Abstract
The mammalian nuclear pore complex (NPC) is comprised of ∼50 unique proteins, collectively known as nucleoporins. Through fractionation of rat liver nuclei, we have isolated >30 potentially novel nucleoporins and have begun a systematic characterization of these proteins. Here, we present the characterization of Nup96, a novel nucleoporin with a predicted molecular mass of 96 kD. Nup96 is generated through an unusual biogenesis pathway that involves synthesis of a 186-kD precursor protein. Proteolytic cleavage of the precursor yields two nucleoporins: Nup98, a previously characterized GLFG-repeat containing nucleoporin, and Nup96. Mutational and functional analyses demonstrate that both the Nup98-Nup96 precursor and the previously characterized Nup98 (synthesized independently from an alternatively spliced mRNA) are proteolytically cleaved in vivo. This biogenesis pathway for Nup98 and Nup96 is evolutionarily conserved, as the putative Saccharomyces cerevisiae homologues, N-Nup145p and C-Nup145p, are also produced through proteolytic cleavage of a precursor protein. Using immunoelectron microscopy, Nup96 was localized to the nucleoplasmic side of the NPC, at or near the nucleoplasmic basket. The correct targeting of both Nup96 and Nup98 to the nucleoplasmic side of the NPC was found to be dependent on proteolytic cleavage, suggesting that the cleavage process may regulate NPC assembly. Finally, by biochemical fractionation, a complex containing Nup96, Nup107, and at least two Sec13- related proteins was identified, revealing that a major sub-complex of the NPC is conserved between yeast and mammals.
Keywords: nuclear pore complex, nucleoporin, proteolytic cleavage, nucleocytoplasmic transport, RNA export
For proteins and RNAs to traffic between the nucleus and the cytoplasm of eukaryotic cells, they must pass through large supramolecular structures in the nuclear membrane known as nuclear pore complexes (NPCs).1 The selection of substrates for transport serves as an important point for controlling cell functions, and considerable interest has been focused on understanding how specific substrates are recognized and targeted to NPCs (for review see Mattaj and Englmeier, 1998; Pemberton et al., 1998). Peptide motifs (or possibly RNA sequences in some cases) present in selected cargo function as nuclear localization signals (NLSs), or nuclear export signals (NESs; for review see Boulikas, 1993). These signals are recognized and bound by soluble receptors known as karyopherins (or importins and exportins), that function to target the substrates to the NPC. The lysine/arginine-rich NLS motifs first identified in the SV-40 large T antigen (Kalderon et al., 1984) and in nucleoplasmin (Robbins et al., 1991) are among the best characterized nuclear transport signals, and all proteins having this type of NLS appear to be transported into the nucleus by a common pathway using a karyopherin heterodimer consisting of karyopherin α and karyopherin β1. However, it is now clear that there are distinct transport pathways, using distinct NLSs and NESs and cognate karyopherins (see Pemberton et al., 1998). However, each of these pathways converges at the NPC where the receptor/substrate complexes bind and are transported across the nuclear envelope in an energy dependent process. The mechanism by which the substrate/receptor complexes are translocated through the NPC is still poorly defined. Efforts to understand the regulation of this process have focused on two areas: the role of the small GTPase Ran and its regulatory factors (for reviews, see Rush et al., 1996; Melchior and Gerace, 1998), and the structure and function of the NPC itself.
At 125 million daltons, the vertebrate NPC is ∼30 times the size of a ribosome (Reichelt et al., 1990). Three-dimensional structures of the NPCs from X. laevis oocytes have been determined at ∼9 nm resolution and reveal a central cylinder (or transporter), surrounded by a spoke-ring complex that serves to anchor the cylinder in the nuclear envelope (Hinshaw et al., 1992; Akey and Radermacher, 1993). Attached to both the cytoplasmic and nucleoplasmic faces of the NPC are filaments that extend 50–100 nm (or more) away from the NPC (Goldberg and Allen, 1996; Cordes et al., 1997). On the nucleoplasmic face of the NPC, these filaments form a basket-like structure that likely has an important role in mRNA export (Ris, 1991; Kiseleva et al., 1996).
The mammalian NPC is composed of ∼50 unique proteins, known collectively as nucleoporins (for reviews see Rout and Wente, 1994; Bastos et al., 1995). In vertebrates, the task of identifying and characterizing the majority of the nucleoporins has only started, as less than one-third have been analyzed at the molecular level. The characterization of this small subset of nucleoporins has, however, provided considerable insights into the structure and function of the NPC. Among the first characterized nucleoporins are a family containing the highly repeated peptide motifs, FXFG and FG (Davis and Blobel, 1986; Sukegawa and Blobel, 1993; Kraemer et al., 1994; Yokoyama et al., 1995; Wu et al., 1995; Hu et al., 1996). Several of these repeat-containing nucleoporins are localized asymmetrically to either the cytoplasmic, or nucleoplasmic sides of the NPC. Nup358 and Nup214, for example, are associated with the filaments on the cytoplasmic face of the NPC (Kraemer et al., 1994; Yokoyama et al., 1995; Wu et al., 1995), whereas Nup98 and Nup153 are located at or near the nucleoplasmic basket (Sukegawa et al., 1993; Radu et al., 1995). p62, on the other hand, forms a complex with three other FXFG/FG-containing nucleoporins, and this complex localizes to both sides of the NPC, near the central transporter (Guan et al., 1995; Hu et al., 1996). Because individual NPCs are presumed to function in both nuclear import and nuclear export, the distribution of these proteins could be an important determinant of the directionality of transport.
Interactions between karyopherin–substrate complexes and components of the NPC have provided insight into how translocation through the NPC may occur. In vitro binding assays have established that karyopherin–substrate complexes bind, in a regulated manner, to the subset of FXFG/FG-containing nucleoporins (Moroianu et al., 1995; Radu et al., 1995; Rexach and Blobel, 1995). This finding, along with the distribution of the repeat-containing nucleoporins along the entire length of the NPC, has suggested that substrates are translocated through the NPC by a series of association and dissociation reactions (Moroianu et al., 1995; Radu et al., 1995; Rexach and Blobel, 1995). These reactions are likely to be coordinated by Ran and its regulators (reviewed in Rush et al., 1996; Melchior and Gerace, 1998), but the exact mechanisms that give rise to vectorial transport through the NPC remain unknown. Furthermore, it is unlikely that the NPC is simply a passive and stationary player in the translocation process. Whereas small particles (<70 Å in diameter) are able to diffuse through the NPC, particles of <250 Å in diameter are transported in a signal-mediated process (Paine et al., 1975; Dworetzky et al., 1988). These findings suggest a regulated dilation, or gating of the NPC during transport. Experiments by Feldherr and Akin (1997) have suggested that a single gate may exist near the center of the central transporter, whereas others have observed potential gates at both the nucleoplasmic and cytoplasmic ends of the transporter that open during RNP export (Kiseleva et al., 1998). In addition to the possible gating of the central transporter, dramatic conformational changes in the nucleoplasmic basket-like structure of the NPC have also been observed during RNP export, again suggesting a dynamic, rather than passive NPC. No data is yet available as to the mechanisms regulating the central transporter or the nucleoplasmic basket, in large measure due to the lack of information concerning the molecular composition and organization of these structures.
To obtain a more complete understanding of the composition and function of the NPC, we have developed a procedure for identifying and purifying nucleoporins from rat liver nuclei. Using this purification procedure, we have isolated ∼30 novel nucleoporins and have begun to characterize them at the molecular level. Here, we report the characterization of a 96-kD nucleoporin, termed Nup96. We demonstrate that Nup96 is synthesized as a 186-kD precursor, and that proteolytic cleavage of this precursor gives rise to two nucleoporins, Nup96 and Nup98. In addition, we show that the previously characterized Nup98, which is produced independently from an alternatively spliced mRNA, is also proteolytically processed. Like Nup98, Nup96 was localized to the nucleoplasmic side of the NPC, possibly associated with the nucleoplasmic basket. Through mutational and functional analysis, we found that proteolytic processing is required for the correct targeting of both Nup98 and Nup96 to the NPC. The apparent yeast homologue of the Nup96-Nup98 precursor, S. cerevisiae Nup145p, is also proteolytically processed to generate two nucleoporins (Dockendorff, 1997; Emtage et al., 1997; Teixeira et al., 1997), indicating that this biogenesis pathway is evolutionarily conserved. Moreover, a complex containing Nup107 (the yeast Nup84p homologue), Nup96 (the yeast C-Nup145p homologue) and two Sec13-related proteins was identified. A similar complex has been described in yeast, indicating that not only the processing of Nup96, but also its incorporation into a specific sub-domain of the NPC, is conserved between yeast and mammals.
Materials and Methods
Isolation and Fractionation of Rat Liver Nuclear Envelopes
Rat liver nuclei were isolated as described (Blobel and Potter, 1966) and stored frozen at −80°C in 100 unit aliquots (1 U = 3 × 106 nuclei). Nuclear envelopes were prepared by a modification of the procedure described by Dwyer and Blobel (1976). Nuclei were thawed and pelleted at 500 rpm in a tabletop microfuge for 1 min. After removing the supernatant, the pellet was resuspended to a final concentration of 100 U/ml by drop-wise addition of cold buffer A (0.1 mM MgCl2, protease inhibitors [0.5 mM phenylmethanesulfonyl fluoride {PMSF}, 1 μg/ml leupeptin, 1 μg/ ml pepstatin A, and 18 μg/ml aprotinin], 5 μg/ml DNase I [Sigma Chemical Co.], and 5 μg/ml RNase A [Sigma Chemical Co.]) with constant vortexing. The nuclei were then immediately diluted to 20 U/ml by addition of ice-cold buffer B (10% sucrose, 20 mM triethanolamine (pH 8.5), 0.1 mM MgCl2, 1 mM DTT and protease inhibitors), again with constant vortexing. The suspension was dounced four times in a glass dounce homogenizer (tight pestle) and incubated at room temperature for 15 min. After the 15-min incubation, the suspension was underlaid with 5 ml of ice-cold buffer C (30% sucrose, 20 mM triethanolamine, pH 7.5, 0.1 mM MgCl2, 1 mM DTT and protease inhibitors) and centrifuged at 4,100 g in a swinging bucket rotor (Sorvall type HB-4) for 15 min at 4°C. After removing the supernatant and sucrose cushion, the pellet was resuspended to a final concentration of 100 U/ml in ice-cold buffer D (10% sucrose, 20 mM triethanolamine, pH 7.5, 0.1 mM MgCl2, 1 mM DTT and protease inhibitors). The suspension was dounced as above, and diluted to 66 U/ml by addition of cold buffer D containing 0.3 mg/ml heparin (Sigma Chemical Co.). The suspension was immediately underlaid with 5 ml of buffer C and pelleted as above. The pellet resulting from this second extraction is operationally defined as the nuclear envelope fraction. After removing the supernatant and sucrose cushion, the pellet was resuspended to a final concentration of 100 U/ml in ice-cold buffer D. The suspension was dounced as above and diluted to 66 U/ml by addition of cold buffer D containing 3% Triton X-100 and 0.075% SDS. The suspension was immediately underlaid with 5 ml of buffer C and pelleted as above. The pellet resulting from this second extraction is operationally defined as the pore complex lamina fraction (PCLF). The PCLF was resuspended in cold buffer D at 100 U/ml, dounced as above, and diluted to 66 U/ml with buffer D containing 0.9% Empigen BB (Calbiochem-Novabiochem). Samples were incubated on ice for 5 min and subsequently pelleted for 15 min at 15,000 g for 15 min. The supernatant fraction, containing nucleoporins minus the nuclear lamins, was precipitated with 10% TCA. Precipitated proteins were resuspended in sample buffer, and the equivalent of 10 U of nuclei were analyzed by Coomassie blue staining.
