Abstract
The hairpin ribozyme acts as a reversible, site-specific endoribonuclease that ligates much more rapidly than it cleaves cognate substrate. While the reaction pathway for ligation is the reversal of cleavage, little is known about the atomic and electrostatic details of the two processes. Here, we report the functional consequences of molecular substitutions of A9 and A10, two highly conserved nucleobases located adjacent to the hairpin ribozyme active site, using G, C, U, 2-aminopurine, 2,6-diaminopurine, purine, and inosine. Cleavage and ligation kinetics were analyzed, tertiary folding was monitored by hydroxyl radical footprinting, and interdomain docking was studied by native gel electrophoresis. We determined that nucleobase substitutions that exhibit significant levels of interference with tertiary folding and interdomain docking have relatively large inhibitory effects on ligation rates while showing little inhibition of cleavage. Indeed, one variant, A10G, showed a fivefold enhancement of cleavage rate and no detectable ligation, and we suggest that this property may be uniquely well suited to intracellular targeted RNA cleavage applications. Results support a model in which formation of a kinetically stable tertiary structure is essential for ligation of the hairpin ribozyme, but is not necessary for cleavage.
Keywords: hairpin ribozyme, ligation, RNA tertiary structure
INTRODUCTION
The hairpin ribozyme is derived from the negative strand of the tobacco ring spot virus satellite RNA (Buzayan et al. 1986b). It catalyzes a reversible, site-specific cleavage and ligation reaction by transesterification (Fig. 1). The secondary structure of the hairpin ribozyme consists of two domains, A and B (Hampel and Tritz 1989), each composed of an internal loop flanked by two helices. These two domains must associate through tertiary interactions (“dock”) to form an active complex (Esteban et al. 1997, 1998; Walter et al. 1998). In nature, the two domains are joined as a four-way helical junction (4WJ) (Buzayan et al. 1986a), of which only the two domains and a two-way helical junction (2WJ) are required to form a functional minimal hairpin ribozyme (Hampel and Tritz 1989; Hampel et al. 1990). The two domains are also capable of interacting when unconnected strands are provided separately. These features provide investigators with the ability to study the ribozyme as a native 4WJ construct, minimal 2WJ, or as separated domains (Butcher et al. 1995; Hampel et al. 1998).
FIGURE 1.
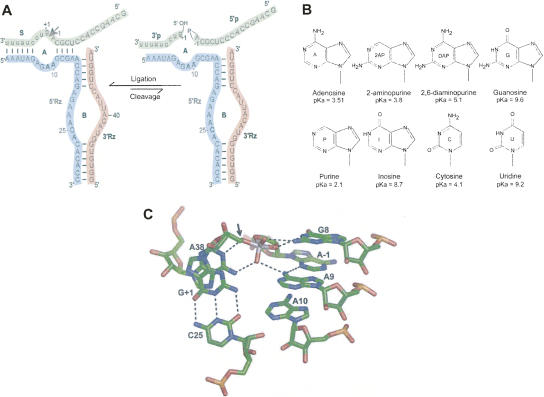
Ribozyme constructs, reactions, and nucleobase variants. (A) Secondary structure of the SV5 hairpin ribozyme used in this study. Loops A and B and individual strands are indicated and color coded. Cleavage site on the substrate (S) is indicated with an arrow and the products (3′p and 5′p) are shown with their respective end groups. Nucleotides at positions 9 and 10 (bold) in the 5′ ribozyme strand (5′ Rz) were individually substituted with nucleobases as shown in B. (B) Structures of the nucleobases used in this study, with their respective N1 pKA values. (C) Model of the active site and the nucleotides surrounding A9 and A10 with potential hydrogen bonds. Adapted from crystal structure, 1M5O (Rupert et al. 2002).
