Abstract
Cajal bodies (also known as coiled bodies) are subnuclear organelles that contain specific nuclear antigens, including splicing small nuclear ribonucleoproteins (snRNPs) and a subset of nucleolar proteins. Cajal bodies are localized in the nucleoplasm and are often found at the nucleolar periphery. We have constructed a stable HeLa cell line, HeLaGFP-coilin, that expresses the Cajal body marker protein, p80 coilin, fused to the green fluorescent protein (GFP-coilin). The localization pattern and biochemical properties of the GFP-coilin fusion protein are identical to the endogenous p80 coilin. Time-lapse recordings on 63 nuclei of HeLaGFP-coilin cells showed that all Cajal bodies move within the nucleoplasm. Movements included translocations through the nucleoplasm, joining of bodies to form larger structures, and separation of smaller bodies from larger Cajal bodies. Also, we observed Cajal bodies moving to and from nucleoli. The data suggest that there may be at least two classes of Cajal bodies that differ in their size, antigen composition, and dynamic behavior. The smaller size class shows more frequent and faster rates of movement, up to 0.9 μm/min. The GFP-coilin protein is dynamically associated with Cajal bodies as shown by changes in their fluorescence intensity over time. This study reveals an unexpectedly high level of movement and interactions of nuclear bodies in human cells and suggests that these movements may be driven, at least in part, by regulated mechanisms.
Keywords: nucleus, Cajal bodies, coilin, GFP, time-lapse microscopy
Introduction
The nuclei of both animal and plant cells are complex organelles and show a high level of structural organization (Spector 1993; Lamond and Earnshaw 1998; Schul et al. 1998). Many critical metabolic activities, including DNA replication, transcription, pre-mRNA processing, and ribosome production, take place and are regulated within nuclei. Chromosomes occupy a large fraction of the nuclear volume in the form of separate “chromosome territories” (Schardin et al. 1985). The degree of chromatin condensation during interphase varies in different regions of each individual chromosome and this in turn can influence the expression of many gene clusters. The interchromatin space also contains several other structures that can be collectively described as “nuclear bodies.” A range of nuclear antigens involved in RNA processing, transcription, ribosome production, and other processes are assembled into structures in the nucleoplasm, including nucleoli, speckles, coiled bodies, gems, and promyelocytic leukemia bodies (Matera 1999). The activities carried out by most of these structures are still unclear. However, recent evidence indicates that the disruption of nuclear body organization may play a role in several forms of human disease and genetic disorders and suggests that nuclear bodies could be important for efficient cell function.
The coiled body is a conserved nuclear structure found in both animal and plant cells. Coiled bodies are usually ∼0.5 μm in diameter, but both their size and the number present per nucleus vary considerably between cell types. A human autoantigen called p80 coilin has been identified and shown to be a marker protein for the coiled body (Andrade et al. 1991; Raska et al. 1991). Immunolabeling studies using anticoilin antibodies show coiled bodies as discrete nuclear foci in the fluorescence microscope and also show a pool of coilin diffusely spread in the nucleoplasm. The name “coiled body” was derived from its appearance in the transmission electron microscope as a tangled ball of dense threads (Monneron and Bernhard 1969). More recently, Gall et al. 1999 have proposed that the name “Cajal body” should be adopted in place of “coiled body,” in honor of their initial discovery by Ramon y Cajal. Therefore, here we refer to the nuclear bodies containing p80 coilin as Cajal bodies.
Cajal bodies contain several nucleolar antigens, including fibrillarin and NOPP140, and small nucleolar (sno) RNPs (Matera 1999). Indeed, they were first identified in neurons and termed nucleolar accessory bodies due to their localization at the nucleolar periphery (Ramon y Cajal 1903). In somatic animal and plant cells, Cajal bodies are often found at the periphery of the nucleolus. They can also be found within nucleoli in human breast carcinoma cells and in liver cells of hibernating dormice (Malatesta et al. 1994). Transient expression in mammalian cells of a mutated p80 coilin protein, where a single serine residue is changed to aspartate, results in Cajal bodies forming within nucleoli, whereas expression of deletion mutants of p80 coilin can disrupt both nucleoli and Cajal bodies (Bohmann et al. 1995b; Sleeman et al. 1998). The p80 coilin protein has also been shown to interact directly with the nucleolar protein NOPP140 (Isaac et al. 1998). These data indicate that at least one of the roles of the Cajal body likely involves a functional interaction with nucleoli.
Recent studies have provided evidence that Cajal bodies may play a role in the transport and/or maturation of both splicing small nuclear (sn)RNPs and nucleolar snoRNPs. Transient expression of GFP-tagged snRNP Sm proteins in mammalian cells shows that when they enter the nucleus, they first concentrate in Cajal bodies before subsequently moving into a speckled pattern (Sleeman and Lamond 1999). Injection of fluorescently labeled U3 and U8 snoRNAs into Xenopus oocytes results in their transient accumulation in Cajal bodies before they move to the nucleolus (Narayanan et al. 1999). Treatment of cells with Leptomycin B, which inhibits export of newly transcribed snRNA to the cytoplasm and hence stems the flow of snRNP production, also causes depletion of snRNPs from Cajal bodies (Carvalho et al. 1999).
It is likely that Cajal bodies can also play other roles in the nucleus. Several gene loci have been found to preferentially colocalize with Cajal bodies (Callan et al. 1991; Frey and Matera 1995; Gall et al. 1995; Smith et al. 1995). These include histone gene clusters and loci encoding the U1, U2, and U3 snRNAs. Interestingly, studies using cell lines containing artificial tandem arrays of the U2 snRNA gene cluster showed that their association with Cajal bodies was dependent on transcription of the locus. Inhibition of transcription abolished association of the U2 genes with Cajal bodies (Frey et al. 1999). It is possible that the association of Cajal bodies with specific gene loci could be part of a feedback mechanism regulating gene expression. Alternatively, it could help to supply newly assembled processing factors to highly expressed genes such as histones. Possible clues concerning the function of Cajal bodies come from their known components. In addition to nucleolar and splicing RNPs, several transcription factors have been localized to coiled bodies in specific cell types (Matera 1999). Based on recent immunolabeling data in Xenopus oocytes, Gall et al. 1999 have proposed that Cajal bodies can function as assembly sites for major transcription machineries or “transcriptosomes.” Presently, it is not clear whether every Cajal body performs all of the functions discussed above, or alternatively, whether different Cajal bodies may be functionally distinct.
Relatively little is known about the dynamic properties of Cajal bodies. Although Cajal bodies can localize to certain gene loci, this localization might be transient; it has been suggested that Cajal bodies might actually be motile structures (Frey and Matera 1995). Boudonck et al. 1999 have observed the movement of Cajal bodies in plant cells that express a GFP-tagged U2 snRNP protein. However, the dynamic properties of Cajal bodies have not been analyzed in animal cells. In this study, we report the characterization of a stable HeLa cell line expressing p80 coilin fused to GFP. Using time-lapse fluorescence microscopy, we show that Cajal bodies in animal cells are highly mobile and can both join and separate from one another and move to and from the nucleolus. The data also suggest that Cajal bodies may include at least two distinct forms with different properties.
Materials and Methods
Plasmid Construct
The GFP-coilin full-length cDNA was amplified from pGFP-coilin (Sleeman et al. 1998) using 5′ primer CFR31 (5′-TCCCCGCGGCTTGCCGCCACCATGGTGAGCAAGGGC-3′) and 3′ primer CRR31 (3′-CTAGTCTAGACCTACTGACGACTGCTACTTGAACA-5′), which contain an SacII and an XbaI site, respectively (bold). The resulting PCR fragment was digested with SacII and XbaI and cloned into the pUHG 10-3 plasmid (Gossen and Bujard 1992) yielding the pTREGFP-coilin plasmid. The SacII–GFP-coilin–XbaI internal portion of plasmid pTREGFP-coilin was sequenced and confirmed to contain the expected sequence. The pUHG 10-3 plasmid contains the Tet-responsive PhCMV promoter. cDNAs inserted downstream of the promoter are responsive to the tTA tetracycline-controlled transactivator protein expressed by the pTet-Off plasmid in the Tet-Off system (Gossen and Bujard 1992). The HeLa Tet-Off cell line (CLONTECH Laboratories, Inc.), stably expressing the tTA from integrated copies of the pTet-Off plasmid, was used for transfection with the pTREhGFP-coilin and pTK-Hyg (CLONTECH Laboratories, Inc.) plasmids. pTK-Hyg allows selection of stably transformed cell lines in the presence of hygromycin.
The fibrillarin full-length cDNA was amplified from the human HeLa Marathon-Ready cDNA library (CLONTECH Laboratories, Inc.) using 5′ primer FCFP-R1 (5′-CCGGAATTCCGGGCTCGCCATGAAGCCAGGA-3′) and a 3′ primer FCFP-R1 (3′-CCGGAATTCCGTCAGTTCTTCACCTTGGGGGGTGGCCT-5′), both containing EcoRI sites (bold). The resulting PCR fragment was digested with EcoRI and cloned into the pEYFP-C1 vector (CLONTECH Laboratories, Inc.) to give rise to the pEYFP-fibrillarin plasmid. The EcoRI–YFP-fibrillarin–EcoRI internal portion of the plasmid was sequenced and contained the expected fibrillarin sequence. Transfections were performed using the Effectene transfection reagent (QIAGEN) according to the manufacturer's protocol.
