Abstract
In healthy mammals, maturation of B cells expressing heavy (H) chain immunoglobulin (Ig) without light (L) chain is prevented by chaperone association of the H chain in the endoplasmic reticulum. Camelids are an exception, expressing homodimeric IgGs, an antibody type that to date has not been found in mice or humans. In camelids, immunization with viral epitopes generates high affinity H chain–only antibodies, which, because of their smaller size, recognize clefts and protrusions not readily distinguished by typical antibodies. Developmental processes leading to H chain antibody expression are unknown. We show that L−/− (κ−/−λ−/−-deficient) mice, in which conventional B cell development is blocked at the immature B cell stage, produce diverse H chain–only antibodies in serum. The generation of H chain–only IgG is caused by the loss of constant (C) γ exon 1, which is accomplished by genomic alterations in CH1-circumventing chaperone association. These mutations can be attributed to errors in class switch recombination, which facilitate the generation of H chain–only Ig-secreting plasma cells. Surprisingly, transcripts with a similar deletion can be found in normal mice. Thus, naturally occurring H chain transcripts without CH1 (VHDJH-hinge-CH2-CH3) are selected for and lead to the formation of fully functional and diverse H chain–only antibodies in L−/− animals.
In the mammalian immune system DNA recombination and surface IgM expression are required for B lymphocyte development. In bone marrow B cells, D to JH rearrangement is completed at the pre–B1 cell stage. This is followed by VH to DJH rearrangement in large pre–B2 cells and VL to JL rearrangement in small pre–B2 cells, indicating sequential differentiation events (1–3). At the pre–B2 cell stage, replacement of surface-expressed surrogate L chain by κ or λ L chain initiates the process of antibody maturation, which is accompanied by cellular migration and class switching. Mature B cells undergo further selection and can differentiate into antibody-secreting plasma cells or memory B cells bearing different isotypes (IgG, IgA, or IgE). Checkpoints during the progression of these regular events ensure that only cells with productive rearrangements advance in differentiation (4). The formation of the B cell receptor (BCR) and its associated chains are regarded as essential to allowing normal B cell development (5). This has been confirmed in mice lacking the H, L, Igα, or Igβ polypeptide of the BCR (6–8).
In Tylopoda or camelids (dromedaries, camels, and llamas), a major type of Ig, composed solely of paired H chains (9), is produced in addition to conventional antibodies of paired H and L chains (10). The secreted homodimeric H chain–only antibodies found in these animals use specific VH (VHH) and γ genes, which results in a smaller than conventional H chain, lacking the constant (C) H1 domain. Interestingly, H chain antibodies are also present in some primitive fish, e.g., the new antigen receptor in the nurse shark and the specialized H chain (COS5) in ratfish (11, 12). Again, these H chain Igs lack the CH1-type domain. However, evolutionary analysis has shown that their genes emerged and evolved independently, whereas H chain genes in camelids evolved from preexisting genes used for conventional heteromeric antibodies (13). H chain antibodies can also be found in humans with H chain disease (HCD), where the H chain–only Ig has part of the VH and/or CH1 domain removed (14).
Intracellullar transport of Ig is dependent on its correct folding and assembly in the endoplasmic reticulum (ER), where a single H chain is chaperoned by noncovalent association with the H chain binding protein BiP or grp78 (15). The BiP–H chain complex is formed by virtue of the KDEL sequence at the carboxy terminus of BiP (16) and the CH1 domain of the H chain. When L chain displaces BiP, Ig can go to the cell surface or be secreted. If CH1 or part of VH is missing, L chain is no longer required to replace BiP, and the H chain can travel unhindered to the cell surface and be secreted, as seen in animals that make H chain–only antibodies and in HCD.
We report that the absence of L chain does not prevent serum antibody production in mice. Quite unexpectedly, we found antibodies in the serum of L chain–deficient mice without any further genetic manipulation. Diverse H chain–only IgG without CH1 is secreted despite compromised B cell development. We show that H chain–only IgGs are produced from transcripts lacking the CH1 exon, and we identify in some somatic cells different genomic deletions that can give rise to these transcripts. The results indicate that L chain–deficient animals could be a useful tool for the production of therapeutic H chain–only antibodies.
RESULTS
IgG expression without L chain
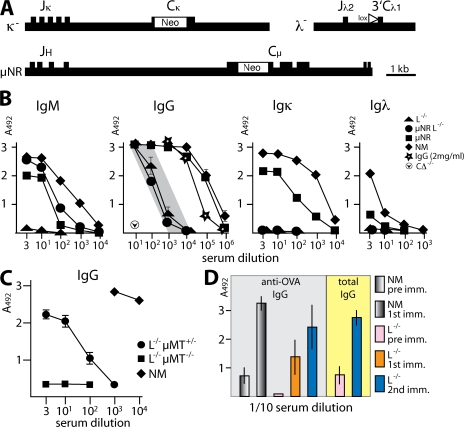
The aim of the initial project was to investigate whether mechanisms for single H chain Ig expression are naturally present in the mouse and are used if production of conventional antibodies is prevented. This was examined by using mice with silenced Igκ and Igλ L chain loci (L−/−) obtained by gene targeting (7). In the L−/− mice, all CL genes are either disrupted (Cκ and Cλ1) or removed (Cλ2, Cλ4, and Cλ3), which prevents the production of functional L chain (Fig. 1 A). Although VL to JL rearrangement is retained and low levels of some truncated transcripts can be detected, no truncated L chain products were identified in serum and cells. Unexpectedly, antibodies were found in these mice with serum IgG levels of at least 20 μg/ml and with some L−/− animals reaching levels >100 μg/ml (Fig. 1 B). This was surprising, as normal IgG cannot be secreted in the absence of L chain, and there is a block in the development of immature bone marrow B cells in these mice (7). The L−/− mice were crossed with μNR mice (17), which express truncated IgM lacking Cμ exons 1 and 2, which prevents chaperone retention of the H chain in the ER. We envisioned that this would allow μ transport to the cell surface and enable B cell differentiation to continue, resulting in higher IgG levels. As predicted, IgM without L chain was secreted in μNRL−/− mice, whereas in L−/− mice, where no provision was made for the transport of H chains to the cell surface, no IgM was detected. However, as shown in Fig. 1 B, IgG levels appear to be unaffected by the dramatic increase in B cell numbers in the μNRL−/− mice.
Figure 1.
Antibody expression in mice without L chain. (A) κ− mice carry an Igκ locus disabled by insertion of neo into Cκ (reference 46); λ− mice carry a Cre-loxP–mediated deletion of ∼120 kb encompassing all Cλ genes (reference 7); and μNR mice have neo inserted into Cμ exons 1 and 2 and express truncated μ H chains (reference 17). (B) The level of H chain Ig in serum from unimmunized mice was titrated in ELISA by binding to antibodies against IgM, IgG, Igκ, and Igλ. In L−/− (κ−/−λ−/−) mice, 20–100 μg/ml H chain IgG without L chain was produced (shaded area). μNRL−/− mice produce a similar level of IgG in addition to truncated IgM. In normal mice (NM), ∼10 mg/ml IgG was produced. Purified IgG (DB3; reference 47) served as a standard, and serum from animals with removed C genes (CΔ mice; reference 45) was used as a negative control. (C) L chain–deficient mice homozygous for μMT (L−/−μMT−/−) do not express H chain IgG in serum, whereas in L−/− (heterozygous) μMT+/− mice, concentrations were similar to those of L−/− mice. At least five mutant mice from separate litters were compared with the SD indicated when greater than ±0.2. (D) Immunizations (1st and 2nd imm.) with OVA show specific antibody responses and an increase in total IgG (pre imm. compared with after immunization). Groups of mice contained at least six animals, and SDs for IgG are shown when individual serum titrations diverged >10%.