The cleavage reaction yields products with 2′, 3′-cyclic phosphate and 5′-OH termini (Buzayan et al. 1986b). The reverse reaction, ligation, is favored, and its rate is at least 10 times faster than that of cleavage (Hegg and Fedor 1995; Esteban et al. 1997; Fedor 1999). Most studies have concentrated on the cleavage reaction of the ribozyme. Less attention has been focused on ligation, which is assumed to be a microscopic reversal of the cleavage reaction (Buzayan et al. 1988a,b; Chowrira et al. 1993; Nesbitt et al. 1997). Docking studies of the ribozyme with cleavage products have proven difficult due to the technical challenges involved. Similarly, residues important for ligation have not been meticulously established. An observation in our laboratory that a ribozyme with an A9G substitution ligated poorly although its cleavage activity was unaffected suggested that certain nucleotides in the ribozyme might be important for ligation but not cleavage. This observation led to the present study.
In order for the ligation reaction to occur, the products must bind the ribozyme, the two ribozyme domains must associate, and the ends of the products must be correctly aligned for an SN2 reaction. Two laboratories have recently solved three-dimensional crystal structures of the hairpin ribozyme, one using the naturally occurring four-way junction ribozyme fused to a binding site for U1A protein and cocrystallized with the protein (Fig. 1C; Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002), the other using two protein-free constructs consisting of the minimal ribozyme–substrate complex (Salter et al. 2006; Torelli et al. 2007). In each structure, the active sites had generally similar conformations. The active site of the hairpin ribozyme is composed of G8, A38, C25, A9, and A10 plus the substrate nucleotides at the reaction sites, G + 1 and A − 1 (Fig. 1C; Berzal-Herranz et al. 1993; Rupert and Ferré-D'Amaré 2001; Salter et al. 2006; Torelli et al. 2007). G8 and A38 are proposed to be directly involved in the reaction chemistry (Fedor 2000; Pinard et al. 2001; Rupert and Ferré-D'Amaré 2001).
A9 and A10 interact less intimately with the reaction site, and their roles in supporting catalysis are less clear. G + 1 flips out from the substrate strand to make an essential interdomain base pair with C25, an interaction that is important for the domain docking (Pinard et al. 1999). The exocyclic 6-amino group of A9 is proposed to form hydrogen bonds with the N3 of A − 1 and with one of the nonbridging oxygens of the scissile bond (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002), while A10 is part of the ribose zipper in which its 2′ OH interacts with the sugar of C25 (Klostermeier and Millar 2001). A10 is located within the active site, but in the crystal structures it is not close enough to make direct interactions with any of the reactants (Fig. 1C; Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002). In the recent crystal structure (Salter et al. 2006; Torelli et al. 2007) the authors propose water-mediated interactions of the exocyclic amino group and the N1 of A10 with A38, and also interactions of the N1 of A9 with the exocyclic amino group of G + 1 through water molecules. A water molecule (W52) that was proposed to mediate an interaction between exocyclic amino groups of A9 and A38 (Salter et al. 2006) is now interpreted as a nonbridging oxygen in the newer crystal structure (2P7E) (Torelli et al. 2007).
Previous studies have generally viewed the ligation reaction as a microscopic reversal of the cleavage reaction in the hairpin ribozyme (Buzayan et al. 1986b; Buzayan et al. 1988a; Chowrira et al. 1993; Nesbitt et al. 1997). However, relatively few studies have specifically addressed the differences between the requirements for cleavage and ligation. Nucleotide analog interference mapping (NAIM) studies of the hairpin ribozyme showed inhibition of the ligation reaction when A9 and A10 were substituted with phosphorothioate nucleotide analogs (Ryder and Strobel 1999). These findings suggested a role for A9 and A10 in catalysis; in particular, A10 was proposed to undergo a catalytically important ionization (Ryder et al. 2001).
The ligation reaction presumably requires, in addition to binding of the products and docking of the two domains, interactions to orient the ribose moiety of A − 1 with its 2′, 3′- cyclic phosphate and that of G + 1 with its 5′-OH, so that their geometry is appropriate for the transesterification reaction. In this work, we explore the requirements for the ligation reaction, in the background of a well-characterized two-way junction hairpin ribozyme, and how they compare to the requirements for cleavage. In the present study, kinetic analysis is supplemented by hydroxyl radical footprinting and native gel electrophoresis, which provide insights into the effects of nucleobase substitutions on tertiary folding and association of the two domains of the ribozyme–substrate complex. Our results indicate that ligation requires a more stably docked complex than cleavage and that the docking of the ribozyme complex is very sensitive to nucleobase substitutions at the A9 and A10 positions. Study of the numerous variants also demonstrated that it is possible to convert a reversible ribozyme into one whose activity is functionally unidirectional, strongly favoring cleavage under typical reaction conditions.