Cell Culture Conditions
l-glutamine was maintained as a stock of 200 mM in ddH2O, geneticin G418 as a stock of 200 μg/ml in DME (no supplements), and tetracycline hydrochloride as a 1 mg/ml stock in ddH2O at −20°C and diluted to appropriate concentrations in tissue culture medium before use. All cells were grown at 37°C in a humidified 5% CO2 atmosphere. Transformed GFP-coilin cell line was grown in DME (GIBCO BRL), supplemented with 10% Tet-approved fetal calf serum (CLONTECH Laboratories, Inc.), 2 mM l-glutamine (GIBCO BRL), 100 U/ml penicillin and streptomycin, 200 μg/ml geneticin G418, and 100 μg/ml hygromycin and 2 μg/ml tetracycline hydrochloride (Gossen and Bujard 1992). The parental Tet-Off HeLa cell line was grown in DME (GIBCO BRL) supplemented with 10% Tet-approved fetal calf serum (CLONTECH Laboratories, Inc.), 2 mM l-glutamine (GIBCO BRL), and 100 U/ml penicillin and streptomycin (GIBCO BRL). The concentration of geneticin G418 was maintained at 200 μg/ml.
Cell Transfection and Establishment of the HeLaGFP-Coilin Cell Line
Transfections were performed after the cells had reached ∼80% confluency using Fugene (Roche) and optimal transfection conditions. 2 d after transfection, hygromycin was added at a concentration of 300 μg/ml. During the time of establishment of the cell line, cells were cultured in the presence of tetracycline hydrochloride at a concentration of 2 μg/ml. The hygromycin concentration used was optimized for each cell line. After a period of 2–3 wk, healthy large hygromycin-resistant colonies were isolated. Colonies were allowed to grow in the absence of tetracycline for 24 h, and the presence of pTREGFP-coilin was confirmed by fluorescence microscopy. GFP-coilin–expressing clones giving lowest background (uninduced expression levels) and correct localization were further subcloned to give single colony clones where ≥90% of the cells were expressing GFP-coilin.
Antibodies, Fixation, and Immunofluorescence
All fixation, permeablization, and immunostaining were performed at room temperature. Cells grown on glass coverslips (no. 1 1/2) were washed in PBS and fixed for 10 min with paraformaldehyde in CSK buffer (10 mM Pipes, pH 6.8, 10 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 2 mM EDTA). Permeablization was performed with 1% Triton X-100 in PBS for 10 min. Cells were subsequently washed in PBS, incubated with 10% goat serum in PBS for 20 min, followed by incubation with primary antibody for 1 h. Three washes with PBS were carried out before incubation with secondary antibody (affinity-purified Texas red–conjugated goat anti–rabbit or goat anti–mouse (Jackson ImmunoResearch Laboratories) for 45 min. Cells were washed in PBS mounted with 0.5% p-phenylenediamine in 20 mM Tris, pH 8.8, 90% glycerol, sealed, and left to dry before examination. The following antibodies were used: rabbit anti-p80 coilin polyclonal serum 204/10 (dilution 1:500) (Bohmann et al. 1995a); mAb Y12 anti-Sm (dilution 1:500) (Pettersson et al. 1984); 72b9 antifibrillarin (dilution 1:10) (Reimer et al. 1987); mouse monoclonal anti–survival of motor neuron (SMN) protein (dilution 1:10) (Young et al. 2000); and mouse anti–SMN-interacting protein (SIP) (Liu et al. 1997; Young et al. 2000) (dilution 1:10).
Microscopy and Image Analysis
Immunostained specimens were examined by using a 40× NA 1.3 or a 100× NA 1.4 Plan-Apochromat objective. Three-dimensional images were recorded on a Nikon DeltaVision Restoration microscope (Applied Precision, Inc.) equipped with a three-dimensional motorized stage and a Photometrics CH350 camera containing a 1401E charge-coupled device (Eastman Kodak Co.). For each nucleus, 20–24 optical sections separated by 0.2 μm were recorded. Exposures were chosen such that images yielded gray scale units between 200 and 2,000, remaining well above the camera dark current but below the 4,096-U maximum.
For live cell imaging, cells were grown on 42-mm glass coverslips (no. 1; Helmut Sauer) in medium containing 2 μg/ml tetracycline. First, we imaged living cells in the presence of tetracycline but saw no difference in the size of Cajal bodies or their dynamics compared with cells that were imaged 4–7 h after inducing GFP-coilin expression by tetracycline-removal. GFP-coilin levels continued to rise up to 24 h after removal of tetracycline (data not shown). We chose to optimize the fluorescence signal but minimize possible effects of overexpression by imaging cells within 4–7 h after induction. HeLaGFP-coilin cells were grown in the absence of tetracycline for up to 2 mo with no detectable effect on viability.
Cells were maintained at 37°C by use of a closed perfusion chamber (Bachofer). Images were collected using the 100× NA 1.4 Plan-Apochromat objective on the DeltaVision microscope. For each nucleus, 20–24 optical sections (depending on the nucleus diameter, typically 10–12 μm) separated by 0.5 μm were recorded. In all cases, the Hg lamp excitation light was attenuated with a 1.0-OD neutral density filter. In addition, all images were recorded using a binning of 2 × 2 on the CH350 charge-coupled device camera, yielding an effective pixel size of 0.102 × 0.102 μm. Three-dimensional images were recorded every 2–3 min over a time period of 1–2.5 h. Each exposure lasted 100 ms.
The three-dimensional motorized stage allowed the imaging of up to eight nuclei in each experiment. In total, time-lapse three-dimensional images of 63 nuclei were recorded. Images were corrected for any fluctuations in Hg lamp power and restored by an iterative constrained deconvolution algorithm using an empirically measured point–spread function (Agard et al. 1989; Swedlow et al. 1997). Time-lapse images were viewed as three-dimensional maximum intensity projections of each time point. All of these manipulations were performed using routines contained within the softWoRx image processing package (Applied Precision, Inc.).
To determine whether our imaging protocol affected cell viability, we identified HeLaGFP-coilin cells that were entering mitosis and imaged them using the protocol described above. All cells completed mitosis and formed new daughter cells that were indistinguishable from others on the coverslip (data not shown). In addition, we subjected 40 interphase cells growing in a perfusion chamber (FCS2; Bioptechs) to our standard 2.5-h imaging protocol and then continued to collect three-dimensional images, once every 30 min, for the next 24–36 h. Of these, four cells entered mitosis as judged by the breakdown of the nuclear envelope and appearance of cell rounding 6.5–12 h after the end of rapid data collection. One of these cells completed mitosis. Therefore, our imaging protocol does not prevent progression to mitosis. However, because we cannot exclude that extended incubation in the perfusion chamber may have effects on cell health, we confined the data recording time to 2–2.5 h in this study.
The shape of the nuclei shown in Fig. 2, Fig. 5, Fig. 6, and Fig. 7 are quite variable, even during the period of data collection. This nuclear morphology is a characteristic of both parental and HeLaGFP-coilin cell lines (see Fig. 2) and is not induced by phototoxicity or imaging. Cells with similar interphase morphology entered and completed mitosis (data not shown).
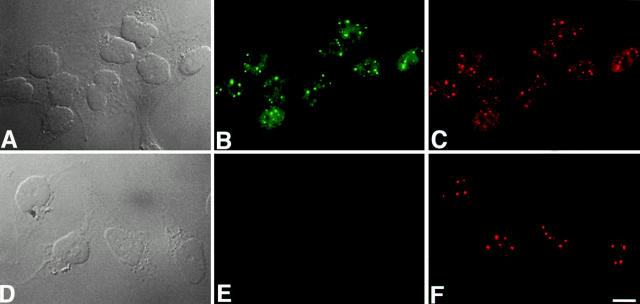
Figure 2.
Coilin localization in parental HeLa and HeLaGFP-coilin cells. Micrographs show both differential interference contrast (DIC) and fluorescence images of the HeLaGFP-coilin cells (A–C) and the parental HeLa cells (D–F). Representative optics sections from deconvolved data sets of the HeLaGFP-coilin cells are presented (B and C) with GFP-coilin in green (B), and counterstaining detecting both endogenous p80-coilin and GFP-coilin in red (C). (A) The DIC image of the same field of cells shown in B and C. Representative optical sections from deconvolved data sets from the parental HeLa cell line are presented (E and F) with endogenous p80 coilin detected with anticoilin antibody in red (F). No GFP fluorescence is detected in the parental HeLa cell line (E). (D) The DIC image of the same field of cells shown in F. For both the parental and HeLaGFP-coilin cells, endogenous p80 coilin and GFP-coilin show an identical pattern, consisting of a diffuse nucleoplasmic signal together with several bright foci of different sizes. Bar, 5 μm.
