Abstract
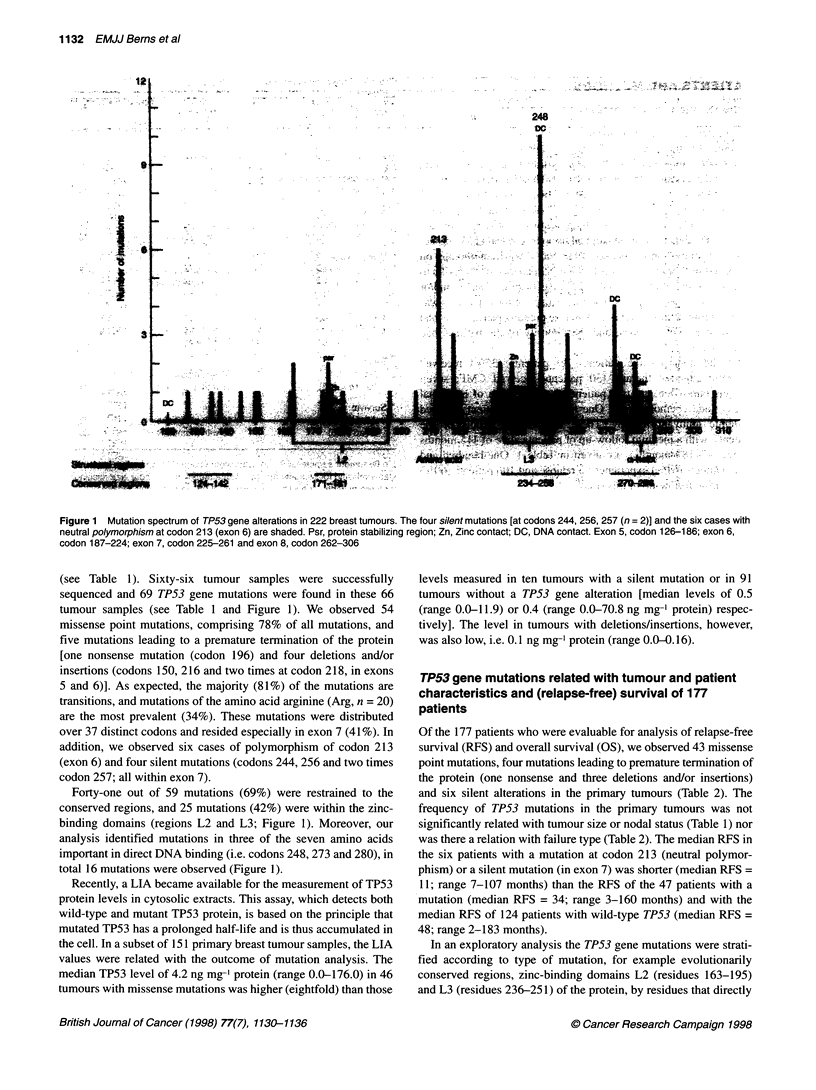
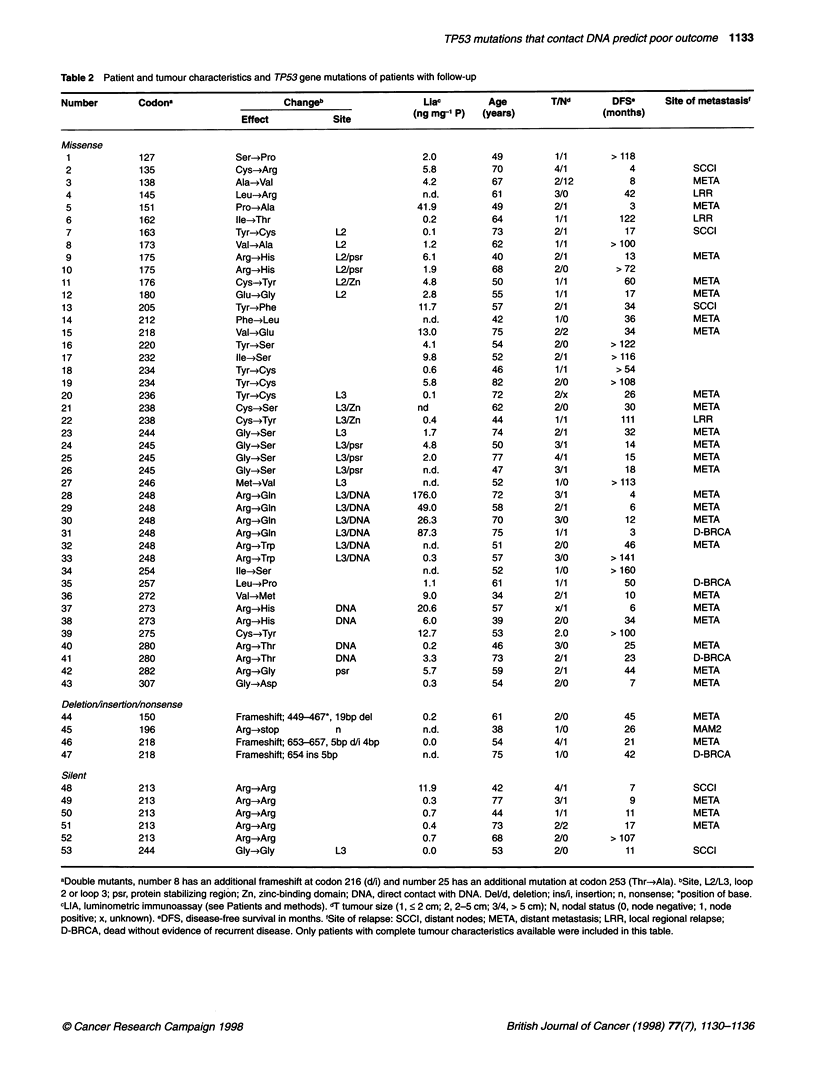
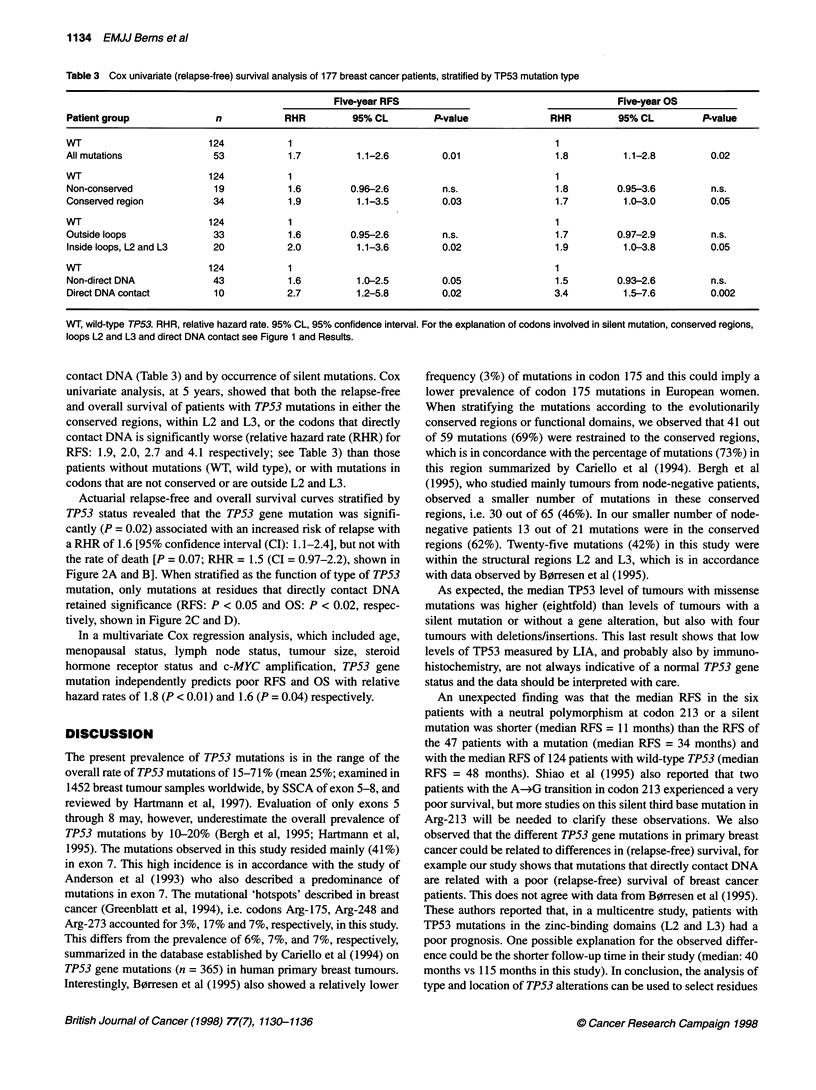
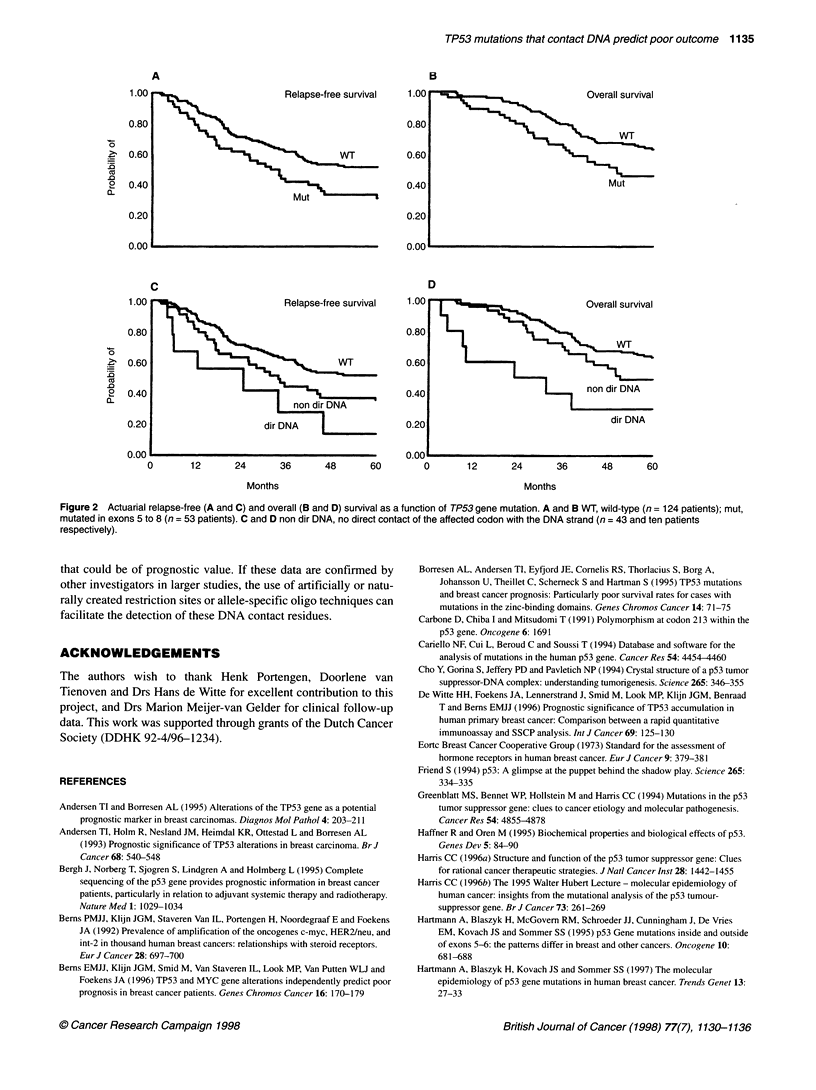
The tumour-suppressor gene TP53 is frequently mutated in breast tumours, and the majority of the mutations are clustered within the core domain, the region involved in DNA binding. We searched for alterations in this central domain of the TP53gene in 222 human breast cancer specimens using polymerase chain reaction-single-strand conformation analysis (PCR-SSCA) followed by sequencing. TP53 gene mutations were observed in 66 tumours (31%), including three tumours that contain two mutations. Fifty-four (78%) of these mutations were missense point mutations, one was a nonsense mutation and four were deletions and/or insertions causing disruption of the protein reading frame, whereas four mutations were either silent or a polymorphism (at codon 213; n = 6). Interestingly, the majority of missense mutations were observed at codon 248. The outcome has been related with patient and tumour characteristics, and with prognosis in 177 patients who were eligible for analysis of both relapse-free and overall survival (median survival for patients alive was 115 months). There was no significant association between the frequency of TP53 mutations and menopausal or nodal status, or tumour size. In a Cox univariate analysis, TP53 gene mutation was significantly associated with poor relapse-free survival (RFS: P = 0.02) but not with overall survival (OS: P = 0.07). In a Cox multivariate analysis, including classical prognostic factors, TP53 gene mutation independently predicted poor RFS and OS (RHR = 1.8 and 1.6 respectively). Unexpectedly, the