Isolation of the Nup96/Nup107 Sub-Complex
The PCLF was isolated as described above and resuspended (1,000 U/ml) in buffer D plus 1% Triton X-100, 0.025% SDS, and 0.1 mg/ml heparin. After resuspension, the samples were immediately spun at 15,000 g for 10 min at 4°C. The supernatant was then loaded onto a 10–40% sucrose gradient (triethanolamine pH 7.5, 0.1 mM MgCl2) and spun at 40,000 rpm in a Beckman SW 41 rotor for 17 h at 4°C. 750-μl fractions were collected, TCA precipitated and analyzed by SDS-PAGE and subsequent silver staining, or immunoblotting.
cDNA Cloning and Plasmid Construction
All clones and constructs were sequenced by the Rockefeller University Protein/DNA Technology Center using an ABI automated DNA sequencer. To isolate cDNAs encoding for Nup96, a 48-nucleotide (nt) ssDNA probe was synthesized based on the region of clone GM2B8 (kindly provided by Dr. Mark Rutherford, University of Minnesota, St. Paul) coding for the sequenced peptide “LVVRHLASDAIINENYD” (Yang et al., 1995). This oligonucleotide probe was used to screen a HeLa cDNA library (kindly provided by Dr. Jian Wu, Rockefeller University, New York). Two classes of cDNA clones, differing by a 225-nt insert, were obtained in approximately equal numbers. All of the isolated clones had incomplete 5′ ends, and 5′ RACE was therefore performed using human fetal liver cDNA (Clontech Laboratories) as template. The 5′ RACE product and the longest clone obtained from the HeLa cDNA library were assembled at an overlapping ACC I site, and the full-length clone was then ligated into the SalI and NotI sites of a modified pAlter-MAX vector (Promega Corp.) containing an in-frame myc epitope tag. Modified myc-pAlter-MAX, containing the myc epitope tag, has been previously described (Matunis et al., 1998).
Myc-tagged Nup96 was generated by PCR using the wild-type Nup98-Nup96 cDNA as template. An oligonucleotide complementary to the 5′ end of the Nup96 ORF (beginning with serine 864) and containing a SalI site was used in conjunction with an antisense oligonucleotide complementary to sequences at the 3′ end of the Nup98-Nup96 ORF and containing a NotI site. The resulting PCR product was digested with SalI and NotI and ligated into the SalI/NotI sites of myc-pAlter-MAX. The myc-tagged, full-length Nup98 was generated using a similar PCR based approach using the Nup98-Nup96 cDNA as template. An oligonucleotide complementary to the 5′ end of the Nup98-Nup96 ORF was used in conjunction with an antisense oligonucleotide complementary to the 3′ end of the Nup98 ORF (ending with proline 912). The oligonucleotide also contained an additional 18 nt coding for the 6 COOH-terminal amino acids of full-length human Nup98 that are not encoded by the Nup98-Nup96 precursor site (see Borrow et al., 1996; Nakamura et al., 1996) and a NotI restriction site. The resulting PCR product was digested with SalI and NotI and ligated into the SalI/NotI sites of myc-pAlter-MAX. The cleaved form of Nup98 (amino acids 1–863) was also generated by PCR using the wild-type Nup98-Nup96 cDNA as template, and the resulting PCR product was cloned into the SalI/NotI sites of myc-pAlter-MAX.
The Nup98-Nup96 precursor double mutant (F863S/Y866R), and the similar Nup98 double mutant, were generated by recombinant PCR using mismatched primers and the myc-tagged Nup98-Nup96 precursor cDNA as template (Higuchi, 1990).
Peptide Sequence Analysis
Proteins from fractionated nuclear envelope were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. The appropriate proteins were excised after identification by staining with Ponceau S. The proteins were extracted or were digested on the membrane with endoproteinase Lys-C. Peptides were separated and sequenced as previously described (Fernandez et al., 1994).
Expression of Recombinant Segment of Nup96 and Antibody Production
For protein production, a DNA fragment corresponding to amino acids 1291–1482 of the Nup98-Nup96 precursor was obtained by PCR and cloned into the NdeI and AvaI sites of the bacterial expression vector pET21a (Novagen, Inc.). E. coli strain BL21 (DE3) pLysE was transformed with the expression vector, and expression of the recombinant protein was induced by adding 0.2 mM isopropyl thio-β-d-galactoside to the culture media. The recombinant protein was purified by Nickel-NTA agarose as described by the manufacturer (QIAGEN Inc.).
For antibody production, rabbits were injected with purified, recombinant protein and serum was collected after an appropriate response had been elicited. Antibodies were purified from the crude serum by affinity chromatography. The recombinant protein was immobilized on Affi-Gel 15 according to the manufacturer (Bio-Rad Laboratories), and serum was incubated with the gel for 12 h. The gel was loaded into a column, and the column was washed with 1 M NaCl in PBS. Bound antibodies were eluted with 0.1 M glycine (pH 2.5), and the eluate immediately neutralized with 1 M Tris (pH 8.0).
SDS-PAGE and Immunoblot Analysis
Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane as previously described (Dreyfuss et al., 1984). Nitrocellulose membranes were blocked with 5% nonfat dry milk in PBS containing 0.05% NP-40, and the blots were probed with affinity-purified Nup96 antibodies diluted 1:300 in PBS containing 2% BSA. Nup107 was detected using an affinity-purified rabbit polyclonal antibody (Radu et al., 1994). Antibodies were detected using luminol-based chemiluminescence.
Immunofluorescence Microscopy and Cell Transfections
HeLa cells were grown in DME supplemented with 10% fetal calf serum. For immunofluorescence, cells were grown overnight on coverslips, washed in PBS, fixed in 2% formaldehyde/PBS for 30 min at room temperature, and permeabilized with −20°C acetone for 3 min. Affinity-purified Nup96 antibodies (diluted 1:300), the anti-myc monoclonal antibody 9E10 (Evan et al., 1985; diluted 1:1,000), and rabbit anti-Nup358 antibodies (Wu et al., 1995; diluted 1:500) were incubated with the cells for 1 h at room temperature. Cells were washed in PBS and incubated with either fluorescein-conjugated goat anti–rabbit (Organon/Teknika), and/or Cy3-conjugated donkey anti–mouse (Jackson ImmunoResearch) antibodies for 30 min at room temperature. Cells were washed again in PBS and mounted in 80% glycerol, 50 mM Tris-HCl (pH 8.0), 0.1% p-phenylenediamine. Samples were analyzed with a Zeiss laser scanning confocal microscope.
For transfections, HeLa cells were grown on 35-mm dishes and were transfected with 2 μg of DNA using lipofectamine (GIBCO-BRL) according to the manufacturer's instructions. After 36 h, cells were assayed either by immunofluorescence microscopy or by immunoblot analysis as described above.
Immunogold Electron Microscopy
Rat liver nuclear envelopes were prepared as described above. Isolated nuclear envelopes were fixed for 15 min in 2.5% formaldehyde in STM (10% sucrose, 20 mM triethanolamine-HCl, pH 7.5, 0.1 mM MgCl2) and centrifuged at 2,000 g for 5 min onto 35-mm tissue culture dishes. The pelleted nuclear envelopes were washed three times with 1% BSA, 68 mM NaCl, 13 mM KCl, 15 mM KH2PO4, 40 mM Na2HPO4, and 0.5 mM PMSF and then incubated with affinity-purified anti-Nup96 antibodies diluted 1:300, and mouse monoclonal antibody 19C7 (specific for RanGAP1; Matunis et al., 1996), or mouse monoclonal antibody 5E10 (specific for Tpr; Matunis, M.J., and G. Blobel, unpublished data) diluted 1:1,000. Bound antibodies were detected with goat anti–rabbit IgG conjugated with 10-nm gold, or goat anti–mouse IgG conjugated with 5-nm gold (Amersham Life Science Inc.).
For in situ labeling of intact cells, HeLa cells grown on 35-mm tissue culture dishes were first permeabilized with digitonin (50 μg/ml) for 5 min at room temperature, fixed with 2% formaldehyde in PBS for 30 min at room temperature, incubated with primary and secondary antibodies as indicated above and processed for thin sectioning and EM as previously described (Pain et al., 1990).
In Vitro Transcription/Translation Assays
All Nup98 and Nup98-Nup96 precursor proteins were synthesized in rabbit reticulocyte lysate transcription and translation extracts, in the presence of [35S]methionine, as described by the manufacturer (Promega Corp.). Proteins were separated by SDS-PAGE and analyzed by autoradiography.
