Abstract
HIV-1 integrase (IN) catalyzes biochemical reactions required for viral cDNA insertion into host cell chromosomal DNA, an essential step in the HIV-1 replication cycle. In one of these reactions, the two ends of the linear viral cDNA are believed to be simultaneously ligated to chromosomal DNA by a tetrameric form of IN. The structure of the full-length IN tetramer is not known but a model consisting of the N-terminal domain and the catalytic core revealed basic residues 186 to 188 at the interface between the two IN dimers. We found that alteration of these residues, in particular changing IN lysine reside 186 to glutamate (K186Q), impairs IN oligomerization in the yeast two-hybrid system and decreases oligomeric forms of IN within virions. When expressed independently of other viral proteins in human cells, IN-K186Q did not concentrate in the nucleus as did wild-type IN. Co-expression of wild-type IN restored the multimerization defects of IN-K186Q, in both the two-hybrid system and in virions, and also rescued the nuclear targeting defects. Virions bearing IN-K186Q were not infectious in a single cycle of replication but when mixed virions containing two different IN mutants were produced, IN-K186Q was capable of complementing the catalytically inactive mutant IN-D116A. Our biochemical and functional data support the crystallographic model in which IN residue K186 lies at the interface between IN dimers and suggest that tetramerization is important, not only for concerted integration, but also for IN nuclear targeting.
INTRODUCTION
Retroviral integration can be subdivided into two steps with distinct kinetics and requirements (reviewed in (Craigie, 2001; Hindmarsh and Leis, 1999)). The process begins with removal of a few nucleotides from the 3′ end of each strand of the linear viral cDNA. The second reaction involves attack of two phosphodiester bonds on each strand of the cellular DNA by the 3′-hydroxyls at the ends of the viral DNA. The resulting recombination intermediate is probably repaired by cellular enzymes (Craigie, 2001; Skalka and Katz, 2005). 3′ end-processing and strand transfer activities can be recapitulated in vitro (Craigie, Fujiwara, and Bushman, 1990). However, IN in solution tends to catalyze integration of only one DNA end into a given target. Only under specific conditions can concerted strand transfer of two viral DNA ends to a target DNA be carried out in vitro (Faure et al., 2005; Li et al., 2006).
Three distinct domains can be identified within HIV-1 IN (Craigie, 2001). The N-terminal domain (NTD, 1–50) contains a zinc-binding domain of the HHCC type (Bushman et al., 1993). The large central catalytic domain (CCD, 51–212) contains amino acids involved in the catalytic site, most notably a non-continuous DDE motif. The C-terminal domain (CTD, 213–288) is highly basic and is involved in the interaction with the DNA substrate (Vink, Oude Groeneger, and Plasterk, 1993). Soluble IN produced in bacteria sediments mostly as a dimer (Vincent et al., 1993). IN also forms tetramers in vitro, but tetramerization is more labile than dimerization, and is favored by high protein concentration or various buffer conditions (Faure et al., 2005; Jenkins et al., 1996; Jones et al., 1992; Lutzke and Plasterk, 1998). IN tetramers can be stabilized in vitro by the use of cross-linking agents (Cherepanov et al., 2003; Engelman, Bushman, and Craigie, 1993; Faure et al., 2005). HIV-1 IN expressed at high concentration in human or yeast cells forms dimers, tetramers and higher-order structures, as seen after stabilization using a cross-linking agent (Cherepanov et al., 2003; Faure et al., 2005). IN does not normally form trimers, implying that the protein:protein interfaces involved in dimer and tetramer formation are different. In other words, the IN tetramer is formed by specific interactions between two IN dimers.
IN 3′ end-processing and DNA end-joining activities correlate with the percentage of IN multimers in solution (Jones et al., 1992). The relevance of IN multimerization to its function was confirmed in mutant complementation experiments: two IN mutants defective for DNA end-joining, one lacking the NTD, the other one lacking the CTD, complemented each other to restore enzymatic activity (Engelman, Bushman, and Craigie, 1993). Similarly, IN lacking a functional zinc-binding motif in the NTD can complement IN with a mutation in the active site to restore DNA end-joining activity (Ellison et al., 1995). Complementation between full-length IN proteins, each bearing a different mutation, can also take place in the viral context to restore infectivity (Fletcher et al., 1997), indicating that IN is active as a multimer in vivo as well as in vitro. The importance of the tetramer for HIV-1 IN activity was demonstrated by isolating monomeric, dimeric and tetrameric IN and showing that only the tetramer was capable of directing concerted integration (Faure et al., 2005). Characterization of integration intermediates blocked by a strand transfer inhibitor revealed IN tetramers bound to the paired end of viral cDNA (Li et al., 2006).
IN-DNA cross-linking experiments were used to show that within the dimer formed by two NTD-truncated IN, one protomer binds DNA ends through its CTD, while the CCD of the other protomer contributes enzymatic activity (Gao, Butler, and Bushman, 2001). Other cross-linking studies showed that within a multimer of full-length IN, one of the protomers is responsible for binding to both viral and target DNA, while the other contributes enzymatic activity leading to DNA transfer (Heuer and Brown, 1998). The division of tasks within a tetramer of IN is less well understood. It is believed, however, that in the concerted integration reaction each dimer of IN catalyzes only one of the two DNA end-joining reactions. This conclusion is inferred from the observation that within a dimer of HIV-1 IN, the two active sites are too remote from each other to attack the target DNA 5bp apart (Podtelezhnikov et al., 2003).