To establish unambiguously whether surface IgM expression is essential to drive H chain IgG expression, we crossed L−/− mice with μMT animals, which carry a targeted disruption of the μ transmembrane exons (6). Serum analysis of heterozygous and homozygous littermates established that H chain–only IgG secretion is only operative when the transmembrane configuration of Cμ is unaltered (Fig. 1 C). In L−/−μMT−/− mice, no serum H chain–only IgG was present, whereas in L−/−μMT+/− mice, serum IgG levels were maintained. This suggests that early B cell differentiation events are essential to produce H chain antibody-secreting cells.
Although B cell differentiation, resulting from the presence of truncated IgM (μNRL−/−), did not increase the level of H chain–only IgG produced by L−/− mice, the amounts could be considerably increased by a conventional immunization regime (Fig. 1 D). This procedure also revealed increased titers of OVA-specific IgG after several encounters with antigen.
Size of secreted mouse H chains
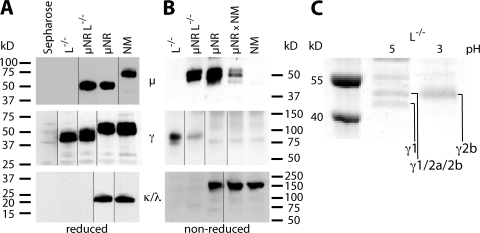
To determine the molecular mass and assembly of these novel mouse H chain–only antibodies, Western blot analysis was performed on serum Ig separated under reducing and nonreducing conditions (Fig. 2).Fig. 2 A shows that γ H chains (44–48 kD), but no μ or L chain, could be detected in L−/− serum. This new type of H chain IgG is smaller than conventional IgG but comparable in size to dromedary IgG (18). H chain–only IgG of the same reduced size is also produced in μNRL−/− mice in addition to H chain–only IgM, which is of the predicted reduced size (17). Separation under nonreducing conditions revealed covalent linkage of two γ H chains with a combined molecular mass of ∼92 kD, implying a homodimeric structure of H chain–only IgG, whereas H chain–only IgM appears to be unlinked (Fig. 2 B). Detailed analysis of gel slices by mass spectrometry, obtained after protein A adsorption of serum protein and separation by SDS-PAGE, revealed IgG2b, IgG2a, and IgG1 H chain fragments from CH2 and CH3 exons but nothing from CH1 (Fig. 2 C). In addition, sequences from five different VH gene families were identified: V7183, VGAM3.8, J558, SM7, and J606.
Figure 2.
Western blot detection of H chain–only Ig. Serum antibodies from L−/−, μNRL−/−, μNR, μNR × NM (a heterozygous μNR animal), and normal mice (NM) were purified by incubation with anti–mouse Ig coupled to sepharose, separated on Ready-Gels, and visualized with antibodies against μ, γ, and κ and λ L chain (references 27, 17). (A) Reducing conditions revealed 44–48-kD bands for γ H chains in L−/− mice and no μ H chain or L chain (κ and λ). μNRL−/− mice showed the same size γ bands in addition to the μ-specific band characteristic of the μNR background (reference 17). (B) Under nonreducing conditions, γ H chains from L−/− and μNRL−/− mice associate as dimers of 88–96 kD. Truncated IgM bands, only found in μNRL−/− and μNR mice, are largely monomeric (reference 17). Pentameric IgM (∼900 kD) does not enter the gel, and the strong signal of conventional IgG above 150 kD is not shown for μNR, μNR × NM, and NM serum. Antibody-coupled sepharose served as a negative control. Black lines indicate where the original gels were spliced. (C) For isotype identification of H chain Ig, serum antibodies from L−/− mice were bound to protein A, eluted at pH 5 and 3, and size separated on SDS-PAGE. γ1, γ2b, and a mixture of γ1/γ2a/γ2b were identified by mass spectrometry in the bands indicated after trypsin digest. Individual isotypes were identified by between five and nine fragments each, with sequences corresponding to hinge, CH2, and CH3 exons but not CH1. For VH sequences, framework and CDR regions were identified for genes from the following families: VH7183 (EVQLVESGGDLVKPGGSLK, NTLYLQMSSLK, LVESGGGLVK, NNLYLQMSSLK, and EVQLVESGGGLVKPGGSLK), VGAM3.8 and/or J558 (ASGYTFTDYSMHWVK), J558 (EVQLQQSGPELVKPGASVK), J558 and/or SM7 (QSGAELVRPGASVK), SM7 (EVQLQPSGAELVKPGASVK and LSCTASGFNIK), and J606 (LLESGGGLVQPGR). Molecular mass size standards are shown.
L chain–independent B cell development
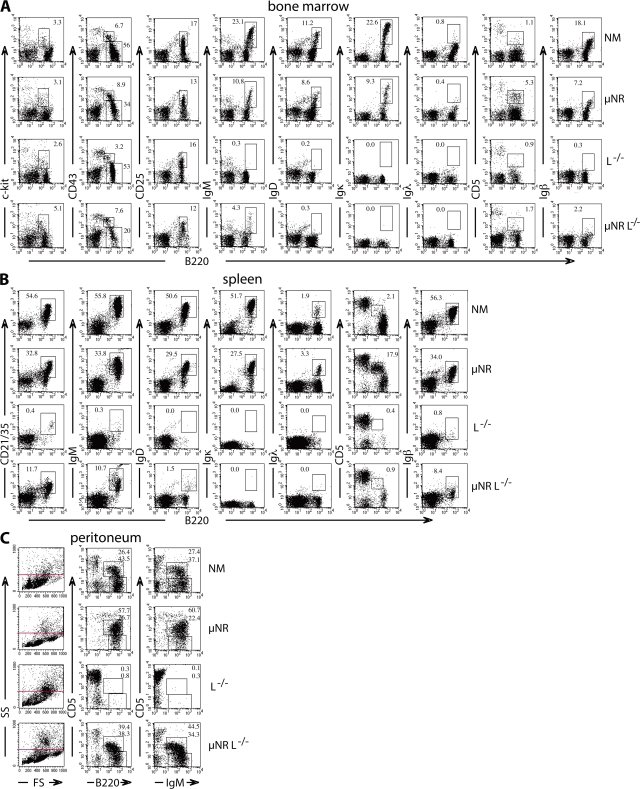
Identification of substantial amounts of diverse H chain–only Ig in the serum of mice lacking L chains prompted extensive analysis of B cell differentiation events using flow cytometry (Fig. 3). Analysis of bone marrow cells from L−/− and μNRL−/− compared with normal and μNR mice showed that developmental progression up to the pre–B1 cell stage, identified by staining for B220 in combination with c-kit, CD43, or CD25, is largely sustained (Fig. 3 A). IgM expression without conventional L chain is not maintained in L−/− mice, whereas truncated IgM in μNRL−/− mice reaches the cell surface, but at a decreased level compared with μNR mice. Similarly IgD is not, or very poorly, expressed on the cell surface without κ or λ L chains. H chain truncation in μNR mice leads to a substantial increase in CD5+ B220+ cells, identified as B1a lymphocytes (17), which is not seen in L−/− mice. Although the early stages of pre–B cell development occur without L chain, B cells expressing solely H chain–only antibodies in L−/− mice cannot be unambiguously identified by cell-surface staining. However, RT-PCR did yield a J/hinge to Cγ membrane sequence from B220+ spleen cells (unpublished data), but we have not yet cloned a complete product from VH to the membrane exon lacking CH1. In contrast, μNRL−/− mice retain cells expressing a BCR without L chain, probably in association with Igβ.
Figure 3.
Generation and maintenance of small numbers of mature B cells in L−/− mice despite a developmental block at the pre–B2 cell to immature transition stage. Flow cytometry analysis of (A) bone marrow, (B) spleen, and (C) peritoneal cells from normal mice (NM), μNR, L−/−, and μNRL−/− mice using antibodies against the following lymphocyte surface markers: c-kit, CD43, CD25, IgM, IgD, Igκ, Igλ, CD5, Igβ, and CD21/35. The profiles are representative for results from at least five different animals each using established lymphocyte gate parameters by plotting FS against SS (reference 20). Numbers represent the percentages of cells in the gates shown.