median relapse-free survival of patients with a polymorphism at codon 213 or with a silent mutation was shorter (median 11 months) than the median relapse-free survival of patients with or without a TP53 gene mutation (median 34 or 48 months respectively). In an exploratory subset analysis, mutations in codons that directly contact DNA were related with the poorest relapse-free (P < 0.05) and overall survival (P < 0.02). These data imply that in the analysis of the prognostic value of TP53, the type of mutation and its biological function should be considered.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen T. I., Holm R., Nesland J. M., Heimdal K. R., Ottestad L., Børresen A. L. Prognostic significance of TP53 alterations in breast carcinoma. Br J Cancer. 1993 Sep;68(3):540–548. doi: 10.1038/bjc.1993.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh J., Norberg T., Sjögren S., Lindgren A., Holmberg L. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat Med. 1995 Oct;1(10):1029–1034. doi: 10.1038/nm1095-1029. [DOI] [PubMed] [Google Scholar]
- Berns E. M., Klijn J. G., Smid M., van Staveren I. L., Look M. P., van Putten W. L., Foekens J. A. TP53 and MYC gene alterations independently predict poor prognosis in breast cancer patients. Genes Chromosomes Cancer. 1996 Jul;16(3):170–179. doi: 10.1002/(SICI)1098-2264(199607)16:3<170::AID-GCC3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Berns E. M., Klijn J. G., van Staveren I. L., Portengen H., Noordegraaf E., Foekens J. A. Prevalence of amplification of the oncogenes c-myc, HER2/neu, and int-2 in one thousand human breast tumours: correlation with steroid receptors. Eur J Cancer. 1992;28(2-3):697–700. doi: 10.1016/s0959-8049(05)80129-7. [DOI] [PubMed] [Google Scholar]