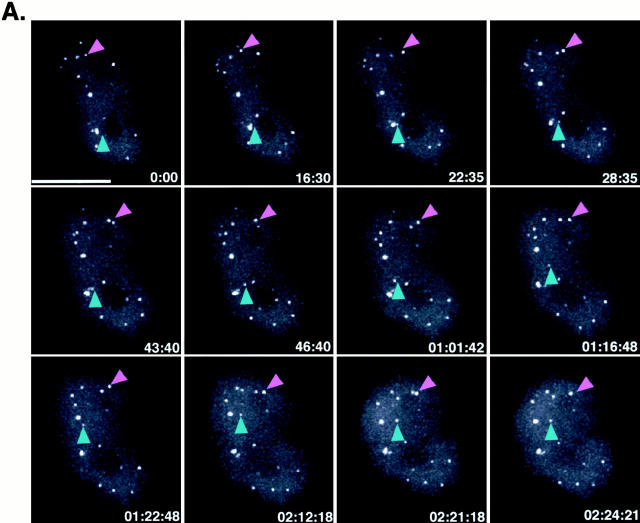
Figure 5.
In vivo time-lapse three-dimensional imaging of HeLaGFP-coilin cells. (A) A time-lapse sequence of the GFP signal from a nucleus of an HeLaGFP-coilin cell. 12 out of a total of 60 time points are shown as a montage. Data were collected every 3 min for 2.5 h. Each image is a maximum intensity projection of 24 optical sections (0.5 μm each) spanning the entire cell nucleus. Both CBs (and mini-CBs) were observed moving. The magenta arrowhead points to a pair of Cajal bodies that fuse in the last time point. Note the changes in intensity that occur in the smaller Cajal body. The blue arrowhead points to a mini-CB that splits off from a larger CB in the second time point and then moves upwards through the nucleoplasm. An animated time-lapse version of this process is available at http://www.jcb.org/cgi/content/full/151/7/1561/DC1. (B) Histogram of Cajal body volumes from a time-lapse data set of an HeLaGFP-coilin nucleus. Volume measurements of all Cajal bodies from a single data set were divided into bins and plotted as a histogram. This analysis verified the presence of specific size classes of Cajal bodies (as shown by the presence of clusters of Cajal body volumes). Bar, 10 μm.
Figure 6.
In vivo time-lapse three-dimensional analysis of an HeLaGFP-coilin cell nucleus showing movement of CBs through the nucleoplasm and separation of mini-CBs from Cajal bodies. 12 three-dimensional time-lapse images out of a total of 22 are shown as a montage. The image at each time point is a maximum intensity projection of 20 optical sections (0.5 μm each) scanning the entire cell nucleus. Data were collected every 3 min over a period of 2.25 h. Note the movement of a CB (magenta arrowheads) through the nucleoplasm from right to left bringing it into close proximity to a Cajal body that was located at the opposite side of the nucleus at time 0.00. A mini-CB (blue arrowhead) can be seen separating from a larger Cajal body located at the nuclear periphery, moving to the nucleolus and returning back to the same Cajal body from where it originated. An animated time-lapse version of this process is available at http://www.jcb.org/cgi/content/full/151/7/1561/DC1. Bar, 15 μm.
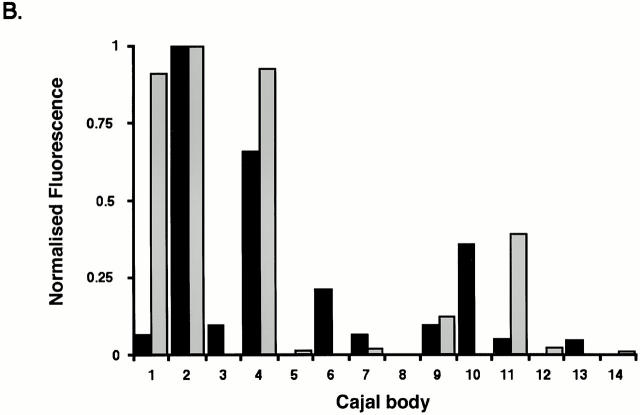
Figure 7.
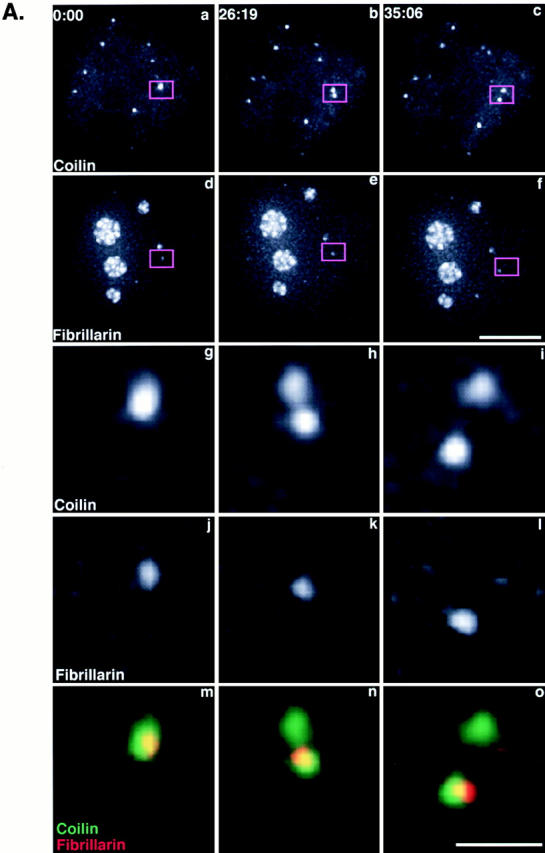
Differential partition of components after separation of a Cajal body. (A) A plasmid containing YFP-fibrillarin was transfected into HeLaGFP-coilin cells (see Materials and Methods). Maximum intensity projections from time-lapse three-dimensional data sets are shown. Top two rows show GFP-coilin (a–c) and YFP-fibrillarin (d–f). The boxed region is enlarged in the bottom three rows to show a CB splitting that generates an unequal distribution of YFP-fibrillarin to the two resulting Cajal bodies (GFP-coilin, g–i; YFP-fibrillarin, j–l; overlay, m–o). An animated time-lapse version of this process is available at http://www.jcb.org/cgi/content/full/151/7/1561/DC1. Bars: (a–f) 5 μm; (g–o) 1 μm. (B) Histogram showing fluorescence from individual Cajal bodies from YFP-fibrillarin–transfected HeLaGFP-coilin cells. The total GFP-coilin and YFP-fibrillarin fluorescence for each Cajal body in a nucleus was summed and corrected for nuclear background nuclear fluorescence. To plot these on the same scale, data were expressed as a fraction of the maximum Cajal body fluorescence for each protein. The relative levels of the two proteins varied significantly between individual Cajal bodies. Black bars, GFP-coilin; white bars, YFP-fibrilarin.
All tracking and quantitative analyses were performed on the full time-lapse three-dimensional data sets. To identify individual Cajal bodies, a combination of an empirical intensity threshold and object size was used. The segmentation algorithm finds “features” by applying a global threshold to each stack of z-sections. Features are defined as sets of contiguous pixels that are above the specified threshold. The threshold was specified as several standard deviations above the mean pixel value of the stack of z-sections. The threshold is recalculated for each time point.
To calculate feature trajectories of Cajal bodies, distances between the centroids of each feature and centroids of features in the subsequent time point were calculated. The feature in the subsequent time point with the nearest centroid was used for the next point in the trajectory. These simple criteria were sufficient to identify and track most Cajal bodies. All derived trajectories were confirmed by visual inspection.
Once each Cajal body in a time-lapse three-dimensional data set was identified and tracked, the total number of pixels of the Cajal body (i.e., its volume), the total intensity inside the Cajal body, and the centroid of the GFP-coilin signal at each time point were recorded. For analysis of Cajal body sizes (see Fig. 5 B), the identification code for each Cajal body in a single time-lapse three-dimensional data set, its centroid, time point, and volume were entered into the data visualization tool SpotFire Pro (SpotFire, Inc.). The range of volumes in each data set was divided into equal sized bins and displayed as a histogram.
To measure GFP-coilin content of individual Cajal bodies (see Fig. 5, Fig. 7, and Fig. 9), we measured the total GFP-coilin fluorescence in each Cajal body throughout each time-lapse three-dimensional data set. For every time point, the total fluorescence intensity for each Cajal body was corrected for any errors in volume determination by subtracting the threshold value that was used for identifying the boundary of the Cajal body (see above), multiplied by the number of pixels (volume) contained in that Cajal body. To display Cajal body GFP-coilin and yellow fluorescent protein (YFP)-fibrillarin content on the same scale, the contents were expressed as a percentage of the maximum to yield the normalized GFP-coilin content.
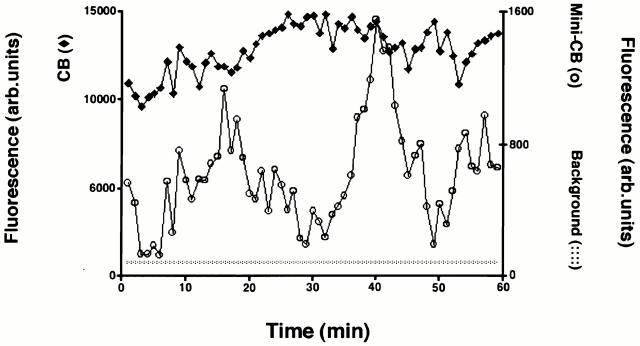
Figure 9.