RESULTS
Choice of ribozyme constructs
In these studies, we used the well-characterized minimal ribozyme construct SV5 (Butcher et al. 1995; Sargueil et al. 1995). The minimal ribozyme (containing a two-way helical junction) was chosen instead of the native ribozyme (containing a four-way helical junction) because the four-way junction is known to counteract the effect of many destabilizing mutations by strongly favoring formation of the docked, active tertiary structure (Fedor 1999; Walter et al. 1999; Zhao et al. 2000; Klostermeier and Millar 2001). Therefore, the minimal construct provides a more sensitive and appropriate platform with which to probe mutational effects on activity and tertiary folding.
Reaction conditions: Cleavage versus ligation
Different reaction conditions were used to carry out cleavage and ligation of the ribozymes. Each reaction was optimized independently and it was observed that higher concentrations of the ribozyme strands were required to saturate the reaction for ligation (Sargueil et al. 2003) while maintaining the same Mg2+ and buffer concentrations.
Cleavage and ligation kinetics of A9 and A10 variants
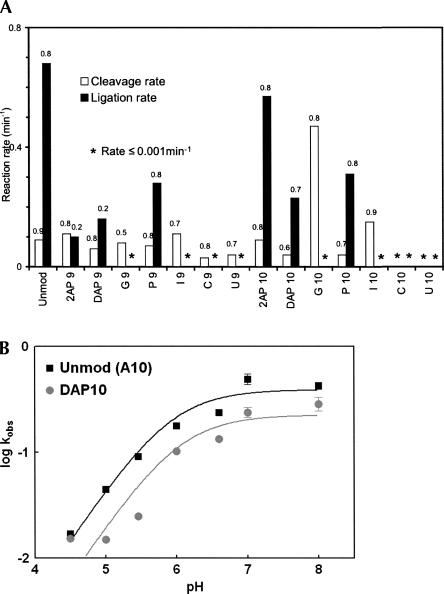
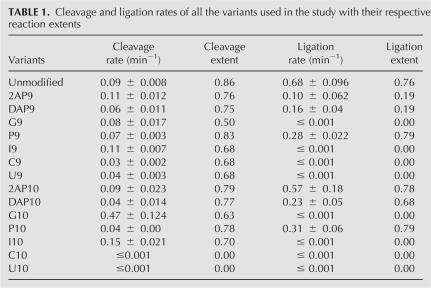
Kinetic assays were conducted to determine the effects of nucleobase substitutions at A9 and A10 on cleavage and ligation rates (Fig. 2A; Table 1). Under our standard assay conditions, the unmodified (SV5) ribozyme exhibits an observed cleavage rate of 0.09 min−1 with 80% cleavage and an observed ligation rate of 0.68 min−1 with 80% ligation. A9 and A10 were independently substituted with G, U, C, 2-aminopurine (2AP), 2,6-diaminopurine (DAP), purine (P), and inosine (I) (Fig. 1B), and cleavage and ligation rates were monitored. In general, ligation rates were more significantly affected than were cleavage rates (Fig. 2A; Table 1).
FIGURE 2.
Cleavage and ligation rates for the hairpin ribozyme variants. (A) Reactions were carried out in the presence of 15 mM MgCl2 and 50 mM HEPES/NaOH at pH 7.0 (cleavage) and pH 7.5 (ligation) at ribozyme concentrations as mentioned in Materials and Methods for optimal rates. Values above each of the bars represent the extent of the reaction. Asterisk denotes catalytic rate ≤0.001 min−1. Experiments were repeated at least three times for each variant, and the experimental variation did not exceed ∼15% for the reaction rates and the amplitudes (Table 1). (B) Ligation rate–pH profile of unmodified, (A10) (black squares) and DAP10 (gray circles) variant. Reaction rates were determined under standard conditions of 50 mM reaction buffer and 15 mM MgCl2 at 25°C as described in Materials and Methods. Buffers used over the pH range include MES buffers for pH 4.5–6.5 and HEPES/NaOH for pH 7–8.