IN has not been crystallized as a tetramer but computational models for tetramerization have been proposed (Podtelezhnikov et al., 2003; Wang et al., 2001). Tetramerization in vitro is greatly enhanced by the addition of zinc, supporting a crucial role for the NTD zinc-binding motif in IN tetramer formation (Zheng, Jenkins, and Craigie, 1996). Accordingly, in the IN tetramer model proposed by Craigie and colleagues (Wang et al., 2001), the protein:protein interface leading to IN tetramerization consists of symmetrical interactions between the NTD of one IN protomer and the CCD of the other one (Fig. 1). In disagreement with this model, however, is the finding that NTD-truncated IN forms tetramers in solution, (Jenkins et al., 1996). Thus, there is no clear consensus on the structure of the IN tetramer.
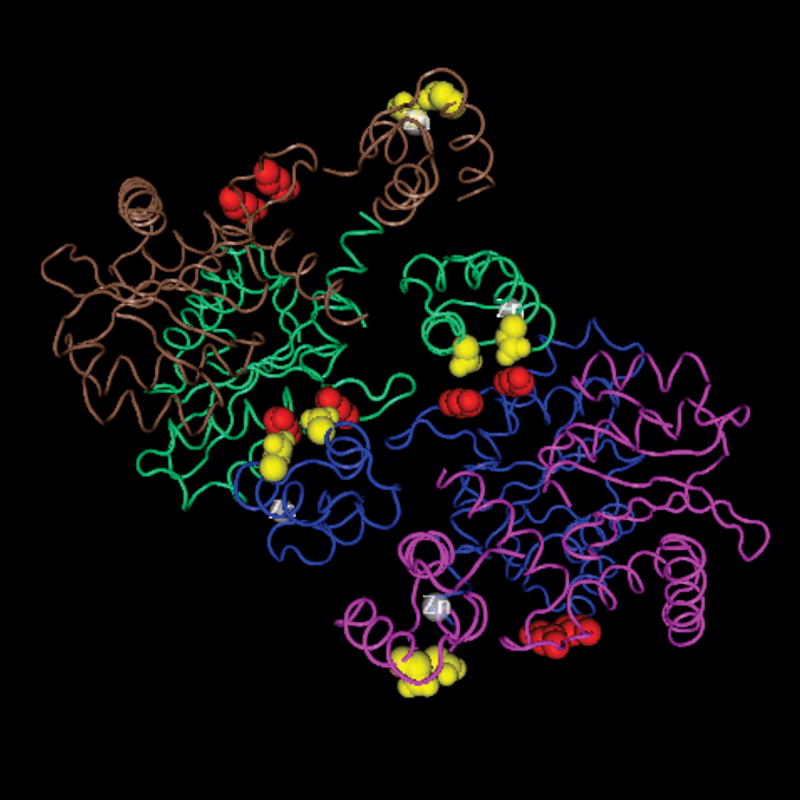
Fig. 1. KRK(186-188) and IN tetramerization.
The image was created in Cn3D, using the coordinates (accession number: 1K6Y) published by Wang and Craigie (Wang et al., 2001). Each of the four monomers of IN (N-terminal and core domains) is shown in a different color (brown, green, blue, purple), and the zinc ion is in white. For each monomer, the side chains of lysine 186 and lysine 188 are shown in red, while the side chains of glutamic acid 11 and aspartic acid 25 are shown in yellow.
Using various experimental approaches, we have examined the behavior of mutations affecting IN residues that are modeled to be points of contact between dimers in the IN tetramer (Wang et al., 2001). Our data show that indeed, mutations in this region interfere with viral replication, IN oligomerization and IN nuclear localization.
MATERIALS AND METHODS
Cells
Human cell lines 293T, HeLa, and Jurkat T cells were maintained as before (Berthoux et al., 2003).
Yeast expression constructs
pSH2 and pGADNOT, constructs for yeast expression of, respectively, the N-terminus region (DNA binding domain) of Escherichia coli LexA, and the activator domain of GAL4, have been described (Hanes and Brent, 1989; Luban et al., 1993). Both pSH2 and pGADNOT harbor BamHI and SalI restriction sites downstream of LexA or GAL4AD, respectively, and these restriction sites were used to construct all the IN fusion proteins shown in this work. HIV-1 IN, wild-type or mutated, was amplified from adequate NL4-3 (Adachi et al., 1986; Tsurutani et al., 2000) or R7/3 (Feinberg, Baltimore, and Frankel, 1991; Wiskerchen and Muesing, 1995) clones, using the oligonucleotides 5′-GACGCGGATCCTTTTTAGATGGAATAGAT (upstream oligonucleotide) and 5′-CACCACTCGAGCTAATCCTCATCCTGTCTACT (downstream oligonucleotide). The activity of the IN mutants in 2-hybrid assays was compared to the relevant wild-type controls. Of note, R7/3 and NL4-3 IN have closely similar sequences and display identical activities in the various assays shown here. All IN mutants were sequenced to verify their identity.
Proviral clones
pNL4-3 constructs containing mutations in the IN coding sequence were a kind gift of T. Masuda, Tokyo Medical and Dental University, Japan) (Tsurutani et al., 2000). These mutations were placed in the context of an envelope-mutated proviral clone, by transfer of a ~4.3 Kb Spe1-Sal1 fragment, containing the mutations, to pNLΔEN (Berthoux, Pechoux, and Darlix, 1999) cut with the same enzymes. pNLIN-HA, also called pFB277 (Bushman and Miller, 1997), encodes an NL4-3-derived, replication-competent HIV-1 isolate with an HA-tagged IN, and was a gift of F. Bushman, Salk Institute of Biological Studies, La Jolla, California. The Spe1-Sal1 fragment containing the coding sequence for IN was subcloned into pBlueScript (Stratagene). PCR products bearing the mutations were cloned in the resulting construct using the unique restriction enzyme sites Swa1 and Nde1. The upstream and downstream oligonucleotides for the PCR were 5′-AGCAGAAGTAATTCCAGCAG-3′ and 5′-TTCCTTGAAATATACATATG-3′. The oligonucleotides used to introduce the point mutations were as follow (the mutated residues are underlined): D64A, 5′-TATGGCAGCTAGCTTGTACACATTTAGAAGG-3′; D116A, 5′-CAGTACATACAGCCAATGGCAGCAATTTCAC-3′; K186Q, 5′-ACAATTTTCAAAGAAAAGGGGGGATTG-3′; KRK, 5′-ATCCACAATTTTGGGGGGATTGGGGGGTAC-3′. Wild-type and mutated Spe1-Sal1 fragments were then transferred from pBlueScript to pNLΔEN (Berthoux, Pechoux, and Darlix, 1999), thus creating pNLIN-HAΔEN clones.