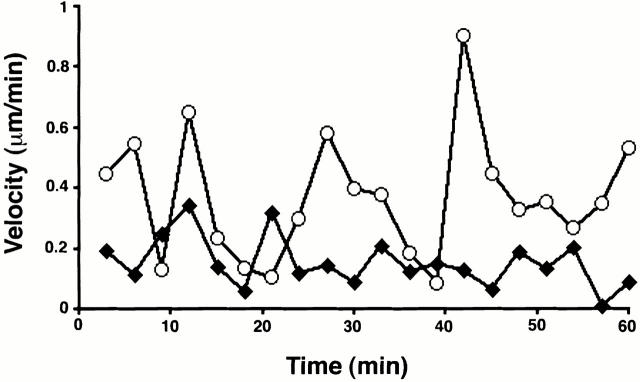
Temporal variations in GFP-coilin content of Cajal bodies. A plot of the GFP-coilin content of a CB (filled diamonds) and mini-CB (open circles) plotted as a function of time. GFP-coilin content was calculated by summing fluorescence within each CB and mini-CB throughout the time-lapse data set. Each measurement of summed fluorescence was corrected for any contribution from background nucleoplasmic fluorescence and errors in volume determination (see Materials and Methods). In addition, the level of background nucleoplasmic fluorescence is plotted (dotted line). The GFP content of the CB is higher than the mini-CB, although both show systematic variations in GFP-coilin content. This indicates that coilin can move between both types of bodies and the diffuse nucleoplasmic pool.
Protein Analysis and Immunoblotting
For preparation of total cell lysates, cells were first washed twice with ice-cold PBS and then lysed in 1 ml (per ∼106–107 cells) of ice-cold 50 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 1% (vol/vol) NP-40, 1% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) SDS, 2 mM EDTA plus complete protease inhibitor cocktail (Roche) for 5 min. The lysates were homogenized and cleared by passage through a QIAshredder unit (QIAGEN) and centrifuged at 4°C for 10 min at 13,000 g.
For preparation of salt- and detergent-fractionated total cell lysates (Capco et al. 1982; Fey and Penman 1988; He et al. 1990), ∼107 cells were trypsinized and centrifuged in tissue culture medium at 1,000 g for 3 min at 4°C. Cells were washed in PBS at 4°C, centrifuged, and the cell pellet was resuspended and incubated in 1 ml of buffer containing 10 mM Pipes, pH 6.8, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, and 0.2 mg/ml PMSF for 5 min at 4°C. This was centrifuged at 1,000 g for 5 min to produce supernatant A and pellet A. Supernatant A was stored for further analysis while pellet A was resuspended and incubated in 1 ml of 10 mM Pipes pH 6.8, 250 mM ammonium sulfate, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.2 mg/ml PMSF for 5 min at 4°C. This was centrifuged at 4°C for 5 min at 1,000 g, giving rise to supernatant B and pellet B. Supernatant B was stored for further analysis, and pellet B was resuspended in 0.5 ml of 10 mM Pipes, pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100 with 400 U/ml of DNase I at 32°C for 50 min. This was centrifuged at 1,000 g, at 4°C for 5 min, giving rise to supernatant C and pellet C. Protein samples from supernatants A–C and pellet C were boiled in SDS sample buffer and analyzed by SDS-PAGE. Approximately equal amounts of protein were loaded in each lane, as estimated by Ponceau staining of proteins transferred to nitrocellulose membrane in pilot experiments. Samples were separated on a 10% SDS polyacrylamide gel and electrophoretically transferred to nitrocellulose membrane (Schleicher & Schuell). The membranes were then incubated with mouse 5P10 anticoilin antibody (1:5,000 dilution) (Almeida et al. 1998), detected using mouse HRP conjugate (1:5,000 dilution) (Pierce Chemical Co.) in PBS containing 5% dry milk, 0.15% Tween 20, and detected via chemiluminescence (Amersham Pharmacia Biotech).
Online Supplemental Material
A time-lapse movie of Fig. 5 A (Video 1) demonstrates the nucleus of an HeLaGFP-coilin cell showing Cajal body dynamics. The time interval between frames is 3 min. Each image is a maximum intensity projection. A time-lapse movie of Fig. 6 (Video 2) demonstrates the nucleus of an HeLaGFP-coilin cell showing Cajal body dynamics and association with the nucleolus. The time interval between frames is 3 min. Each image is a maximum intensity projection. A higher magnification time-lapse movie of Fig. 7 A (Video 3) demonstrates an HeLaGFP-coilin cell nucleus expressing YFP-fibrillarin, showing separation of Cajal bodies and unequal partitioning of YFP-fibrillarin. The time interval between frames is 3 min. Each image is a maximum intensity projection. Time-lapse videos are available at http://www.jcb.org/cgi/content/full/151/7/1561/DC1.
Results
Establishment of an HeLa Cell Line Expressing GFP-Coilin
An HeLa cell line expressing p80 coilin fused to GFP was constructed to facilitate studies on the dynamics of Cajal bodies in living cells. To do this, we used the human p80 coilin cDNA fused at its NH2 terminus to the GFP gene (Sleeman et al. 1998). The resulting fusion protein was inserted into the plasmid vector, pUHD103, in which expression of the fusion protein was under the control of a tetracycline repressible promoter (Fig. 1 A). The resulting vector, pTREGFP-coilin, was transfected into the parental Tet-Off HeLa cell line, and stable transformants that expressed GFP-coilin were isolated (see Materials and Methods). The expression of endogenous p80 coilin and GFP-coilin was analyzed in both the resulting HeLaGFP-coilin and the parental HeLa cell lines (Fig. 1 B). Whole cell protein lysates were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with an anti-p80 coilin mAb. The endogenous p80 coilin protein was detected at similar levels in both cell lines, independent of the presence of tetracycline (Fig. 1 B, lanes 1–4; 80-kD band). A protein of ∼110 kD, corresponding to GFP-coilin, was detected in the HeLaGFP-coilin cell line (Fig. 1 B, lanes 3 and 4) and absent from the parental HeLa cell line (Fig. 1 B, lanes 1 and 2). Removal of tetracycline from the medium for ∼7 h resulted in an increased level of GFP-coilin in the HeLaGFP-coilin cell line (Fig. 1 B, lanes 3 and 4). However, all clones of transformed HeLa cells showed expression of GFP-coilin even when grown in the presence of high levels of either tetracycline or doxycycline (Fig. 1 B, lane 3; other data not shown). The clone selected for further analysis expressed GFP-coilin at higher levels than the endogenous p80 coilin. However, we observed no difference in either the localization or biochemical behavior of GFP-coilin regardless of the level of expression or tetracycline present in the culture medium. All subsequent analyses were performed using HeLaGFP-coilin cells 4–7 h after removing tetracycline from the medium.
Figure 1.
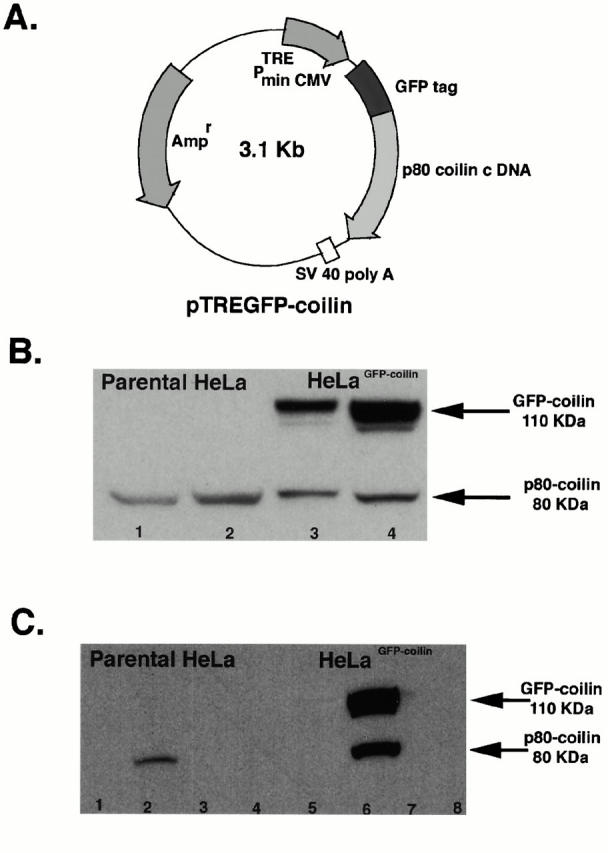
Expression of GFP-coilin. (A) pTREGFP-coilin plasmid. The pTREGFP-coilin plasmid contains the p80 coilin cDNA fused to the 3′ end of hGFP gene under the control of the tetracycline-responsive PhCMV promoter. The PhCMV promoter contains the tetracycline-responsive element (TRE), comprising seven copies of the tetracycline operator sequences. Expression of GFP-coilin from the pTREGFPcoilin plasmid in cells containing the tTA regulatory protein is thereby repressed in the presence of tetracycline. The white box represents the SV40 polyadenylation signal. (B) Analysis of coilin proteins in parental HeLa and HeLaGFP-coilin cell lines. Whole cell lysates were prepared from parental HeLa cells and from the HeLaGFP-coilin cells. 35 μg of total cell protein lysates was loaded in each lane, separated by SDS-PAGE, and detected using an anticoilin antibody. Lane 1, parental HeLa cells, no tetracycline; lane 2, parental HeLa cells cultured with 2 μg/ml of tetracycline; lane 3, HeLaGFP-coilin cells cultured with 2 μg/ml of tetracycline; and lane 4, HeLaGFP-coilin cells, 7 h after removal of tetracycline from the medium. A 110-kD band corresponding to GFP-coilin is detected specifically in the HeLaGFP-coilin cells in lanes 3 and 4. The appearance of GFP-coilin expression at lower levels in lane 3 is a result of incomplete repression by the tTA protein. An 80-kD band corresponding to the endogenous coilin protein is detected in lanes 1–4. (C) Biochemical behavior of endogenous p80 coilin and GFP-coilin. Both the parental HeLa (lanes 1–4) and the HeLaGFP-coilin (lanes 5–8) cells were extracted in buffers containing different salt and detergent conditions (see Materials and Methods). Approximately equal amounts of total protein were loaded in each lane, separated by SDS-PAGE, and detected using an anticoilin antibody. Supernatant A (lanes 1 and 5), supernatant B (lanes 2 and 6), supernatant C (lanes 3 and 7), and pellet C (lanes 4 and 8). In the HeLaGFP-coilin cell extracts both the endogenous p80 coilin and the GFP-coilin proteins are specifically detected in supernatant B (lane 6). The endogenous p80 coilin in the parental HeLa cell extracts is also present specifically in supernatant B (lane 2).