TABLE 1.
Cleavage and ligation rates of all the variants used in the study with their respective reaction extents
Pyrimidine substitutions at positions 9 and 10 affected the ribozyme most severely. Although 70% of the substrate is cleaved by C9 and U9 variants, the observed rate is slower than that of the unmodified SV5 construct. No ligation could be observed in assays with ribozymes containing C9 or U9 (Fig. 2A; Table 1). In contrast, C10 and U10 substitutions inactivate both cleavage and ligation. The latter base has the potential to form a Watson–Crick base pair with cleavage site nucleobase A − 1, thus extending helix 2 and creating a highly unfavorable active site geometry that would interfere with the positioning of the 2′-OH of A − 1 for inline attack on the scissile cleavage site phosphorus. Similarly, the C10 variant has the potential to form a Watson–Crick base pair with the other cleavage site nucleobase G + 1, with potentially profound catalytic and structural effects. In the active ribozyme, G + 1 flips out to make an essential interdomain base pair with C25 (Pinard et al. 1999), in turn positioning its 5′-oxygen for an inline SN2 reaction. A base pair between C10 and G + 1 could inhibit activity by preventing docking of the ribozyme.
Purine analogs at position 9 were much less inhibitory to cleavage, but had dramatic effects on ligation (Fig. 2A; Table 1). Crystallographic studies show that the exocyclic amino group of A9 forms hydrogen bonds with the N3 of A − 1 and one of the nonbridging oxygens of the scissile bond (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002). Substitution of A9 with purine analogs did not significantly affect the observed cleavage rate or the extent of the reaction except for G9, which decreased the extent of the cleavage reaction to 50%. This suggests that the contacts between the exocyclic amine of A9 and the cleavage site are not essential for cleavage in the SV5 construct under the conditions used in these experiments. Substitution with other purines (2AP, DAP, I, and P) at position 9 resulted in no detectable change in the cleavage rate. Strikingly, ligation was much more sensitive to purine substitutions (Fig. 2A; Table 1). The extent of the reaction was reduced fourfold in the ribozymes containing 2AP9 and DAP9 compared to the unmodified SV5 ribozyme (A9), suggesting that the substitution induces a kinetically stable misfold in a majority of the ribozyme–substrate complex population. Together the data argue that the presence of a 2-amino group (2AP and DAP) may interfere with folding and activity, while the high fraction of ligation and relatively fast rate of the ribopurine 9 variant (k = 0.28 min−1, reaction extent = 80%) compared to other active variants argues against an essential role for the 6-amino group in ligation, especially for folding. Interestingly, the presence of a keto group at N6 (G9, I9) not only reduced the rate of ligation, but totally eliminated it. One possibility is that the keto group might be making some unfavorable interactions, causing a misfold or improper orientation of the active site with the products.