Results
Nup96 Is a Novel Nucleoporin Associated with the Nucleoplasmic Side of the NPC
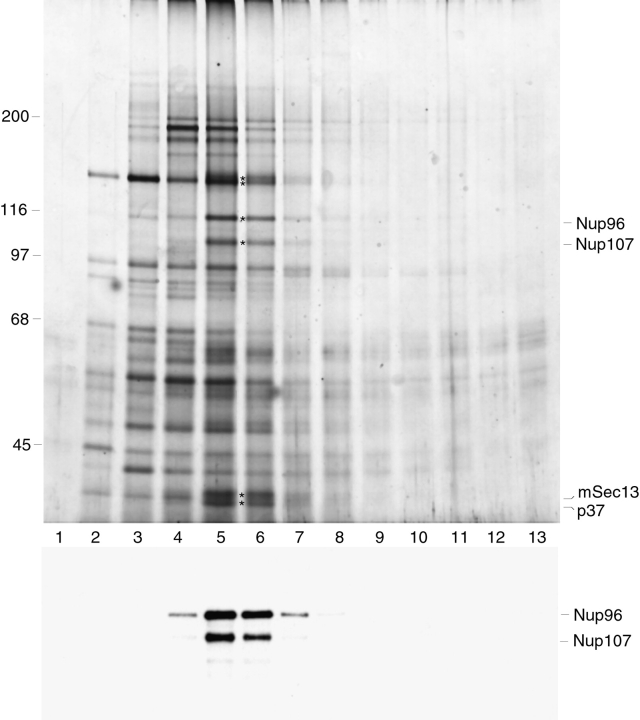
We have developed a novel procedure for purifying the protein components of the mammalian NPC (Matunis et al., 1996). In the final step of this procedure, NPCs are solubilized and released from the nuclear lamina by extraction with the zwitterionic detergent, Empigen BB. The Empigen BB supernatant fraction contains all of the currently known nucleoporins, in addition to ∼30 uncharacterized, candidate nucleoporins (Fig. 1). One of the major uncharacterized proteins, with an apparent molecular mass of 115 kD (indicated by the asterisk in Fig. 1), was analyzed by peptide sequence analysis, and the peptide sequence was used to isolate a full-length cDNA clone encoding this protein (see following sections). Based on its predicted molecular mass of 96 kD, and its association with NPCs, this novel nucleoporin has been termed Nup96. Nup96 runs anomalously at 115 kD by SDS-PAGE.
Figure 1.
Identification of Nup96, a novel nucleoporin with an apparent molecular mass of 115 kD. Nucleoporins were purified from isolated rat liver nuclear envelopes, separated by SDS-PAGE, and stained with Coomassie brilliant blue. A previously uncharacterized protein migrating at 115 kD (indicated by an asterisk) was analyzed by peptide sequence analysis. Additional known nucleoporins are indicated. Numbers at left indicate M r markers.
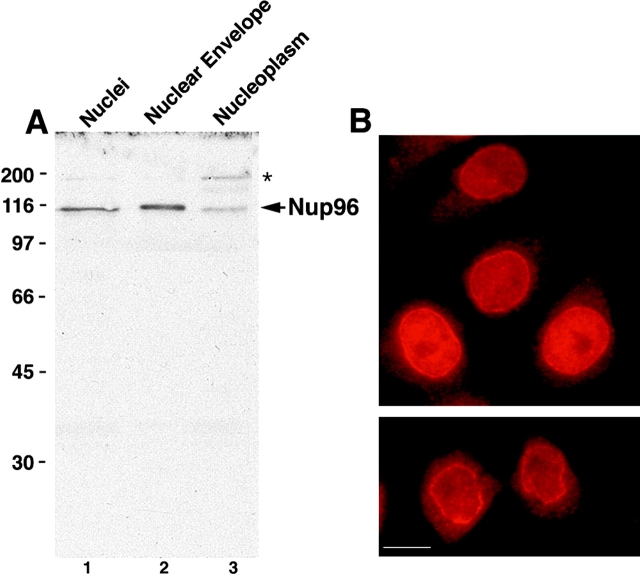
To produce antibodies specific for Nup96, a 22-kD fragment (corresponding to residues 1291–1482 of the predicted full-length protein; see Fig. 5) was expressed in E. coli and injected into rabbits. Affinity-purified antibodies reacted with Nup96, as indicated by immunoblot analysis of SDS-PAGE separated proteins of total nuclei and isolated nuclear envelopes (Fig. 2 A). Fractionation of isolated rat liver nuclei showed that Nup96 is associated predominantly with the nuclear envelope (Fig. 2 A, lane 2), although a significant fraction was also detected in the nucleoplasmic fraction (Fig. 2 A, lane 3), consistent with the immunofluorescence data (see below). In addition to Nup96, the affinity-purified antibodies also recognized a 200-kD protein that fractionated exclusively with the soluble nucleoplasm (Fig. 2 A, lane 3). As Nup96 is synthesized as a 186-kD protein that is proteolytically cleaved in vivo (see following sections), this 200-kD protein could represent the intact precursor. However, a similar band is not detected with antibodies to Nup98 (data not shown), and pulse-labeling experiments indicate that the precursor has a very short half-life (data not shown). Therefore, it is more likely that this band corresponds to a cross-reacting protein unrelated to the Nup98-Nup96 precursor.
Figure 5.
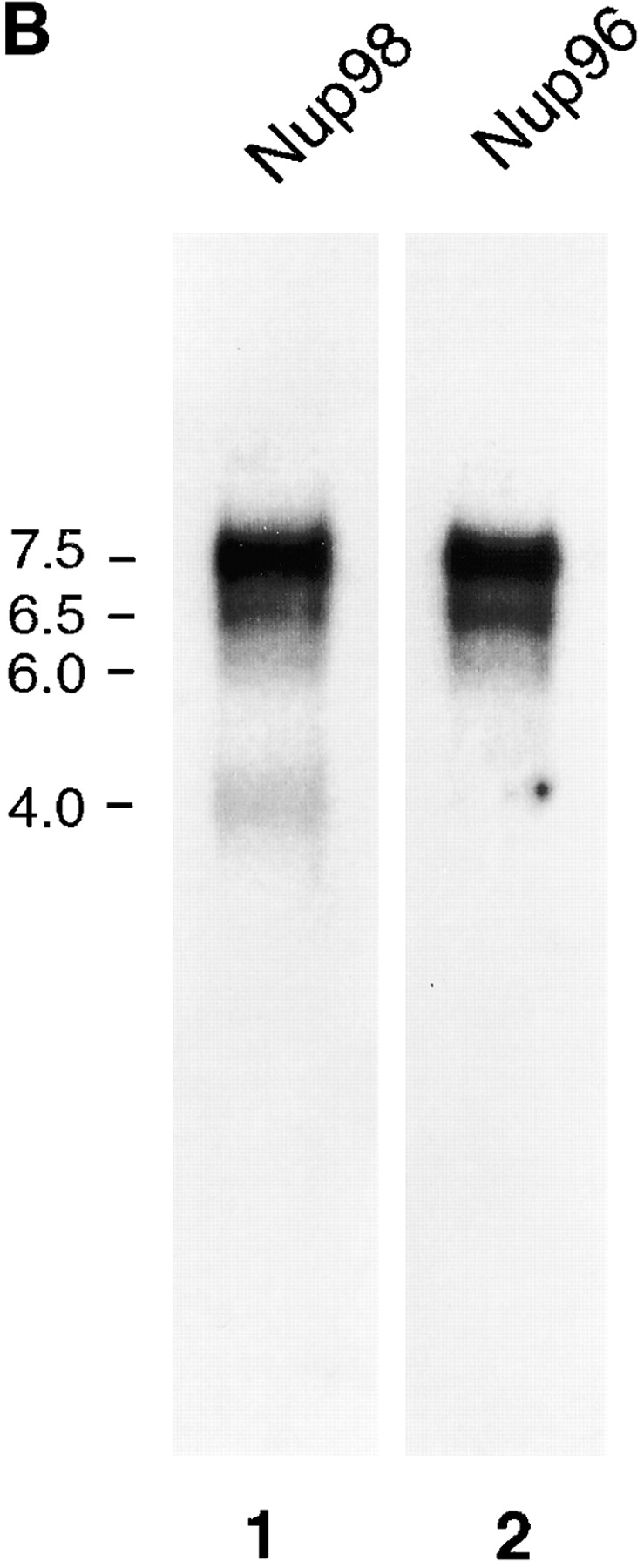
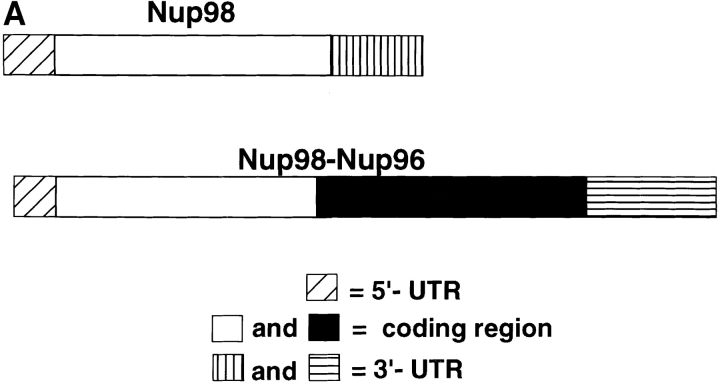
Alternatively spliced mRNAs code for Nup98 and for a Nup98-Nup96 precursor. (A) Schematic representation of Nup98 and Nup98-Nup96 precursor mRNAs. The 5′-UTRs of both mRNAs are identical except for the first 17 nucleotides, which are absent in the Nup98-Nup96 mRNA. The Nup98 coding regions are identical, except that the Nup98-Nup96 mRNA does not contain the last 18 nucleotides coding for the COOH-terminal 6 amino acids of Nup98. The Nup98-Nup96 mRNA has an extended coding region (shown in black) that generates Nup96. Both mRNAs have different 3′-UTRs. (B) Northern blot analysis of Nup98 and Nup98-Nup96 precursor mRNA transcripts. Poly(A)+ RNA was purified from HeLa cells, separated by agarose gel electrophoresis, and transferred to nitrocellulose membranes. Membranes were probed with a Nup98-specific probe (lane 1), or a Nup96-specific probe (lane 2). Approximate size markers (in kb) are indicated on the left.
Figure 2.