We compared the biochemical properties and localization patterns of the GFP-coilin and endogenous p80 coilin proteins to check whether the GFP-coilin fusion protein provided a valid Cajal body marker in this cell line. First, we tested whether extracting nuclei with buffers containing different salt and detergent concentrations would show any differences in the extraction properties of GFP-coilin and endogenous p80 coilin, as might be expected if the GFP tag affected the interaction of coilin with nuclear structures (Fig. 1 C, lanes 1–4). This test showed that both GFP-coilin and endogenous p80 coilin behaved identically (Fig. 1 C, lanes 5–8). The endogenous p80 coilin was also extracted by the same buffer and salt conditions in the parental HeLa cells and the HeLaGFP-coilin cells (Fig. 1 C).
Analysis of the HeLaGFP-coilin and parental HeLa cell lines in the fluorescence microscope showed that only the HeLaGFP-coilin cells expressed a GFP signal (Fig. 2B and Fig. E). Immunostaining fixed HeLaGFP-coilin cells with an anti-p80 coilin mAb showed an identical pattern of labeling to the GFP-coilin signal (Fig. 2B and Fig. C). A similar anticoilin staining pattern was also observed in the HeLaGFP-coilin and parental HeLa cells (Fig. 2C and Fig. F). The morphology of the nuclei shown in Fig. 2 is representative of these cell lines and was observed in both fixed and living cells (see Materials and Methods).
In summary, we conclude that the localization pattern and biochemical properties of the GFP-coilin fusion protein in the HeLaGFP-coilin cell line are equivalent to the endogenous p80 coilin and provide a valid marker for studying Cajal bodies in live cells.
Distinct Classes of Cajal Bodies
The staining pattern of the anticoilin mAb was compared in detail in fixed cells from both the parental and He- LaGFP-coilin cell lines. Three-dimensional images were recorded and deconvolved, and the data were presented as maximum intensity projections of the respective nuclei (Fig. 3). The parental and transformed cell lines both show a similar pattern of nuclear bodies containing coilin. This includes larger bodies, typically 0.4–0.7 μm in diameter, and smaller bodies with a diameter ≤0.2 μm. For the purpose of this study, we will use the term CB as a generic term for all bodies containing p80 coilin. However, because the data presented below indicate differences in the structure and properties of separate classes of CBs, we will refer to the larger bodies (≥0.4) as CBs and the smaller bodies (≤0.2 μm) as mini-CBs to distinguish between them where appropriate (Fig. 3A and Fig. B; arrows indicate CBs, and arrowheads, mini-CBs). Also, coilin shows a diffuse nucleoplasmic distribution, excluding nucleoli, in both cell lines. The number of CBs and mini-CBs was counted in three-dimensional images from 50 separate nuclei from both cell lines (Fig. 3 C). The total number of Cajal bodies and the ratio of CBs to mini-CBs are the same for both cell lines. This demonstrates that the presence of the smaller class of coilin positive bodies is not due to expression of the GFP-coilin fusion protein. This conclusion is supported by our parallel observations that mini-CBs can also be detected in many other mammalian cell lines (data not shown).
Figure 3.
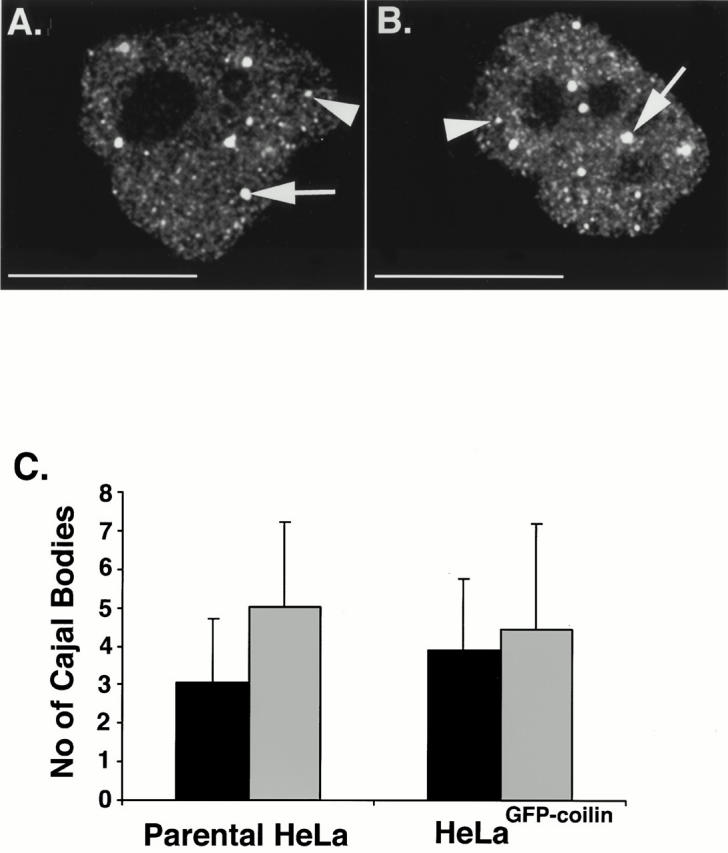
Both parental HeLa and HeLaGFP-coilin cell lines show a similar pattern and number of Cajal bodies. (A and B) Both parental HeLa and HeLaGFP-coilin cells were grown to similar confluencies on glass coverslips using the same tissue culture conditions, fixed, and analyzed in the fluorescence microscope. The HeLaGFP-coilin cells were grown in medium with no tetracycline for ∼7 h before fixation. Maximum intensity projections of deconvolved data sets are shown from fixed parental HeLa (A) and HeLaGFP-coilin (B) cell nuclei immunolabeled using an anticoilin antibody. Both nuclei show a similar diffuse nuclear staining with additional bright Cajal bodies. The Cajal bodies appear to include at least two distinct size classes. The larger bodies, ≥0.4 μm (CBs), are shown by arrows and the smaller bodies, ≤0.2 μm (mini-CBs), are shown by arrowheads. (C) Histogram shows the mean number of Cajal bodies in interphase nuclei of both parental HeLa and HeLaGFP-coilin cell lines. The ratio of CBs (black bars) to mini-CBs (gray bars) is the same in both cell lines. Numbers were determined using three-dimensional maximum intensity projections of all the optical sections of the deconvolved data sets from 50 different nuclei of each cell line. Error bars represent SDs. Bar, 15 μm.
Immunolabeling HeLaGFP-coilin cells with antibodies specific for several nuclear antigens known to localize in Cajal bodies provided further evidence that the CBs and mini-CBs may be distinct structures (Fig. 4; other data not shown). For example, fibrillarin and the Sm snRNP proteins are detected in CBs but not in many of the smaller bodies (Fig. 4, A–D; CBs are marked with magenta arrowheads, and mini-CBs with blue arrowheads). In contrast, antibodies to the SMN and SIP1 proteins label all the bodies that contain coilin (Fig. 4, E–H). The molecular composition of mini-CBs appears to vary, as judged by antibody labeling, which could mean that there are further subclasses among these smaller bodies. In summary, we conclude that the nuclear bodies containing coilin in mammalian cells include a class of smaller structures, typically ≤0.2 μm in diameter, which may also differ in their antigen composition from the larger CBs.
Figure 4.