The cells in μNRL−/− mice that have acquired the expression of an H chain–only BCR may overcome the block in conventional B cell differentiation and be released into the periphery as mature B cells. Proliferation of such cells may explain the distinct B220+CD21/35+ B cell population of splenic lymphocytes in L−/− mice (Fig. 3 B), which may express IgM but little or no IgD. The small distinct population of B220+Igβ+ cells (0.8%) in L−/− mice, which is much increased in μNRL−/− mice (8.4%), may also suggest that mature cells can proliferate and maintain a conventional surface marker profile even without L chain. Further analysis of peritoneal cells (Fig. 3 C) suggested an increase in larger or differently shaped cells not contained in the conventional lymphocyte gate (www.flowjo.com) (19, 20) but evident when plotting forward scatter (FS) against side scatter (SS) to visualize size and shape distribution. Increases in cell size, albeit much less pronounced, are also seen in bone marrow and spleen cell stainings (unpublished data). Interestingly, analysis of cells in the conventional lymphocyte gate showed that, in μNRL−/− mice, the lack of L chain does not appear to affect the generation of CD5+ peritoneal B cells, which are very low in L−/− mice. A reason may be that μNRL−/− mice, despite a lack of L chain, are similar to μNR mice, which assemble a truncated surface receptor unresponsive to stimulation (17).
H chain transcripts lacking CH1 are generated in L−/− and normal mice
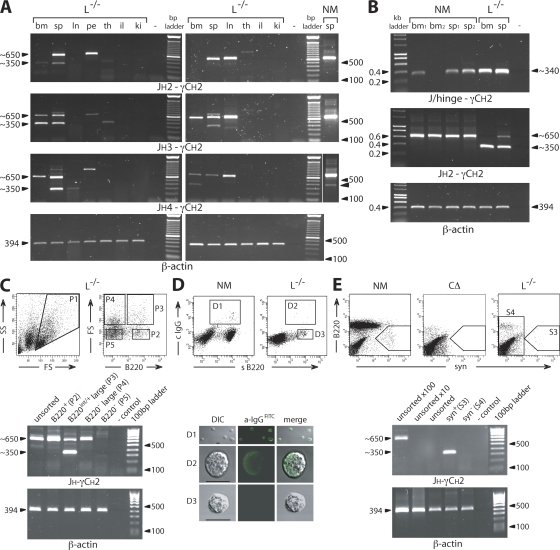
The production of γ H chain transcripts in different tissues from L−/− mice was assessed by RT-PCR using JH to γCH2 amplifications, which in normal animals produces an ∼650-bp band in lymphoid tissue (Fig. 4). In L chain–deficient mice, a prominent novel band of ∼350 bp appears in bone marrow, spleen, and lymph nodes, indicating that γ H chain transcripts of reduced size are generated in these lymphoid organs (Fig. 4 A). The slight variations in product size are caused by the length of the individual J segment and/or Cγ hinge exons used. All JH segments, except JH1, have been readily identified. JH1 amplification did yield bands, but some cross reactivity occurs between the different JH primers, and all sequenced products have thus far not identified this J segment (primers are listed in Table S1, available at http://www.jem.org/cgi/content/full/jem.20071155/DC1). VH usage in the shortened γ H chain transcripts from L−/− mice was determined by RT-PCR with VH-specific primers or linear amplification of complementary DNA (cDNA) ends, followed by cloning and sequencing. The products identified were unusually spliced, linking VHDJH to hinge or CH2, and all lacked the CH1 exon in the Cγ gene (Table I). The V domains showed diverse rearrangements of VH, D, and JH segments, including mutational alterations in VH and nonencoded additions at the VH to D and D to JH junctions (Table II and Fig. S1). The loss of CH1 agrees with the lower molecular mass H chain protein found in serum and the absence of this sequence in mass spectroscopic analysis (Fig. 2). In addition to the lower-size H chain band, a full-size product with CH1 is usually amplified from lymphocyte-containing tissues of L−/− mice (unpublished data).
Figure 4.
Identification of cells that generate H chain products. (A) RT-PCR amplification from JH2, JH3, or JH4 to γCH2 using RNA prepared from total bone marrow (bm), spleen (sp), lymph nodes (ln), peritoneum (pe), thymus (th), ileum (il), and kidney (ki) cells from two L−/− mice and spleen from a normal mouse (NM). γ H chain bands of reduced size (∼350 bp; arrowhead indicated in NM) are present in lymphoid tissue, sometimes accompanied by the full-size product (∼650 bp). β-Actin served as a reference (25 cycles). The thin white line indicates where the original gel was spliced. (B) RT-PCR amplification of bone marrow (bm) and spleen (sp) from normal and L−/− mice using a J/hinge oligo, specific for J to hinge joins that lack CH1 (JH1, JH2, or JH4 and γ2a or γ2b), in combination with the γCH2c oligo. In comparative control reactions, JH2 to γCH2a and β-actin (21 cycles) was amplified. (C) For the analysis of spleen cells by FACS and RT-PCR from L−/− mice, the lymphocyte gate established by FS and SS was set to include large cells (P1). These cells were collected (P2–P4) according to their staining profiles for B220. Large B220+ cells (P3) show a γ H chain RT-PCR band, from JH4 to γCH2, of reduced size (∼350 bp) lacking CH1, whereas other cell fractions contain a normal-size H chain transcript (∼650 bp). PCR reactions were normalized using β-actin. (D) Surface staining for B220 and cytoplasmic staining for IgG showed in confocal images that H chain antibody–producing B cells are of larger size (D2). DIC, differential interference contrast. Bars, 10 μm. (E) Surface staining for syndecan (syn; CD138) identified a population (S3) only expressing H chain transcripts without CH1 in L−/− mice. Syn+ cells from NM, which are lacking in CΔ mice, established the gate for the cell sort. Normalized RT-PCR reactions (32 cycles) were performed with the sensitivity and specificity being verified by increased levels of unsorted spleen cells (10× and 100×). Control reactions without DNA (−) are indicated. The data are representations using different mice in at least three independent experiments giving very similar results.
Table I.
H chain transcripts from L−/− spleen cell populations obtained by RT-PCR and/or cloning
| VH | D | JH | CH | |
|---|---|---|---|---|
| 028 | VH10 | SP2.2 | J4 | γ2b (no CH1) |
| 029 | 7183 | FL16.2 | J3 | γ2b (no CH1) |
| 030 | J558 | FL16.1 | J4 | γ2a (no CH1) |
| 129 | VH10 | SP2.2 | J4 | γ2b (no CH1) |
| 132 | VGAM3.8 | FL16.1 | J2 | γ2b (no CH1, no hinge) |
| 133 | 7183 | SP2.2 | J4 | γ3 (no CH1) |
| 135 | J606 | FL16.1 | J4 | γ2a (no CH1) |
| 208 | SM7 | FL16.1 | J2 | γ2b (no CH1) |
| 213 | J558 | ST4 | J3 | γ2a (no CH1) |
Numbers (left column) refer to the full sequences in Fig. S1, with 028 to 129 from unsorted spleen cells and 132 to 213 from syn+ spleen cells.
Table II.