Nup96 is a component of the mammalian NPC. (A) Immunoblot analysis of fractionated rat liver nuclei. Total nuclei were fractionated into nuclear envelope and nucleoplasm, and equivalent amounts of each fraction (corresponding to 1 × 106 nuclei) were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with antibodies against Nup96. Nup96 is the major protein detected in total nuclei (lane 1), and it fractionates predominantly with the nuclear envelope fraction (lane 2). A significant amount of Nup96, however, is also detected in the nucleoplasm (lane 3). An uncharacterized, cross-reacting protein of 200 kD (indicated by the asterisk), also fractionates with the nucleoplasm. Molecular masses are indicated on the left. (B) Immunofluorescence microscopy. HeLa cells were fixed, permeabilized, and labeled with anti-Nup96 antibodies. When observed at the equatorial plane of the nucleus, a discontinuous rim was apparent, characteristic of NPC staining. Diffuse intranuclear labeling was also apparent. Lower panel shows labeling at telophase. Bar, 13 μm.
By indirect immunofluorescence microscopy Nup96 showed a rim-like staining at the midplane of the nucleus (Fig. 2 B). The nuclear envelope staining became more diffuse and punctate upon focusing near the surface of the nucleus (data not shown). This labeling pattern is characteristic of that observed for other nucleoporins (Davis and Blobel, 1986; Radu et al., 1995), and is further evidence for the association of Nup96 with NPCs. In addition to nuclear envelope staining, a weak intranuclear signal was also observed. This signal may be attributed to the presence of Nup96 within the nucleus, and/or to the 200-kD protein with which the Nup96 antibodies cross-react (see Fig. 2 A, lane 3). Signal was excluded from the nucleoli.
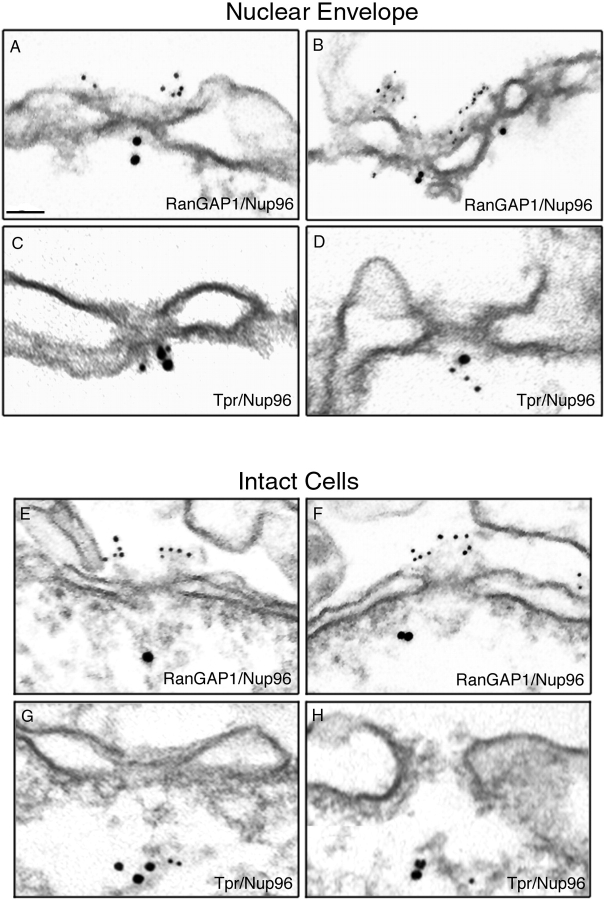
To further sub-localize Nup96 at the level of the NPC, immunogold EM was performed using anti-Nup96 antibodies. When isolated rat liver nuclear envelopes were probed, Nup96 was detected specifically at the nucleoplasmic face of the NPCs (Fig. 3, A–D). Gold particles were distributed over a range from 22 to 50 nm from the mid-plane of the nuclear envelope, with a mean distance of 36 nm (n = 50). The relative position of Nup96 in the NPC was established by double labeling experiments using monoclonal antibodies to RanGAP1 (Fig. 3, A and B) and Tpr (Fig. 3, C and D), markers for cytoplasmic and nucleoplasmic fibers of the NPC, respectively (Matunis et al., 1996; Cordes et al., 1997). Nup96 was located at or near the nucleoplasmic basket of the NPC, on the opposite face compared with RanGAP1, and in close proximity to Tpr.
Figure 3.
Immunolocalization of Nup96 to the nucleoplasmic face of the NPC. Isolated rat liver nuclear envelopes were probed with rabbit anti-Nup96 antibodies (A–D) and the mouse anti-RanGAP1 mAb 19C7 (A and B), or the mouse anti-Tpr mAb 5E10 (C and D). Rabbit antibodies were detected with 10 nm gold-coupled secondary antibodies, whereas mouse antibodies were detected with 5-nm gold-conjugated secondary antibodies. Envelopes were processed for thin sectioning and observed by EM. The cytoplasmic face of the nuclear envelopes (as evident from differences in the morphology of the nuclear and cytoplasmic membranes) are oriented toward the top of each micrograph. Intact HeLa cells were also probed simultaneously with rabbit anti-Nup96 antibodies (E–H) and the mouse anti-RanGAP1 mAb 19C7 (E and F), or the mouse anti-Tpr mAb 5E10 (G and H). Rabbit antibodies were detected with 10-nm gold-coupled secondary antibodies, whereas mouse antibodies were detected with 5-nm gold-conjugated secondary antibodies. The cells were processed for thin sectioning and observed by EM. The cytoplasm of the cells is oriented towards the top of each micrograph. Bar, 0.1 μm.
Because components of the NPC fibers may be lost or the fibers may collapse during isolation of nuclear envelopes, we also examined the localization of Nup96 in intact cells. Cells were permeabilized using digitonin, fixed and labeled with both primary and secondary antibodies before processing and sectioning for EM. Under these conditions, the relative distributions of Nup96 (Fig. 3, E–H), RanGAP1 (Fig. 3, E and F), and Tpr (Fig. 3, G and H) remained the same, confirming the results obtained using isolated nuclear envelopes. However, the distance of Nup96 from the mid-plane of the nuclear envelope was dramatically greater in the intact cells (averaging 100–200 nm), compared with the distance observed with isolated nuclear envelopes (averaging 36 nm). This difference could be due to the collapse of the nucleoplasmic basket and/or attached filaments upon isolation of nuclear envelopes.
Nup96 cDNAs Predict a Nup96/Nup98 Precursor
The internal peptide sequence LVVRHLASDAIINENYD derived from Nup96 was used to search sequence databases for candidate cDNA clones coding for Nup96. A partial cDNA clone was identified, GM2B8, that contained sequences coding for this peptide. GM2B8 was originally derived from a mouse macrophage subtraction library designed to identify messenger RNAs induced by interferon-γ (Yang et al., 1995). Screening a HeLa cell cDNA library with a GM2B8 probe and subsequent 5′ RACE yielded full-length clones coding for Nup96 (see Materials and Methods).
Surprisingly, the longest full-length clone obtained predicted a protein of 1,712 amino acids, with a molecular mass of 186 kD (Fig. 4). This size was in contrast to the expected 115 kD determined by SDS-PAGE (Fig. 1). Analysis of the predicted amino acid sequence of this clone revealed that the NH2-terminal 914 residues are identical to the previously characterized nucleoporin, Nup98 (Radu et al., 1995; Borrow et al., 1996; Nakamura et al., 1996), whereas the remaining 798 COOH-terminal residues are unique and contain the sequenced peptide derived from Nup96. Because this predicted protein contains sequences identical to both Nup98 and Nup96, it will be referred to as the Nup98-Nup96 precursor. Previously identified, complete cDNA clones for Nup98 predict a protein product of 920 amino acids (Radu et al., 1995; Borrow et al., 1996; Nakamura et al., 1996). The Nup98-Nup96 mRNA does not contain the last 18 nucleotides coding for the COOH-terminal 6 amino acids of Nup98. Several lines of evidence suggest that the mRNA coding for the Nup98-Nup96 precursor, and the previously described Nup98 mRNA, are derived from a common, alternatively spliced pre-mRNA. First, the nucleotide sequences in both the 5′ untranslated regions and in the Nup98 coding regions are 100% identical between both mRNAs (Fig. 5 A). Second, Northern blot analysis with a Nup98-specific probe revealed multiple RNA transcripts of ∼4, 6, 6.5, and 7 kb (Fig. 5 B, lane 1). A Nup96-specific probe detected similar 6-, 6.5-, and 7-kb transcripts, but not a 4-kb transcript (Fig. 5 B, lane 2). These data suggest that the larger transcripts code for the Nup98-Nup96 precursor, whereas the shorter 4-kb transcript codes independently for Nup98.
Figure 4.
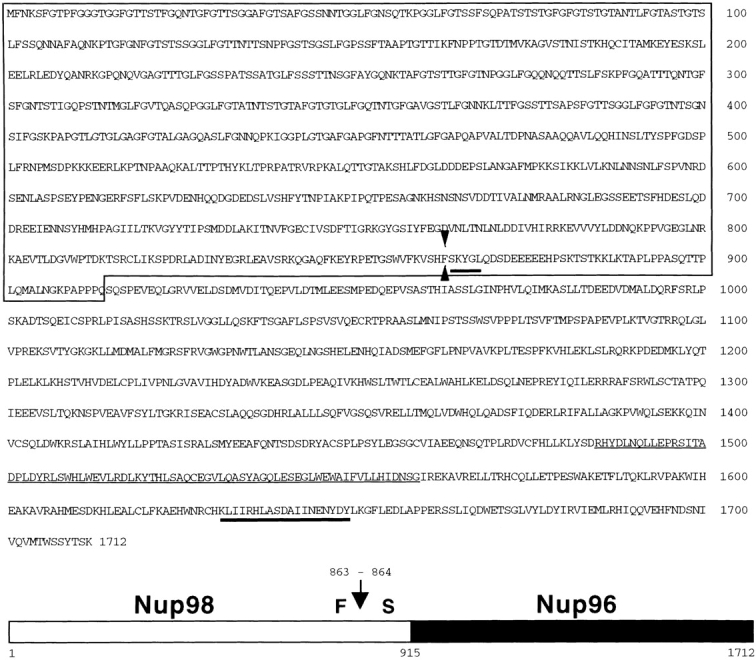
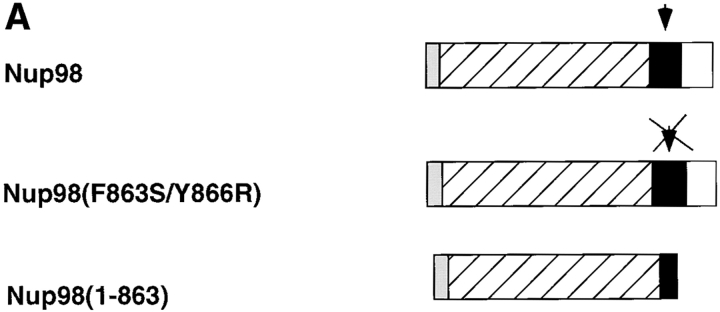
Amino acid sequence and schematic representation of the predicted Nup98-Nup96 precursor. The box indicates the NH2-terminal domain homologous to Nup98. Heavy underlines indicate amino acids identified through direct protein sequence analysis. The thin underline indicates a putative, alternative exon absent from a second class of Nup98-Nup96 precursor cDNAs. Arrow and arrowheads, between phenylalanine 863 and serine 864, indicate the proteolytic cleavage site.