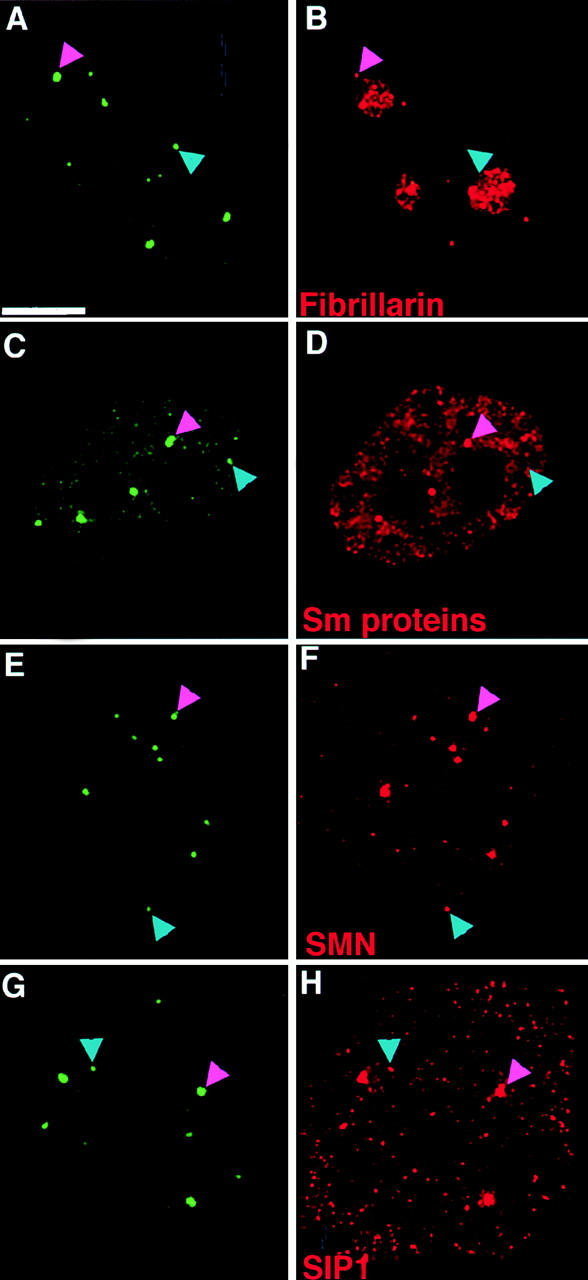
Analysis of Cajal body antigens in CBs and mini-CBs. Fluorescence micrographs of HeLaGFP-coilin cells immunolabeled with antibodies specific for known Cajal body antigens. Maximum intensity projection images of deconvolved data sets are shown with GFP-coilin in green (A, C, E, and G) and either fibrillarin (B), Sm proteins (D), SMN (F), and SIP1 (H) in red. Fibrillarin and the Sm proteins (B and D) are detected in CBs (magenta arrowheads) but not in all of the mini-CBs (blue arrowheads). SMN and SIP1 (F and H) are detected in both CBs (magenta arrowheads) and mini-CBs (blue arrowheads). Bar, 5 μm.
Visualizing Cajal Body Movement by Three-dimensional Time-Lapse Fluorescence Microscopy
Recent studies have emphasized the dynamics of the components of the nucleus (Marshall et al. 1997; Misteli et al. 1997; Bornfleth et al. 1999; Sleeman and Lamond 1999; Phair and Misteli 2000). Moreover, previous studies have demonstrated movement of Cajal bodies in plant cell nuclei (Boudonck et al. 1999). Therefore, the HeLaGFP-coilin cell line was used to investigate the nature and types of movements of Cajal bodies in human cells. Time-lapse three-dimensional recordings were made from 63 separate nuclei. A typical example is shown in Fig. 5 A, which illustrates 12 time points out of 60 total three-dimensional images recorded over a period of 2 h 24 min (see Materials and Methods). Each image corresponds to a two-dimensional maximum intensity projection from 24 separate optical sections spanning the full depth of the nucleus. Examples of CBs and mini-CBs were observed in every recorded data set. In addition, several events demonstrating that Cajal bodies are indeed dynamic structures were seen in all data. In every nucleus, movement of all the Cajal bodies was detected, frequently involving translocation of Cajal bodies through the nucleoplasm (Fig. 5 A and 6; arrowheads). Finally, many examples of Cajal bodies joining and splitting were seen. Quantitative analysis of time-lapse data showed rapid fluctuations in Cajal body velocity. Moreover, the amount of GFP-coilin in individual Cajal bodies often changed dramatically over time. Each of these observations is discussed below.
Visual examination of images of all nuclei confirmed the presence of CBs and mini-CBs in living HeLaGFP-coilin nuclei (Fig. 5 A), as described above in fixed cell nuclei. To test whether the size classes seen in the live images represented distinct families of Cajal bodies, we examined the distribution of Cajal body sizes on a histogram. By analyzing the volumes of Cajal bodies from time-lapse data of single nuclei, we observed a clustering of Cajal body volumes into two or three different classes (Fig. 5 B). The smallest population represents mini-CBs and the larger populations represent CBs. A similar result was seen for all nuclei examined (n = 6). This analysis concurs with our data in fixed parental and HeLaGFP-coilin cells (see Fig. 2 and Fig. 3). However, we note that when we combined measurements of Cajal body volumes of up to 20 separate nuclei, we no longer observed distinct clusters of Cajal body volumes (data not shown). Therefore, combining volume data from many nuclei masked the presence of distinct size classes in individual nuclei. This implies that different nuclei can contain Cajal bodies of different sizes. Indeed, Cajal bodies change in size as cells progress through interphase (Andrade et al. 1993). In summary, we conclude that distinct size classes of Cajal bodies can exist within the nuclei of HeLaGFP-coilin cells.
Joining and Separation of CBs and Movement to and from Nucleoli
The movement of Cajal bodies also involved more complex dynamic behavior. This included the joining of two bodies to form larger Cajal bodies (Fig. 5 A; magenta arrowheads). The formation of larger CBs from the joining together of smaller bodies was a common event seen in most of the nuclei during the 1–2.5-h time period analyzed. The joining events we recorded were often preceded by the movement across the nucleoplasm of one or both Cajal bodies to bring them into close proximity. Cajal bodies destined to join were frequently seen to eschew their closest neighbors in favor of translocating through the nucleoplasm to join with a distant partner. After moving close together, the bodies often remained at a fixed distance for varying time periods before a final rapid movement resulted in joining (Fig. 5 A, magenta arrowheads). Cajal body joining produces a larger body with greater fluorescence intensity than either of the two individual bodies before joining (data not shown).
Separation events were also recorded where the emergent mini-CB first moved away from the larger body and then subsequently returned and rejoined with the original Cajal body (Fig. 6; blue arrowhead). In the example shown in Fig. 6, the emergent mini-CB moves from a larger body located at the edge of the nucleus and travels through the nucleoplasm to the periphery of the nucleolus. It remains at the nucleolar periphery for ∼30 min and then moves back to rejoin with the same CB at the edge of the nucleus. The separation of mini-CBs from larger bodies to form new structures was also observed (data not shown). Although separation of mini-CBs from larger bodies was seen less frequently than CB joining, it was still detected in 25% of the nuclei analyzed. The separation event was usually followed by the movement of the mini-CB away from the larger body. To date, all the separation events we have detected occur in the nucleoplasm and have not shown any obvious association with a specific subnuclear location. This complex behavior indicates that directed mechanisms may be involved in the movement of Cajal bodies.
To analyze the molecular consequences of Cajal body separation in more detail, we transfected HeLaGFP-coilin cells with a plasmid expression vector, encoding YFP-fibrillarin (see Materials and Methods). Fig. 7 A shows a Cajal body (a–c; enlarged in g–i) that splits, resulting in two Cajal bodies with unequal YFP-fibrillarin content (Fig. 7 A, d–f; enlarged in j–l). Analysis of the YFP-fibrillarin in these two Cajal bodies showed that all detectable YFP-fibrillarin was partitioned into one of them. We further analyzed the relative amounts of GFP-coilin and YFP-fibrillarin in 14 individual Cajal bodies from a single nucleus. When plotted on a histogram, we observed no relationship between the content of GFP-coilin and YFP-fibrillarin in individual Cajal bodies (Fig. 7 B). Similar results were obtained from Cajal bodies in seven other nuclei (data not shown). The data in Fig. 7 demonstrate that the amounts of YFP-fibrillarin in Cajal bodies are not correlated with the amounts of GFP-coilin, and that these different molecules might be actively partitioned between different Cajal bodies. These data from live cell imaging confirm the differences in molecular composition for Cajal bodies seen in fixed cells (see Fig. 4). Importantly, the data from live cells also show that the molecular composition of Cajal bodies can change over time.
It is possible that the splitting reactions reported above represent one mechanism for the generation of new Cajal bodies. Also, we have recorded multiple examples in which Cajal bodies appear de novo in the nucleoplasm, and in which Cajal bodies move through the nucleoplasm to join the nucleolar periphery. The time-lapse data are inconsistent with the idea that all Cajal bodies originate from the periphery of nucleoli. We do not exclude that some Cajal bodies may originate at nucleoli. However, at least in the HeLaGFP-coilin cell line, this was not observed as the major mechanism of Cajal body formation.
Differential Movements of Cajal Bodies in Living Cells
An analysis of Cajal body dynamics in living cells provided additional evidence that the smaller mini-CBs represent a distinct class of nuclear bodies. The velocities of individual Cajal bodies varied significantly during the time period of data collection (Fig. 8). Individual Cajal bodies moved at varying rates ≤0.9 μm/min. Separate analysis of mini-CBs showed that they reached higher velocities and altered their velocities over a wider range than the larger CBs (Fig. 8). We next determined the maximum velocity achieved for 14 separate CBs and mini-CBs and expressed this as the mean maximum velocity for CBs and mini-CBs. This showed that the mean maximum velocity for CBs was 0.48 (±0.08) μm/min and 0.65 (±0.13) μm/min for mini-CBs. The findings that individual mini-CBs achieve higher instantaneous velocities than CBs (Fig. 8) and that, as a group, mini-CBs have higher maximum velocities than CBs are consistent with the idea that mini-CBs and CBs may be distinct, if related, structures. The differences in Cajal body migration distance and velocity likely result from differences in interactions between Cajal bodies and other nuclear components that either constrain or promote movement.