Virus-free IN expression constructs
pCEP-INS-FLAG (Cherepanov et al., 2003) encodes a codon-optimized, FLAG-tagged NL43 IN and was a kind gift of Zeger Debyser (Rega Institute, Leuven, Belgium). Mutations were introduced by PCR, using the upstream oligonucleotide 5′-ATGGTGATGCGGTTTTGGCAGTA-3′ and downstream oligonucleotide 5′-ACACCTCCCCCTGAACCTGAAACA-3′. The oligonucleotides introducing the mutations were as follows: D116A, 5′-CAGTGCACACAGCTAACGGCTCCAACTTCAC-3′; K186Q, 5′-ATCCACAACTTCCAGCGAAAGGGCGGCATCG-3′;ΔKRK, 5′-GTTCATCCACAACTTCGGCGGCATCGGTGGCTAC-3′.
Yeast 2-hybrid assays
Pairs of SH2 and GADNOT constructs were transformed into Saccharomyces cerevisiae strain CTY10-5d (MATa ade2 trp1-901 leu2-3,112 his3-200 gal4-gal80-URA3::lexA-lacZ), which contains an integrated GAL1-lacZ gene with the lexA operator (Luban et al., 1993). 2–4 days later, colonies were used to inoculate 2 ml liquid cultures, and after overnight growth β-galactosidase activity was quantitatively measured as follows. Cells from 200 μl of culture were pelleted by centrifugation and resuspended in 25 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 50 mM 2-mercaptoethanol, pH 7.0). 10 μl of chloroform were added and the cells were disrupted by vortex. 200 μl of Z buffer containing 0.1 mg/ml ONPG and 0.0025% weight/vol. SDS were added and the reaction was allowed to proceed at 30 °C for 20 min to 2 hrs. The coloration was stopped by adding 100 μl of a 1M Na2CO3 solution, then the tubes were briefly spun at 10,000 rpm and 200 μl of the supernatant was transferred to 96-well plates and read at OD420 using cell-free reactions as blanks. 200 μl of the cell suspension were read at OD600 in 96-well plates using growth medium as blank, and β-galactosidase activity was computed as Miller units, using the formula (OD420×1000)/(OD600×Time[min]×Vol.[ml]). Each value represents the average β-galactosidase activity in 3 to 5 randomly picked colonies. Error bars are standard deviations.
Infections
To produce HIV-1 virions, 293T cells were transfected as previously described (Berthoux et al., 2003) with 10 μg of wild-type or mutant pNLΔEN plus 5 μg of pMD-G (Zufferey et al., 1997). Virion quantities were normalized prior to infection by using the Perkin-Elmer HIV-1 Capsid (CA(p24)) enzyme-linked immunosorbent assay (ELISA) kit. Jurkat T cells were plated at 50,000 cells in 0.5 ml per well of 24-well plates and were infected with VSV G-pseudotyped NLΔEN. 2 days after infection, cells were trypsinized, fixed for 10 min. in 2% formaldehyde-PBS, washed twice with PBS, then submitted to immune staining of CA(p24) exactly as described before (Berthoux et al., 2003). Flow cytometry analysis was done on a FACScalibur using CellQuest software (Becton Dickinson). Intact cells were identified based on light scatter profiles, and only these cells were included in the analysis. Twenty-five thousand cells per sample were processed, and cells positive for CA(p24) expression were gated and counted as the percentage of total intact cells.
Western blots
To produce HIV-1 virions for western blot analysis, 293T cells were transfected as described (Berthoux et al., 2003) with 10 μg pNLIN-HAΔEN (wild-type or mutated) for each 10-cm plate. Two days later, the supernatants from two plates were grouped, clarified by low-speed centrifugation, filtered (0.45-μm-pore-size Pall Acrodisc), then concentrated by ultracentrifugation through 25% sucrose cushions. Viral pellets were resuspended in 1X TNE (10 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA), and an aliquot was kept for reverse transcriptase assay. The rest was lysed in SDS-containing protein loading buffer without 2-mercaptoethanol. Half of each lysate was transferred to another tube and complemented with 1% vol./vol. 2-mercaptoethanol. Reduced and nonreduced lysates were denaturated by boiling and loaded onto 10% poly-acrylamide gels. After transfer on PVDF membranes, FLAG was detected by the use of a monoclonal mouse antibody (Sigma; dilution 1:2000).
Immunofluorescence microscopy
HeLa cells (2 × 105 per well of 6-well plates) were transfected with pCEP-INs-FLAG (1 μg). 12 hours later, the cells were trypsinized and plated on LabTek Permanox four-chamber slides (5 × 104 per well). The next day, cells were fixed for 15 min in freshly prepared 2% formaldehyde, washed three times in PBS, and then permeabilized with 0.1% Triton X-100. Samples were probed with antibodies against FLAG (mouse monoclonal; Sigma) at a 1:500 dilution, and Caveolin (rabbit polyclonal; Santa-Cruz) at a 1:200 dilution. Fluorescent staining was done using Alexa488-conjugated goat anti-mouse and Alexa594-conjugated goat anti-rabbit antibodies and Hoechst33342 (all from Molecular Probes) to reveal DNA. Pictures were generated using a Nikon TE300 microscope with the Openlab 3.0 software.