- Børresen A. L., Andersen T. I., Eyfjörd J. E., Cornelis R. S., Thorlacius S., Borg A., Johansson U., Theillet C., Scherneck S., Hartman S. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer. 1995 Sep;14(1):71–75. doi: 10.1002/gcc.2870140113. [DOI] [PubMed] [Google Scholar]
- Carbone D., Chiba I., Mitsudomi T. Polymorphism at codon 213 within the p53 gene. Oncogene. 1991 Sep;6(9):1691–1692. [PubMed] [Google Scholar]
- Cariello N. F., Cui L., Beroud C., Soussi T. Database and software for the analysis of mutations in the human p53 gene. Cancer Res. 1994 Aug 15;54(16):4454–4460. [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P. D., Pavletich N. P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994 Jul 15;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Friend S. p53: a glimpse at the puppet behind the shadow play. Science. 1994 Jul 15;265(5170):334–335. doi: 10.1126/science.8023155. [DOI] [PubMed] [Google Scholar]
- Greenblatt M. S., Bennett W. P., Hollstein M., Harris C. C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994 Sep 15;54(18):4855–4878. [PubMed] [Google Scholar]
- Haffner R., Oren M. Biochemical properties and biological effects of p53. Curr Opin Genet Dev. 1995 Feb;5(1):84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- Harris C. C. The 1995 Walter Hubert Lecture--molecular epidemiology of human cancer: insights from the mutational analysis of the p53 tumour-suppressor gene. Br J Cancer. 1996 Feb;73(3):261–269. doi: 10.1038/bjc.1996.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A., Blaszyk H., Kovach J. S., Sommer S. S. The molecular epidemiology of p53 gene mutations in human breast cancer. Trends Genet. 1997 Jan;13(1):27–33. doi: 10.1016/s0168-9525(96)10043-3. [DOI] [PubMed] [Google Scholar]