Figure 8.
Analysis of Cajal body velocities. A plot of the change in the instantaneous velocities over a 1-h time period for a representative CB (filled diamonds) and mini-CB (open circles) from the same nucleus. Positions of Cajal bodies were determined using a three-dimensional segmentation algorithm (see Materials and Methods) and corrected for any motion of the whole nucleus. Mini-CBs achieve higher instantaneous velocities than CBs and show greater variation in their velocity over time.
Variation in GFP-Coilin Association of Cajal Bodies
The GFP-coilin and endogenous p80 coilin proteins are present in a diffuse nucleoplasmic pool and in Cajal bodies (see Fig. 3). To analyze the movement of coilin between Cajal bodies and the nucleoplasmic pool, we measured how the content of GFP-coilin in individual bodies varied with time. Fig. 9 shows the total GFP-coilin content, expressed as fluorescence, of a CB and mini-CB from the same nucleus, analyzed over a time period of 1 h. Each measurement has been corrected for changes in background nucleoplasmic fluorescence (see Materials and Methods). Frequent variations in the GFP-coilin content of individual bodies, up to eight times its initial value, occur over time periods of ≤10 min (Fig. 9; and data not shown). These changes appeared as regular smooth transitions and did not represent random fluctuations in measurements. Many examples of an increase in GFP-coilin content in one Cajal body and a simultaneous decrease in another within the same nucleus were noted (Fig. 9; and data not shown). This excludes the possibility that changes in nuclear GFP-coilin content were caused indirectly by the method of imaging and analysis, or reflected significant changes in GFP-coilin protein concentration in the nucleus. We observe no significant change in the total level of nuclear GFP-coilin during the time period of imaging. The simplest explanation for the finding that the amount of GFP-coilin in Cajal bodies can change dramatically is that GFP-coilin can transit between Cajal bodies and the nucleoplasm.
Discussion
We report here the establishment of a stable HeLa cell line (HeLaGFP-coilin) that expresses the widely used Cajal body marker protein, p80 coilin, fused to GFP. The GFP-coilin fusion protein shows identical localization and biochemical behavior to endogenous p80 coilin in this HeLa cell line and, thereby, provides a useful tool for studying Cajal body dynamics in live human cells. Analysis of 63 separate cells by time-lapse fluorescence microscopy for periods of 1.5–2.5 h showed that all Cajal bodies can move in every interphase nucleus. A range of movements was observed, including translocation through the nucleoplasm, joining together of individual Cajal bodies, and the separation of smaller bodies away from larger ones. Cajal bodies were observed moving to and from the nucleolar periphery and within the nucleolus. These data reveal that Cajal bodies in human cells can be very mobile and show an unexpectedly large range of movements that may be important for their cellular functions. This study also shows that at least two classes of Cajal bodies, which differ in size, molecular composition, and dynamic behavior, can be distinguished. Our results are reminiscent of previous ultrastructural studies of Cajal bodies that reported a variety of Cajal body sizes (Hardin et al. 1969; Hervas et al. 1980; Lafarga et al. 1983; Carmo-Fonseca et al. 1993). We refer to the larger Cajal bodies (diameter ≥0.4 μm) as CBs and the smaller bodies (diameter ≤0.2 μm) as mini-CBs. The two classes were detected in both fixed and living cells and were observed to interact through both joining and separation events.
The movement of Cajal bodies does not occur at constant velocities. Instead, we observe large fluctuations in rates of movement over time as individual bodies move through the nucleoplasm. The fastest instantaneous rates we have measured so far were up to 0.9 μm/min. The most rapid movements were only seen with the smaller mini-CBs. In contrast, the larger CBs generally moved over smaller distances and at slower rates. In at least some cases, the movement of the larger CBs appeared to be restricted within a confined nuclear volume (data not shown). This may reflect the local structure of the nucleus and/or its interactions with other nuclear components. Previous studies have shown that some Cajal bodies are associated with specific gene loci (Callan et al. 1991; Frey and Matera 1995; Smith et al. 1995; Matera 1998; Frey et al. 1999). Associations between Cajal bodies and genes for histones and the U1 and U2 snRNAs were shown to be unstable, suggesting that those Cajal bodies that associate with specific gene clusters might do so transiently and be motile (Frey and Matera 1995). Our data confirm this hypothesis. In addition, chromatin has been found to undergo passive diffusion within constrained volumes in vivo (Marshall et al. 1997; Bornfleth et al. 1999). Whether Cajal body movements are coupled to chromatin dynamics remains to be determined. It is possible that the variations in Cajal body velocities we detect reflect changes in their association with chromatin. However, interactions with chromatin might be only one of several ways Cajal body dynamics are regulated. We consider it unlikely that the complex range of directional movements and interactions of the Cajal bodies observed here could all be explained by passive processes (for example, see Fig. 5 and Fig. 6) (Phair and Misteli 2000). The specific joining and splitting events reported here (see Fig. 7 and Fig. 8) strongly suggest an active, regulated event. This may include an active translocation mechanism. However, directed movement of Cajal bodies could be the result of actively regulated binding to nuclear components combined with passive diffusion within the nucleoplasm. These issues will be addressed in future studies through a detailed quantitative analysis of Cajal body movement and the relationship between chromatin, other nuclear bodies, and Cajal body dynamics.
Our measurements of the GFP-coilin content of Cajal bodies demonstrate another dynamic aspect of Cajal bodies, namely large (up to eightfold) changes in GFP-coilin content. Our observation that the GFP-coilin content of individual Cajal bodies can increase and decrease suggests there may be a constant flux of p80 coilin between Cajal bodies and the nucleoplasm. These changes may reflect Cajal body assembly and disassembly. Alternatively, the structure of the Cajal body may stay intact, but the amounts of GFP-coilin contained in it may change. We are currently examining these issues by simultaneously measuring changes in multiple Cajal body components over time in living cells. Regardless, the evidence presented here suggests that p80 coilin exchanges between a nucleoplasmic pool and the Cajal body.
This study establishes that individual Cajal bodies in live human cells are dynamic and can undergo significant movements and rearrangements during interphase. Previously, Boudonck et al. 1999 have reported the analysis of a GFP-fusion to the plant U2 snRNP protein, B″ in tobacco BY-2 cells and Arabidopsis plants. Although the U2 snRNP particle is found in both Cajal bodies and nuclear speckles in mammalian and plant cell nuclei, in BY-2 cells the B″ protein labels Cajal bodies distinctively. This revealed movement of bodies in 70% of the 60 cells studied by time-lapse confocal fluorescence microscopy over a period of 6–12 h. Rates of movement varied from <1 to ∼10 μm/h. These rates are slower than the fastest movements of Cajal bodies in HeLa cells. However, the fastest movements detected in HeLa cells result from the smaller mini-CBs. It is not clear whether these structures were analyzed in the plant cells because Boudonck et al. 1999 refer to detecting smaller bodies that could not be analyzed due to their rapid photobleaching. In 20% of the plant cell nuclei where movement was observed, a process of “coalescence” of bodies was seen. It is likely that this process is similar to the joining of Cajal bodies to form the larger structures that we report here, although this appears to be a more frequent event in human cells (>70% nuclei). In addition, the joining or coalescence of CBs in plant cells was only observed at the nucleolar periphery, whereas in HeLa cells we also detect joining of bodies in the nucleoplasm. Also, the separation of smaller bodies from larger CBs that we observe in the HeLa cells was not reported in plant cells. Finally, the data from the plant cells show more interactions of bodies within the nucleolus and at the nucleolar periphery than we observe in the HeLa cells. This may occur because the plant cells studied had particularly large nucleoli, which occupied more of the nuclear volume than the nucleoli in the HeLa cells. It has yet to be determined whether these differences reflect fundamental differences between plant and animal cells or possibly differences between the specific cell types analyzed. It will be important to study the dynamics of Cajal bodies in many cell types and organisms to evaluate these differences. Overall, a comparison of the data from plant and animal cells indicates that Cajal bodies in both cases can be highly mobile structures. Further analyses are required to determine whether similar molecular mechanisms are involved in these movements and whether the nuclear bodies are performing the same biological functions in animal and plant cells.
Based on differences we observed here in their size, mobility, and antigen composition, it appears that the Cajal bodies may comprise at least two separate classes of nuclear structures. We also observed that the volumes of large and small Cajal bodies differed between individual nuclei. Previous analysis has shown that Cajal bodies increase in size during S and G2 phases of the cell cycle (Andrade et al. 1993). Therefore, it seems likely that the heterogeneity in Cajal body size seen in this study reflects, at least in part, differences in cell cycle stage in different cells. It will be interesting to analyze in detail how the size and interactions of Cajal bodies are affected by progression through the cell cycle.