RESULTS
Mutations in the KRK(186-188) region affect IN oligomerization
Although full-length HIV-1 IN has not been crystallized, structures have been solved for truncated IN lacking either the C-terminal domain (Wang et al., 2001) (NCBI accession #1K6Y) or the N-terminal domain (Chen et al., 2000) (NCBI accession #1EX4). In each of these published structures, the basic residues at positions 186 to 188 are located far from the IN:IN dimer interface (Fig. 1). Based on their crystallographic data of IN with the C-terminal domain truncated, Wang et al (Wang et al., 2001) propose a model for the structure of IN as a tetramer in which basic residues 186 to 188 are located at the dimer:dimer interface responsible for tetramerization. Specifically, K186 and K188 would engage in polar interactions with acidic residues (E11 and D25, respectively) present within the N-terminal region of one of the IN constituents of the other dimer.
To analyze the role of the KRK motif in IN oligomerization, we constructed a series of IN substitution mutants in the yeast 2-hybrid system. The yeast 2-hybrid system has been used before to investigate IN:IN interactions (Kalpana and Goff, 1993). We fused the reading frames of wild-type (WT) IN and various IN mutants to LexA and GAL4, and transformed pairs of plasmids into the indicator yeast cells, CTY10-5d. We then quantified β-galactosidase activity in transformants (Fig. 2A). Mutations of histidine 183 (to alanine) and of phenylalanine 185 (to lysine) had minor effects on the capacity of the mutated IN to interact with itself. In contrast, substitution of lysine 186 to glutamine or alanine strongly decreased oligomerization of IN (about 6-fold). Substitution of arginine 187 to alanine also decreased IN oligomerization in this assay, though the magnitude of the effect was smaller compared with mutation of the lysine at position 186. Mutation of the lysine 188 to alanine showed only a slight reduction in IN oligomerization (about 1.5-fold). Double or triple substitutions in the KRK motif also strongly decreased IN oligomerization (Fig. 2a). The reduction of IN oligomerization caused by mutations at position 186 was not explained by a decrease in protein expression, as seen by quantitative western blotting analysis (not shown).
Fig. 2. Integrase oligomerization in a yeast 2-hybrid assay.
A, The indicated IN substitution mutations were cloned into the LexA and GAL4AD expression vectors, and the interaction of each mutant protein with itself was evaluated in the two-hybrid system. β-galactosidase activity for three to five transformants is reported in Miller units with standard deviation. Control represents the result obtained in cells transformed with LexA-IN and empty GAL4AD. B is as in A, except that the IN mutants K186Q and ΔKRK fused with LexA were assayed against wild-type IN (WT IN) or the identical IN mutants fused with GAL4AD.
K186Q and ΔKRK IN mutants differ in their capacity to interact with WT IN
K186Q IN mutant viruses are noninfectious (Tsurutani et al., 2000) and show markedly decreased levels of cDNA synthesis in acutely infected cells. The deletion mutant ΔKRK (186-188) is also noninfectious, and also shows a cDNA synthesis defect (Tsurutani et al., 2000). Thus, we compared the level of oligomerization of these two mutants in the yeast 2-hybrid system. We also tested their ability to interact with wild-type IN. As shown in fig. 2B, compared with WT IN, K186Q IN was again markedly defective in its ability to homodimerize (11-fold decrease). ΔKRK IN also showed decreased interaction with itself, although the reduction was much smaller (2-fold). Therefore, the lysine at position 186 is not absolutely required for high levels of IN oligomerization in the yeast. Interaction between K186Q IN and WT IN was 3-times more efficient than K186Q IN homodimerization. Interestingly, ΔKRK showed the opposite phenotype, as interaction between ΔKRK IN and WT IN was 6-times less efficient than interaction of ΔKRK with itself (Fig. 2B). This result suggests that the KRK (186-188) region of IN is important for its oligomerization, but that deletion of this motif in one of the partners can be compensated by introduction of the identical deletion in the other partner.
Analyzing IN oligomerization in virions
Upon denaturation in non-reducing conditions of HIV-1 virions proteins, 2-mercaptolethanol-sensitive IN dimers are observed in western blot analysis (Petit, Schwartz, and Mammano, 2000), although establishment of such disulfide bonds was found to be dispensable for virus infectivity (Bischerour et al., 2003). We first analyzed the capacity of K186Q and ΔKRK IN to engage in 2-mercaptoethanol-sensitive dimers (Fig. 3A). For that, these mutations were introduced in pNLIN-HAΔEN, a version of pNL43ΔEN in which the IN protein bears an HA tag at its C-terminus (Bushman and Miller, 1997). Utilization of this provirus, which encodes a functional IN, allows for easy detection of IN by western blot. We transfected WT or mutant NL43IN-HAΔEN into 293T cells, collected the virions, and prepared protein lysates in the presence or not of 2-mercaptoethanol. Western blot analysis showed the presence of a band whose size was consistent with a dimer of IN, and which disappeared after 2-mercaptoethanol treatment (Fig 3A). Two mutations affecting IN catalytic site residues, D64A and D116A, did not interfere with the formation of the disulfide-bond dimer (Fig. 3A). In contrast, the amount of dimer was reduced for K186Q and ΔKRK IN. However, the extent of decrease in the percentage of dimer for K186Q IN was difficult to assess with accuracy, because the amount of monomeric IN was smaller for K186Q than for WT IN or the other mutants (Fig. 3A). Interestingly, upon analysis of the 2-mercaptoethanol treated samples, the amount of monomeric IN was identical for all mutants and the WT (Fig. 3B). ΔKRK IN weakly formed reduction-sensitive homodimers (Fig. 3A), although this mutant did interact with itself in the yeast 2-hybrid assay (Fig. 2). This might be explained by the fact that the interaction detected in the yeast 2-hybrid assay takes place at the dimer:dimer interface, while the reduction-sensitive disulfide bonds probably form at the level of the monomer:monomer interface. Alternatively, the discrepancy might reflect differences in behavior between IN proteins expressed in yeast and virion-associated IN proteins.