With regard to the three Nup98-Nup96 precursor transcripts detected by Northern blot analysis, two classes of cDNAs were isolated during the screen for Nup96 clones. These cDNAs encode identical proteins, with the exception of a 75–amino acid insert predicted by the longer clone (see Fig. 4). Because the protein containing this 75– amino acid insert comigrates with endogenous Nup96 (see Fig. 7 B), whereas the protein without this insert does not (data not shown), the longer clone was used for subsequent analysis.
Figure 7.
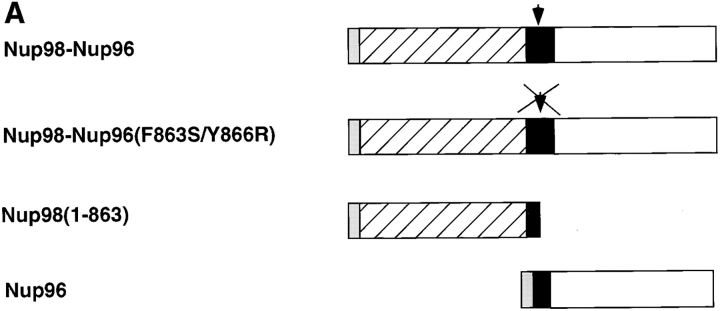
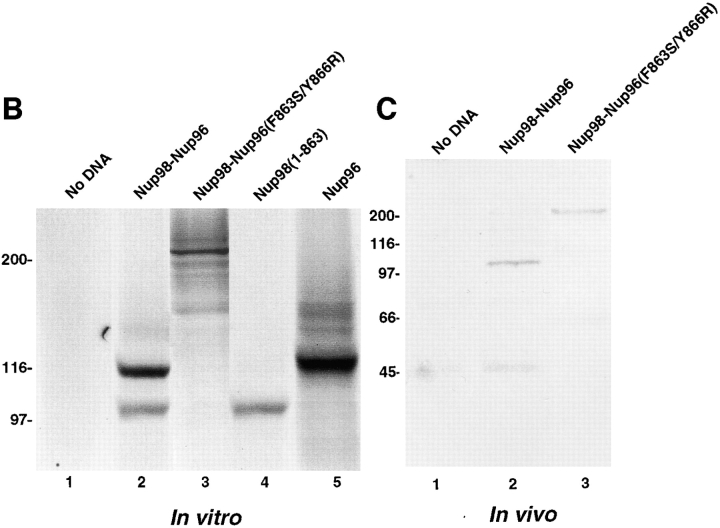
The Nup98-Nup96 precursor is proteolytically processed in vitro and in vivo. (A) Schematic representation of the Nup98-Nup96 precursor protein constructs used in the in vitro and in vivo expression assays. Each protein contains a myc-epitope tag (shown in gray) at its NH2 terminus. (B) In vitro transcription/translation reactions. Protein constructs were transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Reactions were performed with no DNA (lane 1), wild-type Nup98-Nup96 precursor (lane 2), mutant Nup98-Nup96 precursor (F863S/Y866R) (lane 3), Nup98 (1–863; lane 4), and Nup96 (lane 5). Molecular mass markers are indicated on the left. (C) In vivo expression of the wild-type and mutant Nup98-Nup96 precursors. HeLa cells were transfected with no DNA (lane 1), wild-type Nup98-Nup96 precursor (lane 2) and mutant Nup98-Nup96 precursor (F863S/Y866R) (lane 3). 24 h after transfection, cells were lysed in sample buffer and analyzed by immunoblot analysis using an anti-myc epitope antibody.
When the entire Nup98-Nup96 precursor sequence was aligned to all previously known proteins, significant homology was observed with the S. cerevisiae nucleoporin, Nup145p, as well as with an ORF located on chromosome I of Arabidopsis thaliana (these data are available from GenBank/EMBL/DDBJ under accession number U53501), and a protein predicted by the Caenorhabditis elegans cDNA, ZK328.5b (accession number U50193; Bailer et al., 1998). The NH2-terminal domains of the Nup98-96 precursor, Nup145p, and the C. elegans protein, all predict polypeptides with conserved GLFG repeats. The A. thaliana protein does not contain a GLFG-repeat domain. Near the middle of the Nup98-Nup96 precursor, Nup145p, and the C. elegans protein, and near the NH2 terminus of the A. thaliana protein, is a highly conserved domain (∼40% identical between homologues; Fig. 6). This domain is of particular interest, because in S. cerevisiae, Nup145p is posttranslationally cleaved at a site near the COOH-terminus of the domain (after the phenylalanine at position 605), thereby generating two independent nucleoporins, N-Nup145p and C-Nup145p (Dockendorff et al., 1997; Emtage et al., 1997; Teixeira et al., 1997). The high conservation of the cleavage site and surrounding residues between yeast, plant, worm, and mammal suggested that the posttranslational processing pathway may be conserved and functionally important. After the highly conserved cleavage site, all four proteins continue to be homologous over their entire length (Fig. 6). The COOH-terminal domains are rich in leucine and serine and contain a surprising number of conserved tryptophan residues. However, no obvious motifs or homologies to proteins with known functions were identified.
Figure 6.
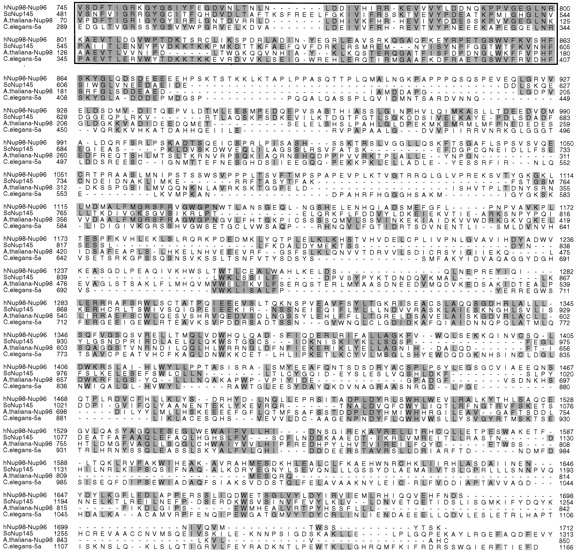
Amino acid sequence alignments of the COOH-terminal portion of Nup98-Nup96 and putative homologues in S. cerevisiae (Nup145p), A. thaliana (accession number U53501), and C. elegans (accession number U50193). The box indicates the highly conserved domain preceding the cleavage site between Nup98 and Nup96. Amino acids are numbered from the presumed initiating methionine in each protein. Identical amino acids are shaded.
Nup98 and Nup96 Are Generated through Proteolytic Processing of the Nup98-Nup96 Precursor
The isolation of cDNA clones for Nup96 that predict a protein product of 186 kD, and the similarity between this protein product and S. cerevisiae Nup145p, prompted us to investigate whether proteolytic processing of the Nup98- Nup96 precursor may also occur. The first approach toward addressing this issue was to determine the NH2-terminal residue of Nup96 by direct amino acid sequence analysis. As indicated in Fig. 4, the NH2 terminus of Nup96 was found to begin with the serine residue at position 864 of the full-length Nup98-Nup96 precursor. This serine is in exact alignment with a serine at the cleavage site in Nup145p, suggesting that the Nup98-Nup96 precursor is proteolytically processed through a mechanism similar to that occurring with Nup145p in yeast.
To further investigate the proteolytic processing of the Nup98-Nup96 precursor and its functional significance, various constructs were designed to express epitope tagged versions of the protein, both in vitro and in vivo (Fig. 7 A). In particular, a mutant of Nup98-Nup96, predicted to be resistant to proteolytic cleavage, was designed using knowledge of mutations made in S. cerevisiae Nup145p (Teixeira et al., 1997). This mutant contains two amino acid substitutions near the predicted cleavage site (F863S and Y866R). Proteolytic processing of the wild-type Nup98-Nup96 precursor was initially assayed using an in vitro transcription/translation system. Consistent with proteolytic cleavage of a precursor protein, two protein products were produced by transcription/translation of the wild-type Nup98-Nup96 precursor in rabbit reticulocyte lysate (Fig. 7 B, lane 2). The lower band comigrated with the myc-tagged Nup98 domain expressed alone (Fig. 7 B, lane 4), whereas the upper band migrated with a similar mobility to the myc-tagged Nup96 domain (Fig. 7 B, lane 5). Although the Nup96 domain has a predicted M r of 96 kD, its mobility by SDS-PAGE is closer to 115 kD. This anomalous mobility is consistent with the mobility of the protein initially identified in the rat liver NPC preparation (Fig. 1).