Also, we have observed heterogeneity in the molecular composition of Cajal bodies in both fixed and living HeLa cells (see Fig. 4 and Fig. 8). This is consistent with previous observations that certain nuclear antigens, such as fibrillarin, are found to colocalize with coilin in some cell types but not in others or in a subset of Cajal bodies in the same nucleus (Raska et al. 1990; Carmo-Fonseca et al. 1991; Raska et al. 1991; Jimenez-Garcia et al. 1994; Alliegro and Alliegro 1998). The idea that Cajal bodies may represent a family of two or more related structures raises the possibility that they could also perform different functions in the nucleus. Several different models have been proposed for possible functions of Cajal bodies including roles in controlling histone and snRNA gene expression, maturation of snRNPs, assembly of macromolecular complexes, and the transport of molecules within the nucleoplasm (Bohmann et al. 1995b; Pellizzoni et al. 1998; Frey et al. 1999; Gall et al. 1999; Narayanan et al. 1999; Sleeman and Lamond 1999). If there are separate classes of Cajal bodies, then it is possible that some of these functions could be specifically carried out by one type of Cajal body. The range of Cajal bodies present may also vary either between cell types or within a given cell according to cell cycle changes or variations in metabolic activity.
Cajal bodies were first described by Ramon y Cajal (1903) as “nucleolar accessory bodies” because of their frequent association with the nucleolar periphery. The time-lapse analyses we have performed indicate that Cajal bodies can move to the nucleolar periphery from the nucleoplasm, consistent with a possible transport function. Although they do not contain rRNA, Cajal bodies do contain some nucleolar antigens, including fibrillarin and NOPP140, and may mediate interactions between the nucleolus and other nucleoplasmic structures. It is possible that both the joining and separation of Cajal bodies may also be connected with some form of transport events.
There is increasing evidence that the nucleus is a highly dynamic cellular compartment. Recent studies using GFP fusions have demonstrated the movement of specific protein molecules to and from nucleoli and speckles (Misteli et al. 1997; Misteli 2000). The data we report here further support the dynamic nature of the nucleus and show that nuclear bodies in animal cells move and interact both with themselves and with nucleoli. Analysis of stable human cell lines expressing either wild-type or mutated forms of fibrillarin fused to GFP also demonstrates movements of Cajal bodies in vivo (Snaar et al. 2000).
It is clearly very important to study the dynamic properties of nuclear bodies in order to characterize their likely functions and interactions with other nuclear components. The fact that Cajal bodies are shown to move position, join, and separate in the same nucleus at different time points means that conclusions based exclusively on single static images could provide an incomplete or misleading picture of their nuclear organization. We have recently isolated stable cell lines in which other nuclear structures apart from Cajal bodies are also labeled with marker proteins fused to fluorescent protein tags. Future studies will be aimed at using these reagents to further characterize the dynamic properties and interactions of animal cell nuclei.
Supplemental Material
Acknowledgments
The authors thank the Advanced Quantitative Light Microscopy course at the Marine Biological Laboratory, Woods Hole, MA, where some of the initial time-lapse microscopy experiments were performed. We thank members of the Lamond lab for help and support during these experiments.
M. Platani was supported by Dame Catherine Cookson and Biotechnology and Biological Sciences Research Council studentships. J.R. Swedlow is a Wellcome Trust Research Career Development Fellow. A.I. Lamond is a Wellcome Trust Principal Research Fellow.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: DIC, differential interference contrast; SIP, SMN-interacting protein; SMN, survival of motor neuron; sn, small nuclear; sno, small nucleolar; YFP, yellow fluorescent protein.
References
- Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Alliegro M.C., Alliegro M.A. Protein heterogeneity in the coiled body compartment. Exp. Cell Res. 1998;239:60–68. doi: 10.1006/excr.1997.3867. [DOI] [PubMed] [Google Scholar]
- Almeida F., Saffrich R., Ansorge W., Carmo-Fonseca M. Microinjection of anticoilin antibodies affects the structure of coiled bodies. J. Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade L.E., Chan E.K., Raska I., Peebles C.L., Roos G., Tan E.M. Human autoantibody to a novel protein of the nuclear coiled bodyimmunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade L.E.C., Tan E.M., Chan E.K.L. Immunocytochemical analysis of the coiled body in the cell-cycle and during cell proliferation. Proc. Natl. Acad. Sci. USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K., Ferreira J., Santama N., Weis K., Lamond A.I. Molecular analysis of the coiled body J. Cell Sci. Suppl. 19 1995. 107 113a [DOI] [PubMed] [Google Scholar]
- Bohmann K., Ferreira J.A., Lamond A.I. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus J. Cell Biol. 131 1995. 817 831b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornfleth H., Edelmann P., Zink D., Cremer T., Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys. J. 1999;77:2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudonck K., Dolan L., Shaw P.J. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol. Biol. Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan H.G., Gall J.G., Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Capco D.G., Wan K.M., Penman S. The nuclear matrixthree-dimensional architecture and protein composition. Cell. 1982;29:847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Pepperkok R., Sproat B.S., Ansorge W., Swanson M.S., Lamond A.I. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Ferreira J., Lamond A.I. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis—evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M.T., Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMNa link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey E.G., Penman S. Nuclear matrix proteins reflect cell type of origin in cultured human cells. Proc. Natl. Acad. Sci. USA. 1988;85:121–125. doi: 10.1073/pnas.85.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R., Matera A.G. Coiled bodies contain U7 small nuclear-RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R., Bailey A.D., Weiner A.M., Matera A.G. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Tsvetkov A., Wu Z., Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev. Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Bellini M., Wu Z., Murphy C. Assembly of the nuclear transcription and processing machineryCBs (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J.H., Spicer S.S., Greene W.B. The paranucleolar structure, accessory body of Cajal, sex chromatin, and related structures in nuclei of rat trigeminal neuronsa cytochemical and ultrastructural study. Anat. Rec. 1969;164:403–431. doi: 10.1002/ar.1091640403. [DOI] [PubMed] [Google Scholar]
- He D.C., Nickerson J.A., Penman S. Core filaments of the nuclear matrix. J. Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas J.P., Villegas J., Crespo D., Lafarga M. Coiled bodies in supraoptic nuclei of the rat hypothalamus during the postnatal period. Am. J. Anat. 1980;159:447–454. doi: 10.1002/aja.1001590408. [DOI] [PubMed] [Google Scholar]
- Isaac C., Yang Y., Meier U. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia L.F., Segura-Valdez M.L., Ochs R.L., Rothblum L.I., Hannan R., Spector D.L. NucleologenesisU3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M., Hervas J.P., Santa-Cruz M.C., Villegas J., Crespo D. The “accessory body” of Cajal in the neuronal nucleus. A light and electron microscopic approach. Anat. Embryol. 1983;166:19–30. doi: 10.1007/BF00317942. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.E. Structure and function in the nucleus. Science. 1998;2880:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Malatesta M., Zancanaro C., Martin T.E., Chan E.K., Amalric F., Luhrmann R., Vogel P.F., Fakan S. Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur. J. Cell Biol. 1994;65:82–93. [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., Sedat J.W. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Matera A.G. Of coiled bodies, gems, and salmon. J. Cell. Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- Matera A.G. Nuclear bodiesmultifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicingnuclear architecture meets nuclear function. J. Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Misteli T., Caceres J.F., Spector D.L. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Monneron A., Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J. Ultrastruct. Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Narayanan A., Speckmann W., Terns R., Terns M.P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka N., Charroux B., Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- Pettersson I., Hinterberger M., Mimori T., Gottlieb E., Steitz J.A. The structure of mammalian small nuclear ribonucleoproteins. Identification of multiple protein components reactive with anti-(U1)ribonucleoprotein and anti-Sm autoantibodies. J. Biol. Chem. 1984;259:5907–5914. [PubMed] [Google Scholar]
- Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. 1903;2:129–221. [Google Scholar]
- Raska I., Ochs R.L., Andrade L.E.C., Chan E.K.L., Burlingame R., Peebles C., Gruol D., Tan E.M. Association between the nucleolus and the coiled body. J. Struct. Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Raska I., Andrade L.E.C., Ochs R.L., Chan E.K.L., Chang C.M., Roos G., Tan E.M. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Reimer G., Pollard K.M., Penning C.A., Ochs R.L., Lischwe M.A., Busch H., Tan E.M. Monoclonal autoantibody from a (New Zealand black × New Zealand white)F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- Schardin M., Cremer T., Hager H.D., Lang M. Specific staining of human chromosomes in Chinese hamster × man hybrid cell lines demonstrates interphase chromosome territories. Hum. Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- Schul W., de Jong L., van Driel R. Nuclear neighboursthe spatial and functional organization of genes and nuclear domains. J. Cell. Biochem. 1998;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E., Lamond A.I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Sleeman J., Lyon C.E., Platani M., Kreivi J.P., Lamond A.I. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res. 1998;243:290–304. doi: 10.1006/excr.1998.4135. [DOI] [PubMed] [Google Scholar]
- Smith K.P., Carter K.C., Johnson C.V., Lawrence J.B. U2 and U1 snRNA gene loci associate with coiled bodies. J. Cell. Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Snaar S., Weismeijer K., Jochemsen A.G., Tanke H.J., Dirks R.V. Mutational analysis of fibrillarin andits mobility in living human cells. J. Cell Biol. 2000;151:653–662. doi: 10.1083/jcb.151.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L. Macromolecular domains within the cell-nucleus. Annu. Rev. Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Swedlow J.R., Sedat J.W., Agard D.A. Deconvolution in optical microscopy. In: Jansson P.A., editor. Deconvolution of Images and Spectra. Academic Press; New York: 1997. [Google Scholar]
- Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.