Junctional diversity of L−/− VH sequencesa
| 3′ VH | N1 | D | N2 | JH | Hinge | 5′ CH2 |
|---|---|---|---|---|---|---|
| 028b TGTGTGAGACA | CTACTATGATTACGAC | GGG | TATGCTATGGACTACTGG // TCCTCAG | AGCCC // CCCAG |
CTCCTAACCTC | |
| 029c TGTGCAAGAG | CGCCGGGGGG | CTACGGCTAC | GTA | TGG // TCTGTAG | AGCCC // CCCAG |
CTCCTAACCTC |
| 030 TGTGC | CCGAAG | CGGT | TTTA | ACTGG // TCCTCAG | AGCCC // CCCAG |
CACCTAACCTC |
| 129 TGTGTGAGACA | T | TACTATGATTACG | GGGGG | TATGCTATGGACTACTGG // TCCTCAG | AGCCC // CCCAG |
CTCCTAACCTC |
| 132 TGTGCAAGA | AGGGGAT | TTACTACGGTGATACCTAC | GA | ACTTTGACTACTGG // TCCTCAG | CTCCTAACCTC | |
| 133 TGTGCAAGACA | TG | TCTACTTTGATTACG | GT | TATGCGACGGACTACTGG // TCCTCAG | AGCCT // CCCAC |
CTGGTAACATC |
| 135 TGTACCAGG | GGAGGTA | AAGGA | ATGGACTACTGG // TCCTCAG | AGCCC // CCCAG |
CACCTAACCTC | |
| 208 TGTAATGCA | GGG | GGTGGTAACTAC | GTGGGGGG | CTTTGACTACTGG // TCCTCAG | AGCCC // CCCAG |
CTCCTAACCTC |
| 213 TGTGCAAGA | AGGGGAG | CAGCTCGG | C | CTTACTGG // TCTGCAG | AGCCC // CCCAG |
CACCTAACCTC |
Mutational differences not found in corresponding germline gene segments from 129, BALB/c, or C57BL/6 mouse strains are underlined.
Clone details are shown in Table I.
Corresponding genomic sequence identified.
In some JH to γCH2 amplifications, the normal mouse spleen cDNA control gave a faint ∼350-bp band (Fig. 4 A, right, arrowhead), which on sequencing was found to lack CH1 (not depicted). This smaller band is frequently obscured because of amplification of an abundance of normal-size products. However, the presence of this band implies that normal mice can generate transcripts, which could produce H chain–only antibodies. To investigate whether H chain transcripts lacking γCH1 are regularly produced in normal mice, we designed oligonucleotides that would only recognize splice products in which a JH segment joins a γ2a or γ2b hinge exon. Surprisingly, in normal mice ∼340-bp JH hinge transcripts are readily found in the spleen and frequently, but not always, found in bone marrow (Fig. 4 B). Sequence analyses revealed a predicted functional product without γCH1 (unpublished data).
Plasma cells produce H chain IgG
Flow cytometry using established separation parameters (e.g., scatter gating) did not reveal any obvious candidates or distinct B cell populations that showed enrichment for the truncated transcripts. Therefore, the lymphocyte gate was extended to include larger or differently shaped cells in the sorting process (20, 21), as antibody production and, conceivably, H chain Ig secretion could be linked to an increase in cell size (Fig. 4, C–E). This, we hypothesized, would clarify whether cells, normally excluded from the conventional lymphocyte gate, would produce a single-size or both a shorter- and normal-length H chain transcript. Gated spleen cells from L−/− mice (P1) were separated into B220-dull large (P4) or average (P5) and B220int/+ large (P3) or average (P2) populations and analyzed by RT-PCR (Fig. 4 C). Diverse H chain products of ∼350 bp were obtained solely from the large B220 intermediate cell population P3, which on sequencing lacked γCH1. Other populations, except small B220− cells, produced the conventional ∼650-bp band. The analysis suggested that only a distinct spleen cell population, large B220int/+ cells, produces H chain–only Ig, which may be accompanied by a full-size product. Cytoplasmic staining (Fig. 4 D) showed that large B220int/+ cells from L−/− mice (D2) are indeed IgG+. The small B220+ cells (Fig. 4 C, D3 = P2) lacking the indicative smaller H chain band but producing full-length transcripts, may either contain reduced amounts or incorrectly folded γ H chain products poorly recognized by anti-IgG.
To understand whether IgG+ B cells from L−/− mice bear the features of conventional antibody secretors and, thus, are the product of normal lymphocyte differentiation events, we performed stainings for syndecan (CD138), which identifies plasma cells, in combination with B220 (22). Syndecan-positive cells (S3) in Fig. 4 E show a unique RT-PCR band characteristic for VHDJH-hinge-CH2-CH3 products, as confirmed by cloning and sequencing (Table II and Fig. S1).
Allelic exclusion and VH gene selection is maintained
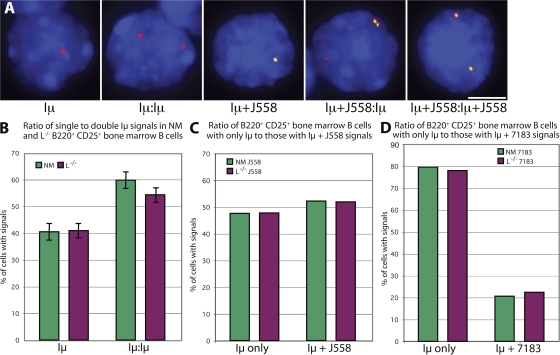
Encouraged by the diversity of the H chain antibodies found, we investigated whether activation of the IgH locus and diversity of VH gene usage is equally operative in L−/− mice compared with normal animals (23, 24). To detect individual IgH alleles, we used RNA–fluorescence in situ hybridization (FISH) with a probe, Iμ, that establishes locus activity (Fig. 5 A). Iμ is a noncoding RNA transcript originating from the IgH intronic enhancer, immediately downstream of the JH genes. It is expressed throughout B cell development and is used as a marker of an actively transcribing allele. In B220+ CD25+ pre–B2 cells from normal mice, a 40:60 ratio (percentages) of detection of Iμ transcripts from one or both IgH alleles, respectively, is observed after VHDJH recombination (23). The 40% of cells with a single Iμ signal represents the proportion of cells in which productive VHDJH recombination has silenced the second, DJH-rearranged allele by allelic exclusion, resulting in loss of Iμ transcription. The 60% of cells with Iμ signals on both alleles represents cells in which nonproductive VHDJH rearrangement on the first allele is followed by productive rearrangement on the second allele, as well as transcription of both types of VHDJH-rearranged alleles. If allelic exclusion were impaired, the ratio would be expected to change to include more cells with double signals. However, in B220+ CD25+ pre–B2 cells from L−/− mice, similar ratios of single to double Iμ signals were observed (Fig. 5 B), suggesting that allelic exclusion of the IgH locus is maintained in these mice. In addition, detection of proportions of VHDJH-rearranged transcripts corresponding to the J558 (Fig. 5 C) or the 7183 (Fig. 5 D) VH gene families on individual alleles were very similar between normal and L−/− mice, indicating that a normal, diverse range of VH genes is used.
Figure 5.
RNA-FISH to assess the transcriptional activity of the H chain alleles and their VH gene usage. Detection with an Iμ probe indicated whether one or both of the IgH loci was actively transcribing, and detection with a J558 or, separately, a 7183 probe revealed the VH gene usage of VHDJH-rearranged alleles. Cells from normal mice (NM) and L−/− mice were analyzed in parallel. (A) Representative signal combinations detected for Iμ (red) and J558 (green) transcripts in sorted B220+ CD25+ L−/− bone marrow cells. Bar, 5 μm. (B–D) Comparison of signal ratios in sorted B220+ CD25+ bone marrow cells from normal and L−/− mice stained for (B) Iμ, (C) Iμ and J558, and (D) Iμ and 7183 transcripts. SD was calculated from four separate experiments, whereas representative plots (C and D) were repeated at least once with similar results.