These data indicate that rabbit reticulocyte lysate contains the necessary factor(s) for the processing of the Nup98-Nup96 precursor. Therefore, the Nup98-Nup96 precursor with mutations near the cleavage site was transcribed and translated using identical conditions. A protein of ∼210 kD was produced using this mutant, a size consistent with the predicted mobility of the uncleaved precursor (Fig. 7 B, lane 3). Taken together, these in vitro data support the conclusion that the Nup98-Nup96 precursor is proteolytically processed to generate two nucleoporins, Nup98 and Nup96. To further demonstrate that the cleavage of the Nup98-Nup96 precursor occurs in vivo, the myc-tagged wild-type and mutant plasmids encoding for proteins were transfected into cells in culture. Immunoblot analysis of lysate from cells transfected with the wild-type protein revealed a major band (only the NH2 terminus of the precursor contains a myc-tag) migrating at ∼100 kD (Fig. 7 C, lane 2), consistent with the predicted size of the myc-tagged Nup98 domain after cleavage. Analysis of lysate from cells transfected with the mutant Nup98-Nup96 precursor, on the other hand, revealed a major protein of ∼210 kD. These results demonstrate that the Nup98-Nup96 precursor is proteolytically processed in vivo.
Synthesis and Processing of the Nup98-Nup96 Precursor Is Required for Proper Targeting of Nup98 and Nup96 to the NPC
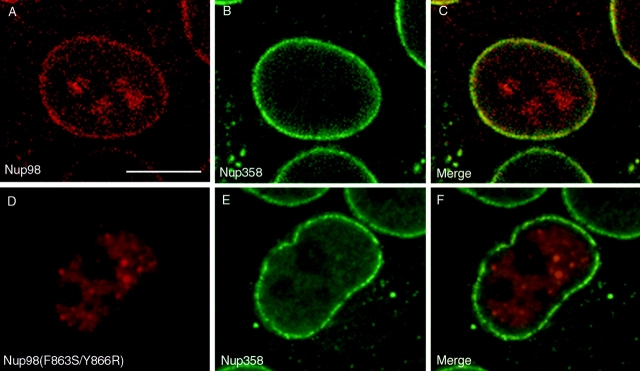
In S. cerevisiae, synthesis and cleavage of the Nup145p precursor is important for some aspects of targeting the resulting nucleoporins (N-Nup145p and C-Nup145p) to the NPC (Fabre et al., 1994; Teixeira et al., 1997). To examine the effects and requirements of Nup98-Nup96 precursor synthesis and processing on Nup98 and Nup96 localization, cultured cells were transfected with the various constructs illustrated in Fig. 7 A, and the localization of the expressed proteins was determined by indirect immunofluorescence confocal microscopy. Cells were double labeled with an anti-myc antibody to localize the transiently expressed proteins, and an antibody to Nup358, a marker for NPCs. When the NH2-terminal tagged, wild-type Nup98- Nup96 precursor was expressed, labeling of the nuclear envelope was observed, indicating targeting of the cleaved Nup98 domain to the NPC (Fig. 8 A). Labeling of nucleoli was also observed in many cells expressing high levels of protein, possibly a consequence of having saturated binding sites for Nup98 at the NPC or a possible intranuclear localization of Nup98 as previously described (Powers et al., 1995). Merging the signals for Nup358 (Fig. 8 B) and Nup98 revealed coincident labeling of the nuclear envelope (Fig. 8 C), confirming NPC targeting.
Figure 8.
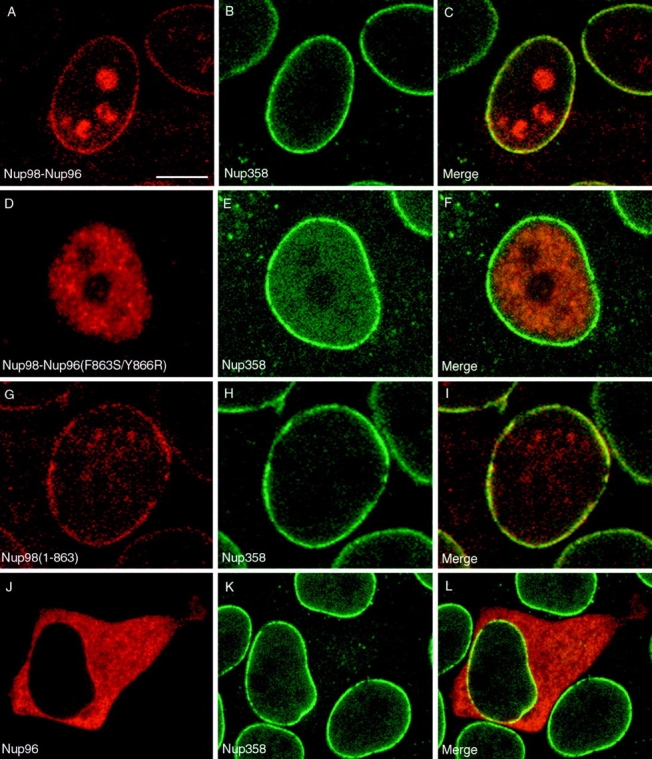
Proteolytic processing of the Nup98-Nup96 precursor is essential for targeting Nup98 and Nup96 to the NPC. HeLa cells were transiently transfected with: (A–C) the wild-type Nup98-Nup96 precursor, (D–F) the mutant Nup98-Nup96 precursor (F863S/Y866R), (G–I) the Nup98 domain (amino acids 1–863), and (J–L) Nup96 (amino acids 864–1712). Localization of the transiently expressed proteins was detected by indirect immunofluorescence and confocal microscopy using an anti-myc epitope antibody followed by a Cy3-conjugated secondary antibody (A, D, G, and J). Cells were also double labeled with an antibody specific for Nup358, which was detected using a secondary antibody conjugated to FITC (B, E, H, and K). Cy3 and FITC images were merged to identify coincident labeling of NPCs (C, F, I, and L). Bar, 10 μm.
The cleavage-deficient mutant of the Nup98-Nup96 precursor was next examined. In this case, only intranuclear labeling was observed, with no evidence of association of the precursor protein with NPCs or nucleoli (Fig. 8 D). Again, cells were double labeled with antibody to Nup358 (Fig. 8 E), and the merged images revealed no detectable overlap between the two signals at the nuclear envelope (Fig. 8 F). This result indicates that cleavage is a prerequisite for NPC localization of Nup98. Expression of the Nup98 domain alone (amino acids 1–863) resulted in a localization that was virtually indistinguishable from the localization observed after expression of the wild-type Nup98-Nup96 precursor, revealing that it could be targeted to the NPC independent of being synthesized as a precursor with Nup96 (Fig. 8, G–I). However, when Nup96 was expressed independently, a diffuse cytoplasmic signal was detected, with very little if any detectable localization at NPCs (Fig. 8 J). Merging the signal obtained with antibody to Nup358 (Fig. 8 K) with the signal for Nup96 revealed some overlap at the nuclear envelope, making it impossible to conclude that the transiently expressed Nup96 was absolutely excluded from NPCs. Relative to Nup98, however, Nup96 is not efficiently targeted to the nucleus, or to NPCs.
Nup98 Is also Proteolytically Processed In Vivo
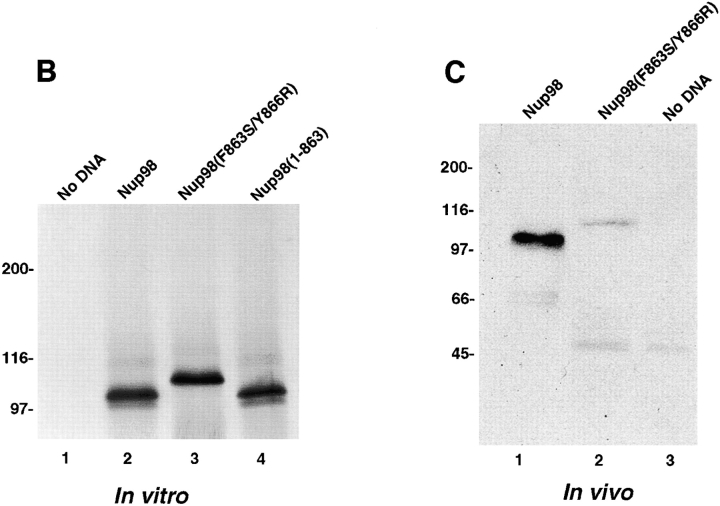
The cleavage site present in the Nup98-Nup96 precursor protein is also present in the protein predicted by the Nup98-specific transcript (Radu et al., 1995; Borrow et al., 1996; Nakamura et al., 1996). To examine whether the independently expressed Nup98 protein is proteolytically processed, mutations were made near the cleavage site, similar to those made in the Nup98-Nup96 precursor (Fig. 9 A). When a myc-tagged wild-type Nup98 was expressed in rabbit reticulocyte lysate, a protein with a relative molecular mass of 100 kD was observed by SDS-PAGE (Fig. 9 B, lane 2). This protein product migrated with a similar mobility to the myc-tagged Nup98 product consisting of amino acids 1–863 (Fig. 9 B, lane 4), suggesting that Nup98 is indeed processed. Expression of the mutant Nup98, resulted in the production of a protein with a relative molecular mass of 110 kD, providing additional evidence that Nup98 is proteolytically cleaved (Fig. 9 B, lane 3). To determine whether cleavage occurs in vivo, the wild-type and mutant Nup98 proteins were transiently expressed in cultured cells. Immunoblot analysis of cell lysates with an anti-myc antibody revealed proteins of 100 and 110 kD in cells transfected with the wild-type and mutant Nup98 proteins (Fig. 9 C, lanes 1 and 2), respectively, indicating that proteolytic cleavage also occurs in vivo. These results indicate that the alternatively spliced Nup98 transcripts each produce identical Nup98 proteins (consisting of amino acid residues 1–863), through proteolytic processing.
Figure 9.
Nup98 is proteolytically processed in vitro and in vivo. (A) Schematic representation of the Nup98 protein constructs used in the in vitro and in vivo expression assays. Each protein contains a myc-epitope tag (shown in gray) at its NH2 terminus. (B) In vitro transcription/translation reactions. Protein constructs were transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Reactions were performed with no DNA (lane 1), wild-type Nup98 (lane 2), mutant Nup98 (F863S/Y866R) (lane 3), and Nup98 (1–863) (lane 4). Molecular mass markers are indicated on the left. (C) In vivo expression of the wild-type and mutant Nup98 precursors. HeLa cells were transfected wild-type Nup98 (lane 1), mutant Nup98 (F863S/Y866R) (lane 2), and with no DNA (lane 3). 24 hours after transfection, cells were lysed in sample buffer and analyzed by immunoblot analysis using an anti-myc epitope antibody.
