4 °C. Beads were washed with buffer D (20 mM HEPES pH 7.9, 20% glycerol, 50 mM KCl, 0.2 mM EDTA, and 0.5 mM DTT). Proteins were eluted from the protein-A sepharose using 400 μL G600 buffer (20 mM HEPES pH 7.9, 600 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, and 5% glycerol). 250 μL of eluate was fractionated on a 15–35% glycerol gradient (in buffer G600) at 50,000 rpm for 17 hrs in a Beckman SW 50.1 rotor. 500 μL fractions were analyzed by SDS-PAGE and Western blotting.
Oocyte Microinjection
Oocytes were dissected under mineral oil to yield GVs. 5 fmol of substrate RNAs were injected in 9.2 nL with blue dextran (20 mg/mL) as an injection marker (dextran was omitted when performing UV cross-linking analysis). For UV cross-linking analysis, RNA with high specific activity (20,000 cpm/fmol) and 20 GVs were used for each timepoint. Immediately post-injection or after 1 hr, the GVs were UV-irradiated (wavelength 254 nm, distance 5 cm, 2400 mJ) at 25°C in a Stratalinker. Irradiated GVs were resuspended in 10 μL of NET-2 buffer and either treated with RNase A for 15 min at 37°C, SDS-PAGE loading dye added, and separated by 12% SDS-PAGE, or the immunoprecipitation portion of the RIP protocol (Supplemental Data) was followed after GVs were resuspended in 100 μL of binding buffer.
For the U7-snRNP-dependent cleavage reactions, injected GVs were immediately placed on dry ice or incubated 1 hr at room temperature. 6 GVs were pooled for each timepoint. Samples were digested with proteinase K and extracted with PCA; 3 GV equivalents of RNA were analyzed by denaturing PAGE. Quantitations are from three independent experiments. The remaining 3 GV equivalents were used to check snRNA knockdown efficiency.
Supplementary Material
Acknowledgments
We thank Y. Huang, A. Krämer, R. Lührmann, R. Reed, and J. Valcarcel for antibodies and plasmids and J. Friend, A. Alexandrov, N. Kolev, K. Tycowski, and S. Vasudevan for critical reading of the manuscript. This work was supported by an NIH Training Grant (to K. F.) and an NIH grant (to J. A. S.). J. A. S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Behrens SE, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Bond UM, Yario TA, Steitz JA. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- Boudonck K, Dolan L, Shaw PJ. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R, Hauri HP, Krämer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- Cass DM, Berglund JA. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry. 2006;45:10092–10101. doi: 10.1021/bi060429o. [DOI] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs JP, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Alwine JC. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BK, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBry RW, Marzluff WF. Selection on silent sites in the rodent H3 histone gene family. Genetics. 1994;138:191–202. doi: 10.1093/genetics/138.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Lührmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouraux MA, Kolkman MJ, Van der Heijden A, De Jong AS, Van Venrooij WJ, Pruijn GJ. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA. 2002;8:1428–1443. doi: 10.1017/s1355838202021076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcarcel J, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Hamm J, Dathan NA, Mattaj IW. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989;59:159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol Cell. 2006;23:673–684. doi: 10.1016/j.molcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hofmann I, Schnolzer M, Kaufmann I, Franke WW. Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol Biol Cell. 2002;13:1665–1676. doi: 10.1091/mbc.01-12-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wimler KM, Carmichael GG. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Ferfoglia F, Huang CJ, Mulhaupt F, Nesic D, Tanackovic G. Structure-function analysis of the U2 snRNP-associated splicing factor SF3a. Biochem Soc Trans. 2005;33:439–442. doi: 10.1042/BST0330439. [DOI] [PubMed] [Google Scholar]
- Krämer A, Legrain P, Mulhauser F, Groning K, Brosi R, Bilbe G. Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res. 1994;22:5223–5228. doi: 10.1093/nar/22.24.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Alwine JC. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol. 2002;22:148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 2002;3:869–874. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Krämer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Knecht S, Krämer A. Molecular characterization of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnik-David H, Moore CL, Sharp PA. Electrophoretic separation of polyadenylation-specific complexes. Genes Dev. 1987;1:672–682. doi: 10.1101/gad.1.7.672. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Krämer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol Biol Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and Cell Cycle Regulation of the Drosophila Histone Locus Body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Schneider C, Reed R, Lührmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J, Shenk T. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Wu CH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci U S A. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.