Acquired genomic alterations of CH1 accomplish H chain–only expression
RT-PCR and sequence analysis of smaller-size H chain bands identified a lack of γCH1 or, in a few cases, both the γCH1 and γ hinge exons (Table II). One way to identify mutations leading to H chain–only antibodies is the derivation of hybridomas. However, despite numerous attempts, we could not obtain IgG-producing hybridomas from L chain–deficient mice, possibly because of the small number of B cells able to fuse. Another approach to gain access to cells expressing IgG is by sorting for syndecan-positive cells. To enrich this starting material with DNA from cells expressing IgG, we set up an assay to amplify switched γ regions. A long-range PCR with a JH and a γCH2 primer, followed by a second nested reaction from 3′ Eμ to γCH2d (Fig. 6, A and B), gave rise to bands whose sizes were consistent with those of switched γ regions. These switched γ regions could be amplified from normal or L chain–deficient DNA from sorted syndecan-positive spleen cells but not from (germline) embryonic stem (ES) cell DNA. Cloning and sequencing of nested PCR products identified conventional exon and intron sequences regarded as functional (not depicted), and shorter sequences with deletions in and around γCH1 (Fig. 6 C and Fig. S1).
Figure 6.
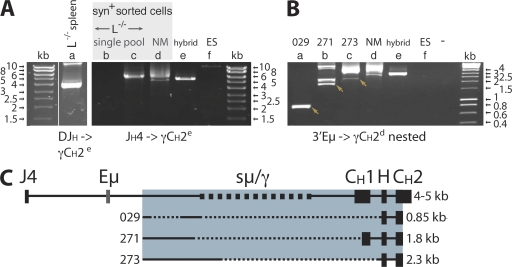
Long-range PCR identified class switch deletions in CH1. DNA preparations from one total spleen and sorted syn+ spleen cells from four L−/− mice were analyzed. (A) PCR amplifications (lanes a–f) from DJH to γCH2e (40 cycles), using a primer (VDJ029) based on the H chain sequences obtained by RT-PCR (left) or from JH4long to γCH2e (20 cycles; right). The thin white line indicates where the original gel was spliced. In the reactions, cell aliquots from one (single) and three (pool) L−/− mice were used. (B) Nested PCR (28 cycles) of first-round products (lanes a–f) from Eμ to γCH2d, with cloned products indicated by arrows. Controls were a γ2a hybridoma (hybrid), ES cell DNA, and amplification without DNA (−). (C) Map of the amplified genomic region from JH4 to Cγ exons CH1, hinge (H), and CH2. Cloning and sequencing of PCR products showed deletions of large parts of the switch region and some or all of CH1. Clone numbers (left) and sizes (right), from 3′ Eμ to γCH2d, are indicated, and sequences are compiled in Fig. S1.
To establish unambiguously whether transcripts that lack CH1 are the result of genomic deletions, we derived forward primers from their D to JH junction sequence. Successful amplification and cloning from a rearranged VH of the 7183 family identified a large deletion removing the μ/γ switch region and CH1 of Cγ2b concluding 107 nucleotides 5′ of the hinge exon (clone 029; Table II; Fig. 6; and Fig. S1). As the deletions that render γCH1 dysfunctional remove a large part of the adjacent switch region, it is possible that the DNA lesions occur during switch recombination (25), leading to alterations in Cγ that permit H chain–only antibody expression.
DISCUSSION
In L chain–deficient mice, B cell development is arrested at the pre–B2 to immature B cell stage in the bone marrow (7). At this transition stage, IgM, comprising a μ H chain covalently linked with a κ or λ L chain in dimeric configuration, should be expressed on the cell surface associated with the coreceptor chains Igα/β. Developmental progression of a compromised BCR lacking any of these chains is normally blocked (6–8, 26), so the finding of IgG in the serum of L chain–deficient mice came as a surprise. Analysis by Western blotting and mass spectrometry indicated that these proteins were lacking the CH1 domain. This was confirmed by RT-PCR, which identified short γ transcripts lacking CH1 (or, in some instances, CH1 and hinge) in the lymphoid organs of L chain–deficient mice. These findings are in agreement with reports for transgenic mice that express H chain–only Ig, where the loss of CH1 appears to be essential (27–29). The shorter nascent-translated H chain cannot form a complex with the H chain binding protein as it lacks the association sites in CH1 (15, 30). This would result in unhindered transport through the ER, allowing surface deposition as well as H chain secretion. The stability of such H chain–only Ig is remarkable, and it can be argued that the lack of CH1 and the loss of chaperone association may prevent degradation of a basically incomplete Ig. Sorting experiments indicated that the main sources of short transcripts were large, syndecan-positive cells (i.e., plasma cells), and thus, the product of normal lymphocyte differentiation, whereas conventional transcripts were abundant in a B220high cell population. However, these results raised the question of how the protein deletions occurred and how antibody-producing cells could be generated in the absence of noticeable BCR-expressing B cells.
Different mechanisms, such as alternative splicing, splice-site mutations, or exon deletions, can lead to exon removal, all of which have been found for Ig genes (31, 32). If expression of H chain IgG were controlled at the transcriptional stage (e.g., by selection of splice products leading to polypeptides, which could be released from the cell), then the rearranged H chain gene should be unaltered. To look for the existence of somatic mutations, we sorted syndecan-positive cells, which are enriched for cells producing transcripts lacking CH1, and extracted the DNA. Long-range DNA-PCR analyses using 5′ primers in the JH and Eμ region and reverse primers in the γCH2 exon ensured that only switched γ genes were amplified. With this approach, we were able to clone three different genomic Cγ deletions in which most or all of the CH1 and the μ/γ switch regions were removed but the hinge exon and Eμ intron enhancer downstream of JH were left intact. With these modifications, transcription levels and splicing from a rearranged VHDJH to the hinge exon should be maintained, producing a truncated protein. A putative mechanism for CH1 deletion suggested by our sequence data is an error during the class-switch process. The switch region upstream of each CH gene is highly repetitive, several kilobases in length, and accommodates repairs to DNA lesions such as double-stranded breaks. The recombination itself, which removes Cμ and juxtaposes the rearranged VHDJH close to a downstream CH gene, occurs between nonhomologous sequences without any consensus motif defining precisely the donor and acceptor breakpoints (33). It is possible that imprecise switching removes all or part of CH1, which would allow Ig surface expression.
However, switching (and presumably faulty switching) occurs from mature IgM-expressing lymphocytes, which are difficult to identify in the spleen of L−/− mice, occurring as a small population of B cells expressing high levels of B220. It is possible that failing to become a mature IgM-expressing B cell initiates early class switching, which may explain why serum IgM is absent in L−/− mice and camelids do not appear to produce H chain–only IgM or VHHDJH-Cμ transcripts (34, 35). This possibility is strengthened by the fact that we can identify in the spleen full-length γ transcripts that are much more abundant and diverse than short transcripts. Presumably, the switch from μ to γ occurs in a large number of cells, but in most cases, this does not allow production of a H chain that can be transported to the cell surface without L chain. Only when faulty switching gives rise to DNA sequences encoding transcripts lacking CH1 would the B cell be selected for survival. The absence of H chain IgG in L−/−μMT−/− mice suggests that a pre-BCR–dependent proliferative stage is required, probably to produce the number of cells required to obtain these specific aberrant switching events. A knock-in gene encoding a mutant μ chain (μNR) (17) was introduced into the genome of L−/− mice, which resulted in expression of a truncated μ H chain on the cell surface in the absence of L chain. This ensured cell survival but did not result in an increased IgG level, which could be seen as puzzling, as μNR mice expressing L chain have a normal, high level of IgG. However, the pre-BCR–dependent proliferative stage is slightly impaired in the μNR mice. Also, in μNR and μNRL−/− mice, the expression of truncated μ significantly increases the number of a particular CD5+ lymphocyte subset, (CD5+ B1a cells), which rarely switch (36, 37). In addition to the deletion of the CH1 region, we have identified CH1 point mutations in some of the switched γ regions from L chain–deficient mice. However, none of them corresponds to a typical splice site mutation, so it is not clear if these changes affect splicing. Further analysis is required to determine whether they cause exon skipping by altering exon recognition by cis-elements involved in the splicing process (38). If this is the case, L chain–deficient mice might provide useful information on the mechanism controlling splice site usage.