Purine analogs at position 10 gave cleavage and ligation results (Fig. 2A; Table 1) that may be less easily interpretable according to existing structural models. The 2AP10 variant cleaves at the same rate as the unmodified A10 ribozyme, and the rates of the P10 and DAP10 variants are both reduced only twofold. In ligation reactions, the 2AP10 variant functions at nearly the same rate as the unmodified SV5 ribozyme, while the P10 and DAP10 variants yield rates around 50% that of unmodified ribozyme. These results suggest that the crystallographic interactions of the 6-amino group of A10 are not essential for activity. Unexpectedly, the ribozyme variant containing G10 cleaves five times faster than the unmodified ribozyme. Since the I10 variant cleaves faster than A10 but not as fast as the G10 ribozyme, the maximal cleavage rate appears to require the presence of both the 2-amino and 6-keto groups of G. The G10 variant was not reported to be an activating mutation in previous work (Shippy et al. 1998). The ribozyme used by Shippy et al. was a cis-acting construct derived from the wild-type hairpin ribozyme sequence. Cleavage was carried out at 37°C in transcription reactions. This combination of construct and conditions may enhance the negative effects of nucleotide variants. In contrast to its enhanced cleavage activity, the ligation reaction of the G10 variant was very strongly inhibited, proceeding at an undetectable rate, as did that of the I10 variant. One possible explanation for lack of ligation by the G10 ribozyme could be that the N1 is unlikely to be ionized, as previously suggested for A10 by studies using the wild-type ribozyme (Ryder et al. 2001). Ligation rates of ribozymes substituted with DAP10 (pKA = 5.1) over a pH range of 4.5–8 were compared with those of A10 (pKA = 3.5). If the protonation of N1 were essential for ligation, the increase in ligation rate would occur at a higher pH for DAP10 than the unmodified ribozyme. However, the ligation rate versus pH plot (Fig. 2B) did not show a significant shift (apparent pKA of the A10 reaction = 5.9, and that of the DAP10 reaction = 6.0); hence, we conclude that in the SV5 ribozyme, the protonation of the base at position 10 is probably not essential and not the reason for failure of ligation in the G10 variant. The reaction catalyzed by the G10 variant of the hairpin ribozyme appears to be shifted very strongly toward cleavage, whereas the unmodified SV5 and native four-way junction ribozymes strongly favor ligation. Biotechnology implications of these findings are discussed below.
Tertiary folding assays
Modifications to ribozymes that inhibit catalytic activity may do so by inhibiting folding to the native tertiary structure or by inhibiting a step (presumably chemical) that follows tertiary structure formation. To distinguish these possibilities, we employ two assays—hydroxyl radical footprinting and native gel electrophoresis. The methods are highly complementary, but each has its advantages and disadvantages.
Hydroxyl radical protection has the advantage of single nucleotide resolution, but may not reveal information about different global tertiary folds. In our hands, the method has a 30-sec experimental dead time, so that any events taking place within this time after the initiation of the reaction are not observed. In the hairpin ribozyme–substrate complex, backbone protection is observed when the two domains dock to form a stable three-dimensional structure (Hampel et al. 1998).
Native gel electrophoresis does not approach the spatial resolution of the footprinting technique, but can resolve distinct tertiary folds, especially those that differ in the disposition of RNA molecules around helical junctions. This latter analysis is particularly informative in the hairpin ribozyme, where docking of the two domains of the ribozyme has been shown to be an essential step in the formation of the active complex (Hampel and Burke 2001).
Tertiary folding of A9 and A10 variants by hydroxyl radical footprinting
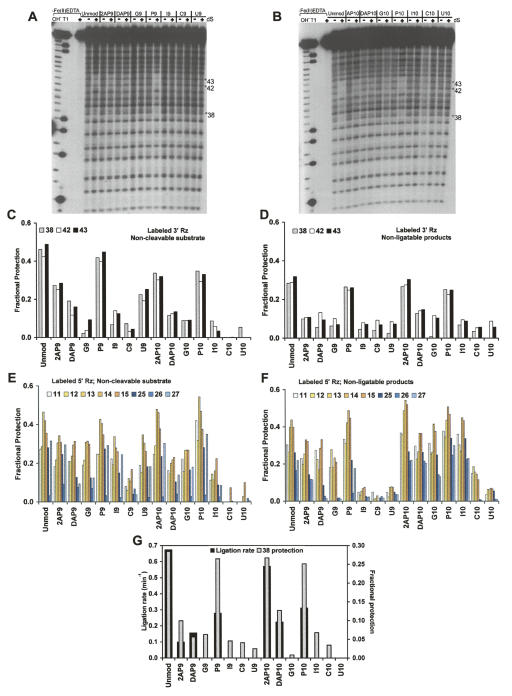
Folding assays were carried out with A9 and A10 variants and appropriate controls, employing noncleavable substrate or nonligatable products, as described in Materials and Methods. Previously, we had demonstrated that unmodified ribozyme in the presence of noncleavable substrate or nonligatable products is protected at positions 11–15 (helix 2), 25–27 (5′ segment of internal loop B), and 38, 42, 43 (3′ segment of internal loop B) (Hampel et al. 1998).