Fig. 3. Detection of reduction-sensitive IN dimers in virions.
A. Virions containing HA-tagged IN (wild-type or mutated as indicated in each lane) were produced, and concentrated by ultracentrifugation. Virion proteins were then prepared in a lysis buffer lacking 2-mercaptoethanol and analyzed by western blot, using an antibody against HA. The positions of molecular weight markers are indicated on the right (sizes in kDa). B. Same as in A except that the proteins were reduced by addition of 2-mercaptoethanol before loading on the gel. In C, virions were produced by cotransfection of two proviral clones, one expressing an HA-tagged version of IN, the other expressing untagged IN. Virion proteins were analyzed as in A. The nature of the bands detected by the HA antibody is shown on the left. D. Same as in C, except that the proteins were reduced by addition of 2-mercaptoethanol before loading on the gel.
Oligomerization of K186Q IN in virions is rescued in the presence of WT IN
In the 2-hybrid experiments, K186Q IN interacted with WT IN more efficiently than ΔKRK did (Fig. 2). We thus investigated interactions between mutant and WT IN proteins in virions. For that, we analyzed disulfide-bond-mediated dimers of wild-type or mutant IN-HA in the presence of wild-type or mutant untagged IN. Virions containing both tagged and untagged IN were produced by cotransfecting 293T cells with pNLIN-HAΔEN and the parent construct pNLΔEN, which expresses an untagged IN. Virions were lysed in the absence (Fig. 3C) or presence (Fig. 3D) of 2-mercaptoethanol, and HA-tagged IN was detected by western blot analysis. In the absence of 2-mercaptoethanol, we detected 2 bands for WT IN-HA (lane 1 in Fig 3C) whose sizes were consistent with IN dimer formation. Presumably, the upper, heavier band consisted of a homodimer of HA-tagged IN, while the lower band was a heterodimer of HA-tagged IN and untagged IN. D116A IN-HA was also capable of forming disulfide-bond dimers (lane 4). When virions contained WT or D116A IN-HA and also contained untagged K186Q IN, disulfide bonds seemed to form more easily between two IN-HA proteins rather than between a tagged and an untagged protein (lanes 2 and 5). When the untagged IN was ΔKRK, the presence of a smear at the level of the dimer bands prevented us to determine whether disulfide bonds were formed between ΔKRK IN and either WT or D116A IN-HA (lanes 3 and 6). Both the monomer and dimer forms of K186Q IN-HA were barely detectable in 2-mercaptoethanol-untreated virions containing both K186Q IN-HA and untagged K186Q IN (lane 7). However, when the virions were lysed in the presence of 2-mercaptoethanol, the amounts of K186Q IN-HA detected were as high as for WT IN-HA and the other mutants (Fig. 3D). Thus, the low levels of K186Q IN-HA seen in Fig. 3C were not the result of a lack of viral particles production, confirming the findings from Fig. 3A. Control experiments using IN monoclonal antibodies (not shown) ruled out the possibility of poor detection of K186Q IN in Fig. 3A and 3C. One possibility is that nonreduced K186Q IN formed abnormal high-molecular-weight disulfide-bonds dimers that did not enter the gel. However, when virions contained a mix of K186Q IN-HA and either WT or D116A IN, near-WT levels of K186Q IN-HA were detected in western blots in nonreducing conditions (lanes 8 and 9 in Fig. 3C). K186Q was detected in the dimeric form as well, which seemed to consist more of homodimers than of heterodimers. Thus, WT or D116A IN corrected the behavior of K186Q IN-HA in our assay, indicating that interactions were taking place between these proteins. As in Fig. 3A, ΔKRK was found to be strongly impaired in its capacity to form disulfide-bond dimers (lane 10 in Fig. 3C). Moreover, in contrast with K186Q IN-HA, the presence of untagged WT or D116A IN did not reestablish a WT-like phenotype for this mutant (lanes 11 and 12). Thus, the phenotype of ΔKRK IN was distinct from that of K186Q IN in this experiment.