To determine whether the processing of Nup98 influenced its targeting to the NPC, indirect immunofluorescence confocal microscopy was performed on cells transfected with the wild-type and mutant proteins using the double labeling approach indicated above. The wild-type, cleaved Nup98, was efficiently targeted to the NPC, as demonstrated in Fig. 10, A–C. Again, labeling of nucleoli was observed in cells expressing high levels of processed Nup98. When mature Nup98 (residues 1–863) was transiently expressed, localization to the NPC was also observed (Fig. 8, G–I), indicating that mature Nup98 is imported into the nucleus and targeted to the NPC. In contrast, mutant uncleaved Nup98 was found to accumulate strictly in the nucleoplasm, with no detectable association with NPCs (Fig. 10, D–F). In addition, uncleaved Nup98 was also absent from nucleoli. These data indicate that, as with the Nup98-Nup96 precursor, proteolytic processing of the Nup98 precursor is essential for proper targeting to the NPC.
Figure 10.
Proteolytic processing of Nup98 is essential for NPC targeting. HeLa cells were transiently transfected with: (A–C) wild-type Nup98, (D–F) mutant Nup98 (F863S/Y866R). Localization of the transiently expressed proteins was detected by indirect immunofluorescence and confocal microscopy using an anti-myc epitope antibody followed by a Cy3-conjugated secondary antibody (A and D). Cells were also double labeled with an antibody specific for Nup358, which was detected using a secondary antibody conjugated to FITC (B and E). Cy3 and FITC images were merged to identify coincident labeling of NPCs (C and F). Bar, 10 μm.
Nup96 Fractionates with a Complex Containing Nup107 and Two Sec13-Related Proteins
To determine whether Nup96 interacts with other nucleoporins to form specific sub-domains of the NPC, we carried out subfractionation with isolated rat liver nuclei. We identified a sub-complex of the NPC that contains Nup96 and at least five additional proteins (Fig. 11). This complex was released from the pore-complex-lamina fraction using heparin and detergent, and then analyzed by sucrose gradient centrifugation. One of the proteins found to cosediment in the gradient with Nup96 had an apparent molecular mass of 105 kD, similar to the expected mass of Nup107 (Radu et al., 1994). Immunoblot analysis of the sucrose gradient fractions confirmed that Nup96 and Nup107 cosedimented, peaking in fractions 5 and 6 (Fig. 11).
Figure 11.
Identification of a complex containing Nup96, Nup107, and Sec13. Rat liver nuclei were fractionated as described in Materials and Methods. A fraction containing soluble NPCs was loaded onto a 10–40% sucrose gradient and centrifuged. Gradient fractions were collected and analyzed by SDS-PAGE followed by silver stain analysis (top), or immunoblot analysis with antibodies specific for Nup96 and Nup107 (bottom). Fractions, numbered from the top of the gradient, are indicated. Asterisks between lanes 5 and 6 (top) indicate proteins that sediment as a complex. Some of the proteins comprising this complex are indicated on the right.
In yeast, a similar complex has been observed between C-Nup145p and Nup84p, the putative homologues of Nup96 and Nup107, respectively (Siniossoglou et al., 1996). This complex also contains a number of additional proteins, including Nup120p, Nup85p, Sec13p, and a Sec13p-related protein, Seh1p. Two of the proteins that we found to cosediment with Nup96 had molecular masses that closely matched the molecular mass of Sec13p (∼35 kD). Therefore, we obtained peptide sequences for these two proteins (which are indicated by the asterisks in Fig. 11), to determine their molecular identity. Two internal peptide sequences obtained from the larger of the two proteins indicated that it was identical to the human homologue of yeast Sec13p, a protein referred to as mSec13 or Sec13Rp (Shaywitz et al., 1995; Tang et al., 1997). Sequence obtained from the second, lower molecular mass protein (referred to as p37) indicated that it was a novel protein with limited homology to Sec13p. Full-length clones coding for this protein will have to be isolated before a complete comparison can be made with Sec13, and with the Sec13-related protein (Seh1p) found to copurify with the homologous yeast complex. In summary, we have identified a complex released from mammalian NPCs that contains Nup96, Nup107, mSec13, and a novel Sec13-related protein.
Discussion
We are using a novel nuclear envelope fractionation procedure to characterize more completely the protein components making up the mammalian NPC. Based on this fractionation procedure and separation of the isolated proteins by reverse phase column chromatography, we estimate that less than one-third of the mammalian nucleoporins have been characterized at the molecular level. This factor contributes significantly to our lack of understanding about how the NPC is assembled, and how it functions to regulate nucleocytoplasmic transport. The fractionation procedure that we have developed yields nucleoporins and NPC-associated proteins in high purity and yield, and has allowed us to identify a significant number of novel proteins. The first NPC-associated protein that we characterized using this fractionation procedure was the SUMO-1 modified form of RanGAP1 (Matunis et al., 1996). Here, we have reported the characterization of a second protein, a new nucleoporin of 96 kD termed Nup96.
The most intriguing aspect of Nup96 is its unusual pathway of biogenesis. Through analysis at a number of levels, we have demonstrated that Nup96 is synthesized as a precursor that is proteolytically cleaved in vivo. Cleavage of this precursor generates not only Nup96, but also the GLFG-containing nucleoporin, Nup98. Nup98 can also be produced independently of Nup96, by what appears to be an alternatively spliced mRNA that does not include the Nup96 open reading frame (Radu et al., 1995; Borrow et al., 1996; Nakamura et al., 1996). Whereas it was not previously recognized that this predicted Nup98 protein is proteolytically processed, we have shown that it is processed like the Nup98-Nup96 precursor. Evidence for the synthesis and processing the Nup98-Nup96 precursor and the Nup98 precursor in vivo are: (a) Northern blot analysis that supports the existence of the precursor mRNAs, and isolation of cDNAs that predict the precursor proteins; (b) NH2-terminal peptide sequence of Nup96 that supports the cleavage of the Nup98-Nup96 precursor between phenylalanine 863 and serine 864; (c) mutagenesis of the cleavage sites that yield the uncleaved precursors, both in vitro and in vivo; (d) homology with S. cerevisiae Nup145p, which undergoes posttranslational in vivo cleavage at an identical site (Dockendorff, 1997; Emtage et al., 1997; Teixeira et al., 1997).
The Nup98-Nup96 precursor is the apparent vertebrate homologue of S. cerevisiae Nup145p. The NH2 terminus of both proteins, Nup98 and N-Nup145p, are ∼20% identical over their entire length and contain highly conserved GLFG repeats. N-Nup145p is not essential, but it may have RNA-binding activity and function in RNA transport in yeast (Fabre et al., 1994; Emtage, 1997; Teixeira et al., 1997). In vertebrates, Nup98 is located on the nucleoplasmic side of the NPC, at or near the basket, consistent with a possible role in RNA export (Radu et al., 1995). Further supporting such a role, antibodies against Nup98 inhibit export of multiple classes of RNAs when injected into the nuclei of Xenopus oocytes (Powers et al., 1997). The COOH-terminal domains of both precursors, Nup96 and C-Nup145p, are also 20% identical over their entire length, but contain no obvious motifs or homologies to proteins of know function. Studies in yeast, nonetheless, indicate a role for C-Nup145p in mRNA export (Dockendorff, 1997; Emtage, 1997; Teixeira et al., 1997). Like Nup98, Nup96 also localizes to the nucleoplasmic side of the NPC, at or near the basket, consistent with a potential role in mRNA export. Our data obtained from in situ labeling of intact cells indicate that Nup96 may be located near the distal end of the nucleoplasmic basket, in proximity to the site where Tpr associates with the NPC.
Of particular interest is the domain around the Nup98-Nup96 cleavage site, as it is especially conserved among protein homologues in yeast, plant, worm, and man. A similar domain is also present at the extreme COOH-terminus of two additional yeast GLFG repeat-containing nucleoporins, Nup100p and Nup116p (Wente et al., 1992; Radu et al., 1995; Teixeira et al., 1997). In particular, the ∼50 amino acids preceding the cleavage site, and the ∼12 amino acids after the cleavage site, are ∼40% identical between yeast and human (relative to 20% identity between other regions of the proteins), suggesting that this region may be important for the cleavage process. Surprisingly, however, a large portion of this highly conserved domain is not required for proteolytic processing of Nup145p in vivo (Emtage et al., 1997). This raises the possibility that the domain preceding the cleavage site may have another function, a function that could possibly be regulated by the cleavage process. This domain includes a conserved RNP1 consensus site that has been suggested to contribute to the RNA-binding properties of N-Nup145p (Fabre et al., 1994). It will be interesting to determine whether the RNA-binding properties of either Nup98 or N-Nup145p are affected by proteolytic cleavage. The conserved residues surrounding the cleavage site itself, H-F-S, do not resemble the consensus site for any known protease, and the exact mechanism for the cleavage process remains to be elucidated. It is also unclear when, or where the cleavage event occurs, although pulse labeling experiments indicate that the Nup98-Nup96 precursor is cleaved within five minutes of being synthesized (data not shown).
Whereas the physiological relevance of producing both Nup98 and Nup96 through proteolytic cleavage of a precursor is not known, the conservation of this pathway from yeast to human suggests a potentially important function. Posttranslational proteolytic cleavage of precursor proteins is widely used to regulate many different cellular functions. Processing of many neuropeptides, hormones and certain plasma proteins is used to regulate the formation of mature, active factors without the need for de novo protein synthesis (Resnick and Zasloff, 1992; Seidah and Chretien, 1997). Protelolytic cleavage can also function to indirectly activate proteins by regulating their localization within the cell. Examples include the signal-mediated nuclear targeting of Notch-1 from the plasma membrane (Schroeter et al., 1998), and relocalization of the sterol regulatory element binding proteins from the nuclear membrane to the nucleus in response to sterol levels (Brown and Goldstein, 1997). Other possible functions of precursor synthesis and proteolytic cleavage include protein folding, as has been suggested for certain ribosomal protein-ubiquitin precursors (reviewed by Johnson and Hochstrasser, 1997), and as a mechanism to strictly control protein stoichiometry. Another important role for regulated proteolytic cleavage is in the assembly and maturation of large molecular complexes, best exemplified by virus particle assembly (reviewed in Krausslich and Welker, 1996). By this analogy, proteolytic cleavage of nucleoporins could be related to the orderly assembly of the NPC. As an example, the Nup98-Nup96 precursor could be inserted into a newly synthesized pore and cleaved only after the appropriate cleavage factor is assembled. Cleavage may then result in conformational changes, or exposure of binding sites on Nup98 and/or Nup96, that allow other sets of proteins to bind in an orderly sequence.