Evidence that secretion of H chain–only Ig in L chain–deficient mice is antigen dependent comes from the increased titers of specific antibody after immunization. Also, several functional VH sequences were found, which harbored mutations that can be attributed to somatic hypermutation (39). We investigated whether specific alterations compensate for the lack of L chain association, as found in adapted camelid VHH exons (34). From the alignment of VHH sequences with the V regions of mouse H chain antibodies, it was found that this was not the case. None of the hallmark amino acids found in VH to VHH substitutions in framework 2 (Val37Phe, Gly44Glu, Leu45Arg, and Trp47Gly) (40, 41) were seen, and in one case, the reverse was found with an Arg to Leu change at position 45 (Kabat numbering). Some camelid H chain Igs bear a long CDR3, and it has been suggested that CDR3s encompassing a more extensive D segment and/or substantial N sequence additions may be an advantage to compensate for the smaller antigen-binding area of H chain Ig compared with the conventional H–L Ig (34, 35, 40, 41). A longer CDR3 was not found with the mouse H chain antibodies, but it should be noted that antigen-specific dromedary H chain antibodies with shorter CDR3 (seven amino acids) have also been identified (42).
Expression of H chain–only IgG in L−/− mice appears to differ from human HCD. HCD are monoclonal B cell lymphoproliferations secreting mutant H chain not associated with L chain. It has been hypothesized that these proliferations are caused by expansion of cells that express altered H chains because they have previously lost the ability to produce L chains (although there are cases in which free L chains are produced by tumor cells) (43). The results we have obtained show that the absence of L chains leads to the selection of cells producing mutant H chain lacking CH1 when normal competing B cells are absent. However, unlike in HCD, it appears from the Western blot analysis and the sequence data from L−/− mice that gross alterations in the VH regions are not present in the majority of the cells. In addition, we have not observed the lymphoproliferations that occur in HCD in L chain–deficient mice.
The modifications observed in L chain–deficient mice produce a domain configuration comparable to those that have been identified in both camelids and cartilaginous fish, representing in the former a relatively recent adaptation (13) and in the latter a possible remnant of the primordial antibody structure that preceded the heterodimeric association of H and L chains (11). In lower vertebrates, H chain dimers have been recognized that lack a classical CH1 domain, which is important to provide the cysteine residue that forms the disulphide linkage with the L chain (11, 12). The evolution of Ig domains, as well as their multiplication and diversification to permit functional interaction, vividly illustrates the ongoing selective pressure on antibody genes. Specific alterations (in the case of the L−/− mice, the removal of CH1) prevented Bip association without affecting H chain dimerization, an essential requirement to secure the antibody structure for immune protection (44).
In conclusion, our study has shown that in mouse B cells the removal of CH1 permits the cellular release of fully functional IgG antibodies without L chain and the development of a diverse H chain–only antibody repertoire. Mouse VH genes can be expressed as H chain antibodies without acquiring VHH-specific changes and maintain their inherited sequence characteristics and lengths (41). A B cell repertoire with somatic hypermutation would be of great importance for the production of H chain–only monoclonal antibodies in mice. The problems with generating hybridomas from L chain–deficient mice using whole organs (e.g., spleen) may be caused by the small numbers of activated lymphoblasts present. This could be overcome by increasing the cell population of H chain–only Ig-producing progenitors. Because somatic alterations leading to CH1 deletion, caused by its very low frequency, are strong limiting steps in H chain–only IgG production, deleting a γ CH1 exon in the germline of L chain–deficient mice should allow H chain–only monoclonal antibodies with defined specificities to be readily produced.
MATERIALS AND METHODS
Mice.
The derivation of Igκ- and Igλ-deficient (L−/−, with Cκ disrupted by neomycin gene (neo) insertion and an ∼120-kb region from Cλ2 to Cλ1 removed by targeted integration and Cre-loxP deletion), Cμ truncation (μNR, lacking Cμ1 and Cμ2 by targeted integration of neo), C deletion (CΔ, lacking an ∼200-kb region from Cμ to 3′of Cα removed by targeted integration and Cre-loxP deletion), and μMT mice has been previously described (7, 17, 45, 6). L−/− and μNR animals were crossbred to homozygosity. 12–28-wk-old mice were analyzed and compared with littermates or age-matched controls. Animals were housed in the Babraham barrier facility, and procedures were performed under project license PPL 80/1872, which was approved by the UK Home Office.
ELISA and immunization.
Serum antibodies were analyzed as previously described (46) on Falcon plates coated with 10 μg/ml of anti–mouse IgM, Igκ (Sigma-Aldrich), or IgG (The Binding Site). Biotinylated detection antibodies were anti–mouse IgM (Sigma-Aldrich), IgG (GE Healthcare), Igκ (Invitrogen), and Igλ (BD Biosciences). To determine the antibody concentration, purified IgG (DB3) was used, as previously described (47). Immunizations were performed with 100 μg OVA in CFA (s.c.) and, subsequently, 50 μg OVA in FA (i.p.) 30 and 14 d later, respectively. Ig secretion was identified by ELISA on plates coated with 10 μg/ml OVA (Sigma-Aldrich).
Western blot analysis.
Serum Ig was incubated with anti–mouse μ, γ, and α H chain–specific Ig–coupled sepharose, separated on Ready-Gels (Bio-Rad Laboratories) and transferred to nitrocellulose membranes, as previously described (27). Filters were incubated with biotinylated anti–mouse μ (Sigma-Aldrich), γ (GE Healthcare), and κ (Invitrogen) and λ (BD Biosciences) L chain–specific Ig. This was followed by incubation with streptavidin-biotinylated horseradish peroxidase solution (GE Healthcare) and visualization of bands using a chemiluminescent substrate (SuperSignal West Pico; Thermo Fisher Scientific). Protein molecular mass standards were supplied by Bio-Rad Laboratories and Fermentas.
Flow cytometry.
Bone marrow, spleen, and peritoneal cell suspensions were prepared, and multicolor analyses were performed on a flow cytometer (FACSCalibur; BD Biosciences). Cells were stained, in combination, with labeled anti–mouse Ig recognizing CD45R (either PE, or allophycocyanin or biotin [BIO] conjugated; B220), PE-conjugated anti–c-kit (CD117), BIO-conjugated anti-CD43, BIO-conjugated anti-CD25, FITC-conjugated anti-IgD, PE-conjugated anti-Igκ, FITC-conjugated anti-Igλ, PE-conjugated anti-CD5 (Ly-1), FITC-conjugated anti-CD79b (Igβ), FITC-conjugated anti–mouse CD21/35 (all from BD Biosciences), and FITC-conjugated anti-IgM (Invitrogen). Reactions with BIO-conjugated antibodies were subsequently incubated with TRI-COLOR–conjugated streptavidin (Caltag). CellQuest software (BD Biosciences) was used for the analysis.
For sorting on a FACSAria (BD Biosciences), spleen cells were stained with PE-conjugated anti-CD45R and, separately, FITC-conjugated anti-CD45R and BIO-conjugated anti-CD138 (syndecan-1; BD Biosciences), followed by incubation with PE-Cy5.5–conjugated streptavidin (eBioscience). For the analysis of cytoplasmic Ig, spleen cells stained for surface CD45R were incubated with Fc-specific FITC-conjugated anti-IgG (Sigma-Aldrich) using a fix and perm cell permeabilization kit (Caltag). IgG-positive cells were collected and viewed on a confocal microscope (LSM 510 META; Carl Zeiss, Inc.), and images were obtained using LSM 3.2 software (Carl Zeiss, Inc.).
RT-PCR analysis.