K186Q IN intracellular localization defect is corrected by the presence of WT IN
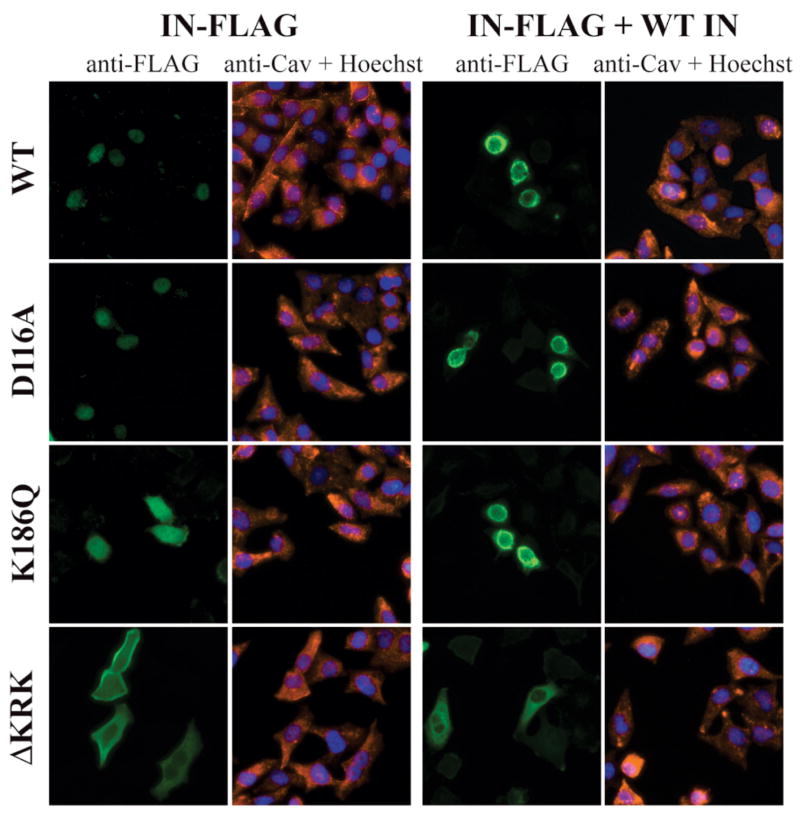
K186Q and ΔKRK IN both show multimerization defects in yeast 2-hybrid experiments and in virions. We investigated whether this could result in altered intracellular distribution of the virus-free protein. We expressed a codon-optimized (“stabilized”) version of IN, bearing a C-terminal FLAG (Cherepanov et al., 2003) in HeLa cells, and performed immunofluorescence (IF) microscopy with an anti-FLAG antibody (Fig. 4). As expected, both WT and D116A IN-FLAG were nuclear (Petit, Schwartz, and Mammano, 1999; Pluymers et al., 1999; Tsurutani et al., 2000). K186Q IN-FLAG was found both in the nucleus and in the cytoplasm (Fig. 4). ΔKRK IN-FLAG, in contrast, was mostly cytoplasmic, and in some cells accumulated at the plasma membrane level. We then analyzed whether co-expression of untagged WT IN could modify the distribution of mutant IN proteins. When WT IN-FLAG was co-transfected in HeLa cells with untagged WT IN, the localization was slightly modified. IN-FLAG was still nuclear, but part of the signal was concentrated at the level of the nuclear membrane (Fig. 4). This was most probably the result of IN overexpression. In the presence of untagged WT IN, both D116A and K186Q IN-FLAG showed similar distribution as WT IN-FLAG. In contrast, ΔKRK IN-FLAG, expressed in the presence of untagged IN, was diffuse cytoplasmic, with a slight concentration around the nucleus. Thus, K186Q IN, but not ΔKRK IN, had a WT-like localization pattern in the presence of WT IN, suggesting that an interaction was taking place between K186Q and WT IN proteins.
Fig. 4. Intracellular localization of virus-free IN.
FLAG-tagged IN (wild-type or mutated, as indicated on the left) was expressed in HeLa cells by transient transfection. In the right panels, FLAG-tagged IN was coexpressed with untagged wild-type IN by cotransfection of the two plasmids (1:1 ratio). IN was detected by indirect immune fluorescence microscopy, using an antibody against the FLAG epitope (green signal). The cells were counterstained with an antibody directed against caveolin, a plasma membrane marker (red signal), and with Hoechst33342 to reveal DNA (blue signal).
Virions containing a mixture of K186Q and D116A IN are infectious
K186Q and ΔKRK IN both show a multimerization defect (Fig. 2 and Fig. 3) and a nuclear transport defect (Fig. 4). Both mutations cause a total loss of infectivity, as seen in Fig. 5 and in published work (Tsurutani et al., 2000). We reasoned that the IN enzymatic site, comprising the D64-D116-E152 conserved motif, might still be intact and functional in these two mutants, since the enzymatic site is located at the opposite of the KRK motif in the published IN 2-domain structure shown Fig. 1. Mutations in the DDE motif, such as D116A, result in defects specific to the integration step, and abrogate infectivity (Fig. 5) (Engelman and Craigie, 1992; Kulkosky et al., 1992; Wiskerchen and Muesing, 1995). Interestingly, at the highest dose of virus used, D116A viruses seemed to be mildly infectious. Transient expression of HIV-1 proteins has been observed for mutants of the DDE motif (Wiskerchen and Muesing, 1995), and is thought to be due to expression from nonintegrated viral DNA (Nakajima, Lu, and Engelman, 2001; Poon and Chen, 2003; Wu and Marsh, 2001; Wu and Marsh, 2003). To analyze whether replication of D116A IN could be restored in the presence of either K186Q or ΔKRK IN, we produced viruses by cotransfection of the corresponding mutant NL43ΔEN plasmids. The infectivity of VSV G-pseudotyped viruses containing both ΔKRK and D116A IN was not rescued to any obvious extent (Fig. 5). In contrast, viruses containing a mixture of K186Q and D116A IN were infectious. The infectivity of these virions was reduced about 10-times compared to WT virions. Therefore, K186Q and D116A blocked HIV-1 replication at distinct steps and could complement each other to restore infectivity. Of note, such functional complementation between mutants in the DDE motif and other point mutants in the CCD has been observed before (Lu et al., 2005).
Fig. 5. Viruses containing both K186Q IN and D116A IN are infectious.
Jurkat T cells were infected with serial dilutions of VSV G-pseudotyped wild-type NLΔEN virus, with virus bearing the indicated mutations in IN, or with viral particles containing a mixture of 2 or 3 different mutated IN. The latter viruses were produced by cotransfection of the corresponding mutated pNLΔEN proviral clones. Viruses were normalized by p24 ELISA. A. Jurkat T cells were infected with a single dose of virus, corresponding to 12.5 ng of p24. The percentage of cells expressing p24 was determined 2.5 days later by flow cytometry and is indicated for each data panel. In B the experiment was performed using multiple doses of the viruses, and the percentage of cells expressing p24 is plotted against the amount of input virus.