- Hartmann A., Blaszyk H., McGovern R. M., Schroeder J. J., Cunningham J., De Vries E. M., Kovach J. S., Sommer S. S. p53 gene mutations inside and outside of exons 5-8: the patterns differ in breast and other cancers. Oncogene. 1995 Feb 16;10(4):681–688. [PubMed] [Google Scholar]
- Kinzler K. W., Vogelstein B. Life (and death) in a malignant tumour. Nature. 1996 Jan 4;379(6560):19–20. doi: 10.1038/379019a0. [DOI] [PubMed] [Google Scholar]
- Levine A. J., Momand J., Finlay C. A. The p53 tumour suppressor gene. Nature. 1991 Jun 6;351(6326):453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Prives C. How loops, beta sheets, and alpha helices help us to understand p53. Cell. 1994 Aug 26;78(4):543–546. doi: 10.1016/0092-8674(94)90519-3. [DOI] [PubMed] [Google Scholar]
- Shiao Y. H., Chen V. W., Scheer W. D., Wu X. C., Correa P. Racial disparity in the association of p53 gene alterations with breast cancer survival. Cancer Res. 1995 Apr 1;55(7):1485–1490. [PubMed] [Google Scholar]
- Velculescu V. E., El-Deiry W. S. Biological and clinical importance of the p53 tumor suppressor gene. Clin Chem. 1996 Jun;42(6 Pt 1):858–868. [PubMed] [Google Scholar]
- de Witte H. H., Foekens J. A., Lennerstrand J., Smid M., Look M. P., Klijn J. G., Benraad T. J., Berns E. M. Prognostic significance of TP53 accumulation in human primary breast cancer: comparison between a rapid quantitative immunoassay and SSCP analysis. Int J Cancer. 1996 Apr 22;69(2):125–130. doi: 10.1002/(SICI)1097-0215(19960422)69:2<125::AID-IJC10>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]