RNA was isolated from tissue or sorted cells using TRIzol (Invitrogen) and reverse transcribed at 42°C with Omniscript reverse transcriptase (QIAGEN). PCR reactions with JH and γCH2a primers were set up using KOD Hot Start DNA polymerase (Novagen) at a final MgSO4 concentration of 0.8 mM. The cycling conditions were 94°C for 2 min, and 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 15 s, followed by 72°C for 10 min. Amplification with VH-specific primers was as described, but with a 52°C annealing temperature for Vgeneric, V3609, and VS107/J606. RT-PCR products were either sequenced directly or cloned by adding a 3′ A overhang and using a TA cloning kit (Invitrogen). β-Actin PCR was performed as described, but with an annealing temperature of 61°C. The J/hinge to γCH2c PCR was set up as described, but with a touchdown program: 94°C for 2 min, and 21 cycles of 94°C for 15 s, 69°C (−0.33°C per cycle) for 30 s, and 72°C for 10 s, followed by 25 cycles of 94°C for 15 s, 62°C for 30 s, and 72°C for 10 s. For linear amplification of cDNA ends, first-strand cDNA synthesis and primer extension was performed on 1 μg of total RNA, as described (SMART mRNA amplification kit; BD Biosciences). Double-stranded cDNA was purified using the Wizard SV Gel and PCR Clean-Up System (Promega), and linear amplification of the 5′ ends was performed using KOD and nested γCH2 primers (γCH2b and γCH2a), as previously described (48). The product from the fourth round of amplification was separated by agarose gel electrophoresis, and bands between 700 and 1,200 bp were excised and purified using the Wizard kit. The purified fragments were cloned as described and sequenced. All primer sequences used are listed in Table S1. Agarose gels were run with size markers in kilobases (Hyperladder 1; Bioline) and/or basepairs (100 bp per DNA ladder; Invitrogen).
Genomic DNA analysis.
Genomic DNA was prepared as previously described (27), with the addition of linear acrylamide to aid the precipitation of small amounts of DNA from the sorted cells. Long PCR was performed with Platinum PCR Supermix High Fidelity (Invitrogen). Reactions were set up with DNA from 1–2 × 103 sorted cells (∼10 ng), equivalent amounts of ES cell or hybridoma DNA, and 100 nM of each primer. The reactions with unsorted spleen DNA contained ∼100 ng DNA. An initial denaturating step of 94°C for 1 min was followed by 94°C for 15 s and 68°C for 15 min. A first-round PCR of 20–36 cycles from VDJ or JH4long to γCH2e was followed by a nested second-round PCR of 15–30 cycles from 3′ Eμ to γCH2d, γ2aCH2long, or γ2bhingelong. Any bands obtained were cloned as described in the previous section and sequenced.
Mass spectrometry.
Coomassie-stained bands were destained, reduced, carbamidomethylated, and digested overnight with 10 ng/μl of sequencing grade modified trypsin (Promega) in 25 mM NH4HCO3 at 30°C. The resulting peptide mixtures were separated by reversed-phase liquid chromatography on a Vydac C18 column (0.1 × 100 mm, 5-μm particle size), with a gradient of 0–30% acetonitrile over 30 min, containing 0.1% formic acid, at a flow rate of 500 nl/min. The column was coupled to a nanospray ion source (Protana Engineering) fitted to a quadruple-TOF mass spectrometer (Qstar Pulsar i; Applied Biosystems). The instrument was operated in information-dependent acquisition mode, with an acquisition cycle consisting of a 0.5-s TOF scan over the m/z range 350–1,500, followed by two 2-s MS/MS scans (triggered by two or three positive ions) recorded over the m/z range 100–1,700. Proteins were identified by database searching of the mass spectral data using Mascot software (Matrix Science).
RNA-FISH.
Sorted cells were fixed on slides and analyzed by two-color RNA-FISH. Methods for the analysis, with probes for FISH generated previously, have been previously described (23). Images were visualized using two microscopes (BX40 and BX41; Olympus). Experiments were performed two to four times, and at least 100 nuclei or 200 alleles were counted each time.
Online supplemental material.
Table S1 shows the primer sequences used for PCR. Fig. S1 lists the VH cDNA and genomic Cγ H chain Ig sequences obtained. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071155/DC1.
Supplemental Material
Acknowledgments
We thank Martin Turner, Geoff Butcher, and Michael Neuberger for critical reading of the manuscript, and Simon Andrews for help with the sequence analyses.
The research was supported by the Babraham Institute and the Biotechnology and Biological Sciences Research Council.
The authors have no conflicting financial interests.
Abbreviations used: BCR, B cell receptor; BIO, biotin; C, constant; cDNA, complementary DNA; ER, endoplasmic reticulum; ES, embryonic stem; FISH, fluorescence in situ hybridization; FS and SS, forward and side scatter, respectively; HCD, H chain disease.
X. Zou and M.J. Osborn contributed equally to this work.
References
- 1.Hardy, R.R., C.E. Carmack, S.A. Shinton, J.D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro–B and pre–pro–B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osmond, D.G., A. Rolink, and F. Melchers. 1998. Murine B lymphopoiesis: towards a unified model. Immunol. Today. 19:65–68. [DOI] [PubMed] [Google Scholar]
- 3.Kearney, J.F. 2004. Development and function of B-cell subsets. In Molecular Biology of B-cells. T. Honjo, F.W. Alt, and M.S. Neuberger, editors. Elsevier Academic Press, Amsterdam. 155–160.
- 4.Melchers, F., and P. Kincade. 2004. Early B cell development to a mature, antigen-sensitive cell. In Molecular Biology of B-cells. T. Honjo, F.W. Alt, and M.S. Neuberger, editors. Elsevier Academic Press, Amsterdam. 101–126.
- 5.Pike, K.A., and M.J.H. Radcliffe. 2002. Cell surface immunoglobulin receptors in B cell development. Semin. Immunol. 14:351–358. [DOI] [PubMed] [Google Scholar]
- 6.Kitamura, D., J. Roes, R. Kuhn, and K. Rajewsky. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 350:423–426. [DOI] [PubMed] [Google Scholar]
- 7.Zou, X., T.A. Piper, J.A. Smith, N.D. Allen, J. Xian, and M. Brüggemann. 2003. Block in development at the pre B-II to immature B-cell stage in mice without Igκ and Igλ light chain. J. Immunol. 170:1354–1361. [DOI] [PubMed] [Google Scholar]
- 8.Pelanda, R., U. Braun, E. Hobeika, M.C. Nussenzweig, and M. Reth. 2002. B cell progenitors are arrested in maturation but have intact VDJ recombination in the absence of Ig-α and Ig-β. J. Immunol. 169:865–872. [DOI] [PubMed] [Google Scholar]
- 9.Hamers-Casterman, C., T. Atarhouch, S. Muyldermans, G. Robinson, C. Hamers, E.B. Songa, N. Bendahman, and R. Hamers. 1993. Naturally occurring antibodies devoid of light chains. Nature. 363:446–448. [DOI] [PubMed] [Google Scholar]
- 10.Padlan, E.A. 1994. Anatomy of the antibody molecule. Mol. Immunol. 31:169–217. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg, A.S., A.L. Hughes, J. Guo, D. Avila, E.C. McKinney, and M.F. Flajnik. 1996. A novel “chimeric” antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur. J. Immunol. 26:1123–1129. [DOI] [PubMed] [Google Scholar]
- 12.Rast, J.P., C.T. Amemiya, R.T. Litman, S.J. Strong, and G.W. Litman. 1998. Distinct patters of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics. 47:234–245. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen, V.K., C. Su, S. Muyldermans, and W. van der Loo. 2002. Heavy-chain antibodies in Camelidae; a case of evolutionary innovation. Immunogenetics. 54:39–47. [DOI] [PubMed] [Google Scholar]
- 14.Alexander, A., M. Steinmetz, D. Barritault, B. Frangione, E.C. Franklin, L. Hood, and J.N. Buxbaum. 1982. γ Heavy chain disease in man: cDNA sequence supports partial gene deletion model. Proc. Natl. Acad. Sci. USA. 79:3260–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas, I.G., and M. Wabl. 1983. Immunoglobulin heavy chain binding protein. Nature. 306:387–389. [DOI] [PubMed] [Google Scholar]
- 16.Munro, S., and H.R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 48:899–907. [DOI] [PubMed] [Google Scholar]
- 17.Zou, X., C. Ayling, J. Xian, T.A. Piper, P.J. Barker, and M. Brüggemann. 2001. Truncation of the μ heavy chain alters BCR signalling and allows recruitment of CD5+ B cells. Int. Immunol. 13:1489–1499. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen, V.K., X. Zou, M. Lauwereyes, L. Brys, M. Brüggemann, and S. Muyldermans. 2003. Heavy-chain only antibodies derived from dromedary are secreted and displayed by mouse B-cells. Immunology. 109:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roederer, M., W. Moore, A. Treister, R.R. Hardy, and L.A. Herzenberg. 2001. Probability binning comparison: a metric for quantitating multivariate distribution differences. Cytometry. 45:47–55. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro, H.M. 2003. Practical Flow Cytometry. Fourth edition. John Wiley & Sons Inc., New York. 736 pp.