DISCUSSION
Significance of IN oligomerization in yeast cells
Mutations in the 186-188 region decrease IN:IN oligomerization in a yeast 2-hybrid assay (Fig. 1). The IN CCD domain, that contains the KRK motif, has been crystallized, either alone (Dyda et al., 1994) or with the CTD (Chen et al., 2000) or the NTD (Wang et al., 2001). Although the exact location of the KRK motif varies between the various models, all place it away from the protein:protein interface responsible for dimer formation. However, Wang and colleagues propose that K186 and K188 are present at the dimer:dimer interface. These structural data suggest that in the yeast 2-hybrid system used here, the IN:IN interaction affected by mutations in the 186-188 region is the one leading to tetramerization, not dimerization.
Phenotype of K186Q and ΔKRK mutant viruses
Protein sequence alignment shows that the IN KRK motif is conserved among lentiviruses found in primates and other species (Engelman and Craigie, 1992). Both K186Q and ΔKRK mutations abrogate infectivity (Lu et al., 2004; Petit, Schwartz, and Mammano, 2000; Tsurutani et al., 2000). Production and maturation of virions are not affected by these mutations, but low levels of HIV-1 cDNA accumulate in acutely infected cells (Lu et al., 2004; Tsurutani et al., 2000), a phenotype which we have confirmed (data not showed). Analysis of disulfide-bond-mediated IN dimers in virions (Fig. 3) suggested that IN oligomerization was disturbed by mutations in the KRK motif, in agreement with the yeast 2-hybrid data. We could not analyze the multimerization of virus-free IN-FLAG in mammalian cells after treatment with the crosslinking agent DTSSP (Cherepanov et al., 2003), as in our hands, this treatment resulted in nonspecific dimer formation for all of the IN mutants (not shown). GFP-IN fusions of K186Q and ΔKRK IN were previously found to be nuclear (Tsurutani et al., 2000). FLAG-tagged K186Q IN was nuclear in one study (Lu et al., 2004) while it was both cytoplasmic and nuclear in another one (Petit, Schwartz, and Mammano, 2000). In our hands, FLAG-tagged K186Q distributed in both cytoplasm and nucleus, while ΔKRK IN was totally defective for nuclear targeting (Fig. 4). The explanation for this discrepancy is unclear. IN localization could also be dependent on its intracellular concentration, or on other factors such as the timing of analysis relative to DNA transfection. Our data indicate that the K186Q mutation does not abrogate the catalytic potential of the enzyme, since it can rescue infectivity of D116A mutant IN in mixed virions (Fig. 5).
In summary, IN oligomerization defects are associated with reduced HIV-1 cDNA levels and reduced IN nuclear targeting. It is conceivable that abnormal oligomerization of IN along the nascent viral cDNA may hinder completion of reverse transcription, thus explaining the low levels of viral cDNA observed for mutants of the KRK region. Alternatively, IN tetramerization might be required for correct protection of the nascent viral cDNA against cellular DNases (Nermut and Fassati, 2003). The reduction in cDNA levels, however, does not account for the full abrogation of infectivity caused by K186Q and ΔKRK mutations. Thus, subsequent steps of replication must be inhibited as well. The viral cDNA synthesized in replication complexes containing K186Q and ΔKRK IN is correctly transported to the nucleus, as judged by the presence of normal amounts of 1-LTR and 2-LTR circles (Lu et al., 2004; Petit, Schwartz, and Mammano, 2000). It is not clear yet what the different viral determinants for nuclear targeting of the replications complexes are, but cis sequences in the HIV-1 cDNA, like the central PPT, are probably involved (Zennou et al., 2000). It is thus possible that mutations in the KRK domain, by preventing nuclear targeting of IN (Fig. 4), causes it to dissociate from the viral cDNA when the latter translocates to the nucleus. In that model, HIV-1 cDNA would be present in the nucleus but would not be able to integrate due to a lack of integrase proteins. A second, non-excluding possibility for the lack of infectivity of K186Q and ΔKRK is that these mutations prevent concerted integration, which would be consistent with the tetramerization defect (Faure et al., 2005; Li et al., 2006). Finally, one could speculate that mutations in the 186-188 region might disrupt the structure of the nearby LEDGF-binding domain (residues 168 to 171) (Cherepanov et al., 2005a; Cherepanov et al., 2005b), thus interfering with IN nuclear transport and function (Ciuffi et al., 2005; Llano et al., 2006; Llano et al., 2004). However, K186Q interacted with LEDGF as efficiently as the WT control in a 2-hybrid assay (data not shown).
Interactions between mutant and WT IN
Several lines of evidence suggest that WT IN can interact with K186Q IN but not with ΔKRK IN. In yeast cells, K186Q IN interacted with WT IN better than it interacted with itself, while ΔKRK IN interacted with WT IN less efficiently than with itself (Fig. 2). The analysis of disulfide-mediated IN dimerization in virions showed that WT IN was able to modify multimerization of K186Q IN, while it had no effect on the inability of ΔKRK IN to form disulfide-mediated dimers (Fig. 3). Finally, WT IN was able to correct the cellular distribution of K186Q IN but not of ΔKRK IN (Fig. 4). Consistently, virions containing a mixture of K186Q IN and D116A were infectious, while virions containing a mixture of ΔKRK IN and D116A IN were not (Fig. 5). Presumably, in tetramers containing both K186Q and D116A IN proteins, two heterodimers (each containing one K186Q IN and one D116A IN) are associated through interactions between the D116A proteins, while the K186Q proteins catalyze the two integration reactions (one for each dimer). That complementation between ΔKRK and D116A does not occur could be due to the inability of ΔKRK to form dimers with either itself or with D116A IN. This is suggested by the lack of apparent interaction between WT IN and ΔKRK IN in HeLa cells (Fig. 4). Alternatively, the KRK mutation could abrogate activity of the enzymatic site.