Our data demonstrate that proper processing of both the Nup98-Nup96 precursor and the Nup98 precursor is required for efficient NPC association, indicating that there is an ordered series of events leading to the association of these nucleoporins with the NPC. Mutations in the cleavage site that prevent processing lead to accumulation of both proteins in the nucleoplasm, but not at NPCs. In the case of Nup98, this result suggests that the COOH-terminal 58 amino acids (that would normally be removed) either tether Nup98 to intranuclear sites, or mask the NPC-targeting domain. Interestingly, in addition to not being targeted to NPCs, unprocessed Nup98 was also absent from nucleoli when over expressed. Although the significance of processed Nup98 appearing in nucleoli is not clear, it has previously been suggested that Nup98 may not be absolutely confined to NPCs (Powers et al., 1995). These data raise the intriguing possibility that Nup98 could shuttle between nucleoli and NPCs.
In addition to cleavage being important, our studies also demonstrate that proper targeting of Nup98 and Nup96 to the NPC is influenced by the synthesis of the precursor protein. In particular, we found that targeting of Nup96 to the NPC is dependent on its being synthesized as a precursor. When expressed independently, Nup96 accumulates in the cytoplasm, suggesting that its nuclear import may be Nup98-mediated (Nup98 expressed alone is imported into the nucleus). Nup98-mediated import of Nup96 could occur in one of two ways. Either the Nup98-Nup96 precursor could be imported into the nucleus before proteolytic processing, or Nup98 and Nup96 could remain associated after proteolytic processing in the cytoplasm, and then be imported as a complex. At present, there is no data to suggest that Nup98 and Nup96 remain associated with each other once they are inserted into the NPC. The sub-complex of nucleoporins containing Nup96, Nup107, and mSec13 that we isolated does not contain Nup98, and experiments in yeast indicate that the analogous sub-complex also lacks N-Nup145p (Teixeira et al., 1997).
To date, relatively few sub-complexes of the mammalian NPC have been identified. And although strikingly similar at a morphological level (Yang et al., 1998), no homologous sub-complexes of mammalian and yeast NPCs have previously been identified. This has raised the notion that the molecular structure of the NPC may be less well conserved than its morphological structure. We have identified a complex of mammalian nucleoporins that contains Nup96, Nup107, mSec13, at least one Sec13-related protein, and at least two additional proteins of ∼150 kD. This complex is highly similar to the Nup84p complex of yeast, and therefore represents one of the first homologous complexes of nucleoporins identified between yeast and mammals. There is also evidence for homology between a yeast nucleoporin complex containing Nup159p, Nup82p, and Nsp1p and a vertebrate complex containing Nup214/CAN, Nup88, and possibly p62 (Macaulay et al., 1995; Bastos et al., 1997; Fornerod et al., 1997; Belgareh et al., 1998). The identification of these complexes suggests that the molecular structure of the NPC may be more highly conserved than was anticipated. Similarly, homologous sub-complexes will likely be identified in the future, as more mammalian and yeast nucleoporins are identified and characterized. In the immediate future, further purification of this complex will allow us to identify several additional nucleoporins, and possibly obtain information about its structural features. Of particular interest is the presence of the Sec13-related proteins in this complex. Whereas the exact role that Sec13 may have at the NPC has been previously discussed (Siniossoglou et al., 1996), our combined data suggest that this function is likely to be exerted on the nucleoplasmic side of the NPC, given the localization Nup96. It will be interesting to investigate the role that Sec13 may have in NPC biogenesis using Xenopus nuclear envelope reconstitution systems.
What are the functions of Nup98 and Nup96 once assembled into the NPC? As indicated, both Nup98 and Nup96 localize to the nucleoplasmic side of the NPC, at or near the basket. The nucleoplasmic basket appears to have an active role in binding RNP complexes and mediating their export from the nucleus. Electron micrographs of Balbiani ring RNPs being transported through the NPC show intimate interactions between the basket and the RNP, implying that the proteins in this structure actively engage the particles during transport (Kiseleva et al., 1996). It is not yet known whether the nucleoporins at the basket bind the RNA itself, and/or the hnRNP proteins that accompany the RNA during transport (Nakielny et al., 1997). As implied from their localization, and the functional studies of Nup145p in yeast and Nup98 in Xenopus, Nup98 and Nup96 are likely to be important factors mediating the docking and translocation of RNPs through the NPC. The challenge ahead will be to understand how these nucleoporins interact with RNPs as they are transported through the NPC, and how these interactions regulate the transport process.
Acknowledgments
We thank Michael P. Rout and Aurelian Radu for helpful discussions and critical reading of the manuscript. We thank Jian Wu for the anti-Nup358 antibody and Daniel R. Nussenzveig for advice in the confocal microscopy. We also thank Helen Shio for expert technical support in preparing and analyzing EM samples, members of the Rockefeller University Protein/DNA Technology Center for DNA and protein sequence analysis, and Dr. Mark Rutherford for the cDNA clone, GM2B8.
This work was supported by the Howard Hughes Medical Institute. M.J. Matunis was supported by an American Cancer Society-Amgen Fellowship (PF-4195).
Abbreviations used in this paper
- NES
nuclear export signal
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- nt
nucleotide
- PCLF
pore complex lamina fraction
Footnotes
Dr. Matunis' present address is The Johns Hopkins University, School of Hygiene and Public Health, Department of Biochemistry, Baltimore, MD 21205.
References
- Akey CW, Radermacher M. Architecture of the Xenopusnuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO (Eur Mol Biol Organ) J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos, R., N. Pante, and B. Burke. 1995. Nuclear pore complex proteins. Int. Rev. Cytol. 162B:257–302. [DOI] [PubMed]
- Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole CN, Doye V. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol Biol Cell. 1998;9:3475–3492. doi: 10.1091/mbc.9.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Borrow J, Shearman AM, Stanton VP, Jr, Becher R, Collins T, Williams AJ, Dube I, Katz F, Kwong YL, Morris C, et al. The t(7;11)(p15; p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin Nup98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Nuclear localization signals (NLS) Critical Rev Eukar Gene Expr. 1993;3:193–227. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN. C-terminal truncations of the yeast nucleoporin Nup145pp produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol. 1997;17:906–920. doi: 10.1128/mcb.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Adam SA, Choi YD. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984;4:415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworetzky S, Feldherr C. Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J Cell Biol. 1988;106:575–584. doi: 10.1083/jcb.106.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N, Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage JLT, Bucci M, Watkins JL, Wente SR. Defining the essential functional regions of the nucleoporin Nup145pp. J Cell Sci. 1997;110:911–925. doi: 10.1242/jcs.110.7.911. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC. Nup145pp is required for nuclear export of mRNA and binds homopolymeric RNA in vitrovia a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Akin D. The location of the transport gate in the nuclear pore complex. J Cell Sci. 1997;110:3065–3070. doi: 10.1242/jcs.110.24.3065. [DOI] [PubMed] [Google Scholar]
- Fernandez J, Andrews L, Mische SM. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., J. van Deursen, S. van Baal, A. Reynolds, D. Davis, K. Gopal Murti, J. Fransen, and G. Grosveld. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur. Mol. Biol. Organ.) J. 16:807–816. [DOI] [PMC free article] [PubMed]
- Goldberg MW, Allen TD. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J Mol Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, R. 1990. Recombinant PCR. In PCR Protocols. A Guide to Methods and Applications. M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White, editors. Academic Press, Inc., San Diego, CA. 177–183.
- Hinshaw JE, Carragher BO, Miligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore proteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Daneholt B, Allen TD. RNP export is mediated by structural reorganization of the nuclear pore basket. J Mol Biol. 1996;260:304–311. doi: 10.1006/jmbi.1996.0401. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich H-G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Melchior F, Gerace L. Two-way trafficking with ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, Chen SJ, Willman CL, Chen I-M, Feinberg AP, et al. Fusion of the nucleoporin gene Nup98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Fischer U, Michael WM, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- Pain D, Murakami H, Blobel G. Identification of a receptor for protein import into mitochondria. Nature. 1990;347:444–449. doi: 10.1038/347444a0. [DOI] [PubMed] [Google Scholar]
- Paine P, Moore L, Horowitz S. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Blobel G, Rosenblum J. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz F, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG containing nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate nucleoporin, Nup98, is an essential component of RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Wazniak R. Nup107 is a novel nuclear pore complex protein that contains a leucine zipper. J Biol Chem. 1994;269:17600–17605. [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick N, Zasloff MA. Novel proteases with unusual specificities. Curr Biol. 1992;4:1032–1036. doi: 10.1016/0955-0674(92)90136-z. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Ris H. The 3D-structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull. 1991;21:54–56. [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Rout M, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Rush MG, Drivas G, D'Eustachio P. The small GTPase Ran: how much does it run? . Bioessays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Chretien M. Eukaryotic protein processing: endoproteolysis of precursor proteins. Curr Opin Biotech. 1997;8:602–607. doi: 10.1016/s0958-1669(97)80036-5. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Orci L, Ravazzola M, Swaroop A, Kaiser CA. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J Cell Biol. 1995;128:769–777. doi: 10.1083/jcb.128.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt E. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Tang BL, Peter F, Krijnse-Locker J, Low SH, Griffiths G, Hong W. The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol Cell Biol. 1997;17:256–266. doi: 10.1128/mcb.17.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Siniossoglou S, Podtelejnikov S, Benichou JC, Mann M, Dujon B, Hurt E, Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145pp: a novel biogenesis pathway for nucleoporins. EMBO (Eur Mol Biol Organ) J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, ran-gtp binding sites, zinc fingers, a cyclophilin a homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yang S-D, Schook LB, Rutherford MS. Differential expression of novel genes by bone marrow-derived macrophage populations. Mol Immunol. 1995;32:733–742. doi: 10.1016/0161-5890(95)00035-d. [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds ran/tc4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]