- 21.Gass, J.N., K.E. Gunn, R. Sriburi, and J.W. Brewer. 2004. Stressed-out B cells? Plasma-cell differentiation and the unfolded protein response. Trends Immunol. 25:17–24. [DOI] [PubMed] [Google Scholar]
- 22.Sanderson, R.D., P. Lalor, and M. Bernfield. 1989. B lymphocytes express and loose syndecan at specific stages of differentiation. Cell Regul. 1:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolland, D.J., A.L. Wood, C.M. Johnston, S.F. Bunting, G. Morgan, L. Chakalova, P.J. Fraser, and A.E. Corcoran. 2004. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 5:630–637. [DOI] [PubMed] [Google Scholar]
- 24.Corcoran, A.E. 2005. Immunoglobulin locus silencing and allelic exclusion. Semin. Immunol. 17:141–154. [DOI] [PubMed] [Google Scholar]
- 25.Xu, Z., Z. Fulop, Y. Zhong, A.J. Evinger, H. Zan, and P. Casali. 2005. DNA lesions and repair in immunoglobulin class switch recombination and somatic mutation. Ann. NY Acad. Sci. 1050:146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, J., M. Trounstine, F.W. Alt, F. Young, C. Kurahara, J.F. Loring, and D. Huszar. 1993. Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int. Immunol. 5:647–656. [DOI] [PubMed] [Google Scholar]
- 27.Zou, X., J.A. Smith, V.K. Nguyen, L. Ren, K. Luyten, S. Muyldermans, and M. Brüggemann. 2005. Expression of a dromedary heavy chain-only antibody and B cell development in the mouse. J. Immunol. 175:3769–3779. [DOI] [PubMed] [Google Scholar]
- 28.Janssens, R., S. Dekker, R.W. Hendriks, G. Panayotou, A. van Remoortere, J.K. San, F. Grosveld, and D. Drabek. 2006. Generation of heavy-chain-only antibodies in mice. Proc. Natl. Acad. Sci. USA. 103:15130–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brüggemann, M., J.A. Smith, M.J. Osborn, D. Corcos, X. Zou, V.K. Nguyen, and S. Muyldermans. 2006. Heavy-chain-only antibody expression and B-cell development in the mouse. Crit. Rev. Immunol. 26:377–390. [DOI] [PubMed] [Google Scholar]
- 30.Hendershot, L.M. 1990. Immunoglobulin heavy chain and binding protein complexes are dissociated in vivo by light chain addition. J. Cell Biol. 111:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kan, J.L., and M.R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monk, R.J., S.L. Morrison, and C. Milcarek. 1981. Heavy-chain mutants derived from γ2b mouse myeloma: characterization of heavy-chain messenger ribonucleic acid, proteins, and secretion in deletion mutants and messenger ribonucleic acid in γ2a mutant progeny. Biochemistry. 20:2330–2339. [DOI] [PubMed] [Google Scholar]
- 33.Kinoshita, K., J. Tashiro, S. Tomita, C.G. Lee, and T. Honjo. 1998. Target specificity of immunoglobulin class switch recombination is not determined by nucleotide sequence of S region. Immunity. 9:849–858. [DOI] [PubMed] [Google Scholar]
- 34.De Genst, E., D. Saerens, S. Muyldermans, and K. Conrath. 2006. Antibody repertoire development in camelids. Dev. Comp. Immunol. 30:187–198. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, V.K. 2002. Generation of heavy chain antibodies in Camelids. Ph.D. thesis. Free University of Brussels, Brussels, Belgium. 131 pp.
- 36.Tarlinton, D.M., M. McLean, and G.B. Nossal. 1995. B1 and B2 cells differ in their potential to switch immunoglobulin isotype. Eur. J. Immunol. 25:3388–3393. [DOI] [PubMed] [Google Scholar]
- 37.Herzenberg, L.A. 2000. B-1 cells: the lineage question revisited. Immunol. Rev. 175:9–22. [PubMed] [Google Scholar]
- 38.Cartegni, L., S.L. Chew, and A.R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Natl. Rev. Genet. 3:285–298. [DOI] [PubMed] [Google Scholar]
- 39.Odegard, V.H., and D.G. Schatz. 2006. Targeting of somatic hypermutation. Nat. Rev. Immunol. 6:573–583. [DOI] [PubMed] [Google Scholar]
- 40.Muyldermans, S., C. Cambillau, and L. Wyns. 2001. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 26:230–235. 11295555 [Google Scholar]
- 41.Wu, T.T., and E.A. Kabat. 1992. Possible use of similar framework region amino acid sequences between human and mouse immunoglobulins for humanizing mouse antibodies. Mol. Immunol. 29:1141–1146. [DOI] [PubMed] [Google Scholar]
- 42.Lauwereys, M., M. Arbadi Ghahroudi, A. Desmyter, J. Kinne, W. Hölzer, E. de Genst, L. Wyns, and S. Muyldermans. 1998. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 17:3512–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cogné, M., C. Silvain, A.A. Khamlichi, and J.L. Preud'homme. 1992. Structurally abnormal immunoglobulins in human immunoproliferative disorders. Blood. 79:2181–2195. [PubMed] [Google Scholar]
- 44.Litman, G.W., J.P. Rast, M.J. Shamblott, R.N. Haire, M. Hulst, W. Roess, R.T. Litman, K.R. Hinds-Frey, A. Zilch, and C.T. Amemiya. 1993. Phylogenetic diversification of immunoglobulin genes and antibody repertoire. Mol. Biol. Evol. 10:60–72. [DOI] [PubMed] [Google Scholar]
- 45.Ren, L., X. Zou, J.A. Smith, and M. Brüggemann. 2004. Silencing of the immunoglobulin heavy chain locus by removal of all 8 constant region genes on a 200 kb region. Genomics. 84:686–695. [DOI] [PubMed] [Google Scholar]
- 46.Zou, X., J. Xian, A.V. Popov, I.R. Rosewell, M. Müller, and M. Brüggemann. 1995. Subtle differences in antibody responses and hypermutation of λ light chains in mice with a disrupted κ constant region. Eur. J. Immunol. 25:2154–2162. [DOI] [PubMed] [Google Scholar]
- 47.Deverson, E., C. Berek, M.J. Taussig, and A. Feinstein. 1987. Monoclonal BALB/c anti-progesterone antibodies use family IX variable region heavy chain genes. Eur. J. Immunol. 17:9–13. [DOI] [PubMed] [Google Scholar]
- 48.Yu, Y.P., F. Lin, R. Dhir, D. Krill, M.J. Becich, and J.H. Luo. 2001. Linear amplification of gene-specific cDNA ends to isolate full-length of a cDNA. Anal. Biochem. 292:297–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.