Anti-integrase inhibitors
Successful integration of HIV-1 involves protein:protein and protein:DNA interactions, all of whom are required and could be individually targeted by drugs. Indeed, our data suggest that a single mutation specifically affecting tetramer formation can totally disrupt infectivity. A better knowledge of the structural determinants for IN multimerization would likely reveal avenues for drug development.
Acknowledgments
The authors wish to thank Takao Masuda, Zeger Debyser and Frederic Bushman for reagents. L.B. was supported by a fellowship from AmFAR (106524-35-RFHF) and a scholarship from the Elizabeth Glaser Pediatric AIDS Foundation. This work was also supported by the National Institutes of Health (USA) Grant RO1AI36199 and the Swiss National Science Foundation Grant 3100A0-113558 to J.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Pechoux C, Darlix JL. Multiple effects of an anti-human immunodeficiency virus nucleocapsid inhibitor on virus morphology and replication. J Virol. 1999;73(12):10000–9. doi: 10.1128/jvi.73.12.10000-10009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Towers GJ, Gurer C, Salomoni P, Pandolfi PP, Luban J. As(2)O(3) enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J Virol. 2003;77(5):3167–80. doi: 10.1128/JVI.77.5.3167-3180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischerour J, Leh H, Deprez E, Brochon JC, Mouscadet JF. Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J Virol. 2003;77(1):135–41. doi: 10.1128/JVI.77.1.135-141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993;90(8):3428–32. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Miller MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71(1):458–64. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A. 2000;97(15):8233–8. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A. 2005a;102(48):17308–13. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278(1):372–81. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005b;12(6):526–32. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11(12):1287–9. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Craigie R. HIV integrase, a brief overview from chemistry to therapeutics. J Biol Chem. 2001;276(26):23213–6. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62(4):829–37. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266(5193):1981–6. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Ellison V, Gerton J, Vincent KA, Brown PO. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270(7):3320–6. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. Embo J. 1993;12(8):3269–75. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66(11):6361–9. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33(3):977–86. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A. 1991;88(9):4045–9. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, 3rd, Soares MA, McPhearson S, Hui H, Wiskerchen M, Muesing MA, Shaw GM, Leavitt AD, Boeke JD, Hahn BH. Complementation of integrase function in HIV-1 virions. Embo J. 1997;16(16):5123–38. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Butler SL, Bushman F. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. Embo J. 2001;20 (13):3565–76. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57(7):1275–83. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Heuer TS, Brown PO. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37(19):6667–78. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- Hindmarsh P, Leis J. Retroviral DNA integration. Microbiol Mol Biol Rev. 1999;63(4):836–43. doi: 10.1128/mmbr.63.4.836-843.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271(13):7712–8. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;267(23):16037–40. [PubMed] [Google Scholar]
- Kalpana GV, Goff SP. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc Natl Acad Sci U S A. 1993;90(22):10593–7. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12(5):2331–8. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. Embo J. 2006;25(6):1295–304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An Essential Role for LEDGF/p75 in HIV Integration. Science. 2006;314(5798):461–4. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78(17):9524–37. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Limon A, Devroe E, Silver PA, Cherepanov P, Engelman A. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J Virol. 2004;78(23):12735–46. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Limon A, Ghory HZ, Engelman A. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. J Virol. 2005;79(4):2493–505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73(6):1067–78. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Lutzke RA, Plasterk RH. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol. 1998;72 (6):4841–8. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N, Lu R, Engelman A. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated dna recombination: definition of permissive and nonpermissive T-cell lines. J Virol. 2001;75(17):7944–55. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermut MV, Fassati A. Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J Virol. 2003;77(15):8196–206. doi: 10.1128/JVI.77.15.8196-8206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Schwartz O, Mammano F. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J Virol. 1999;73(6):5079–88. doi: 10.1128/jvi.73.6.5079-5088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Schwartz O, Mammano F. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J Virol. 2000;74(15):7119–26. doi: 10.1128/jvi.74.15.7119-7126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluymers W, Cherepanov P, Schols D, De Clercq E, Debyser Z. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology. 1999;258(2):327–32. doi: 10.1006/viro.1999.9727. [DOI] [PubMed] [Google Scholar]
- Podtelezhnikov AA, Gao K, Bushman FD, McCammon JA. Modeling HIV-1 integrase complexes based on their hydrodynamic properties. Biopolymers. 2003;68(1):110–20. doi: 10.1002/bip.10217. [DOI] [PubMed] [Google Scholar]
- Poon B, Chen IS. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J Virol. 2003;77(7):3962–72. doi: 10.1128/JVI.77.7.3962-3972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12(Suppl 1):971–8. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- Tsurutani N, Kubo M, Maeda Y, Ohashi T, Yamamoto N, Kannagi M, Masuda T. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J Virol. 2000;74(10):4795–806. doi: 10.1128/jvi.74.10.4795-4806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent KA, Ellison V, Chow SA, Brown PO. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67(1):425–37. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C, Oude Groeneger AM, Plasterk RH. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21(6):1419–25. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. Embo J. 2001;20(24):7333–43. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskerchen M, Muesing MA. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69(1):376–86. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293(5534):1503–6. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J Virol. 2003;77(19):10376–82. doi: 10.1128/JVI.77.19.10376-10382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101(2):173–85. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Zheng R, Jenkins TM, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci U S A. 1996;93(24):13659–64. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15(9):871–5. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]