For the A9 and A10 variants, we observe poor correlation between the extent of protection and the cleavage rates (Figs. 2A, 3A–C,E; Table 1). For example, comparing the cleavage rates with the protections in the 3′ Rz strand (Fig. 3C), we find that ribozymes with I9 and I10 cleave faster than wild type, but no backbone protection is seen at any position. Also, G10, which cleaves extremely fast, shows no protection at all. P10, with a slow cleavage rate of 0.04 min−1 shows more protection than I10, which cleaves at the rate of 0.15 min−1. The protections in the 5′ Rz strand also do not correlate with the cleavage rates (Fig. 2A). Note that previous studies had identified variants with robust cleavage activity that lacked the ability to form a stable, docked structure (Walter et al. 2001). These results are interpreted as complexes that can form a transient docked state, resulting in only a small fraction of molecules residing in the docked state at any point in time. However, this small fraction of docked complexes is apparently sufficient for supporting the cleavage reaction at significant rates, and such states have been physically identified through single-molecule fluorescence resonance energy transfer studies (FRET) (Walter et al. 2001).
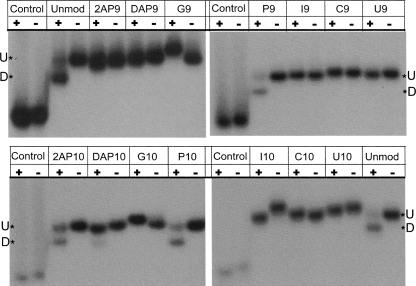
FIGURE 3.
Hydroxyl radical footprinting assay. Polyacrylamide gel analysis of 5′-end-labeled 3′ ribozyme in the absence (−) or presence (+) of noncleavable substrate with different 5′ ribozyme strands containing nucleobase variants; A9-substituted 5′ Rz (A) and A10- substituted 5′ Rz (B), reacted with Fe(II)-EDTA reagent. (C–F) Fractional protection seen in 3′ Rz (C,D) and 5′ Rz (E,F) strands of the various mutants with noncleavable substrate (C,E) and nonligatable products (D,F). Vertical scale indicates fraction protected. For the unmodified ribozyme, 3′ Rz protection is seen at positions 38, 42, and 43 and 5′ Rz protection is seen at 11–15 and 25–27 (Hampel et al. 1998). Protection and loading controls were taken into account while calculating fractional protection, as described in Materials and Methods. Values represented are the average of at least three experiments. (G) Comparison of ligation rates of ribozyme variants with degree of backbone protection at position 38 in the 3′ Rz with nonligatable products.
Protection data were obtained for ligation complexes with nonligatable products (Fig. 3D,F). These data reveal a much stronger relationship between biochemical activity and tertiary complex formation (Fig. 3G). Importantly, significant levels of protection were observed in the case of the P9, 2AP10, and P10 variants along with the wild-type control; these are the variants that show significant ligation activity (Fig. 2A; Table 1). All the other variants show levels of protection that are either undetectable or barely detectable. These results indicate that the ligation reaction is more sensitive to tertiary structure formation and/or stability than is the cleavage reaction.
Tertiary folding of A9 and A10 variants by native gel electrophoresis
Native gel analysis was used as a complementary method to examine global tertiary folding by qualitatively determining the ability of each variant to fold into the docked, active tertiary complex. A construct (LB-S) in which a noncleavable substrate analog (containing 2′-deoxy A − 1) is linked to residue A50 of the 3′ ribozyme fragment (3′ Rz) via a pentacytidine linker was used to differentiate between the docked and undocked ribozyme–substrate complex (Hampel and Burke 2001). As a negative control for docked complex formation, the G + 1A mutant that cannot form the essential G + 1·C25 interdomain base pair was used (Hampel et al. 1998; Walter et al. 1998). The ribozyme–substrate complex can resolve into two bands—the docked complex that migrates with higher mobility and the undocked complex with lower mobility that comigrates with the negative control (Walter et al. 1998). Cleavage reactions in the presence of T4 RNA ligase demonstrated that helices 2 and 3 are coaxially stacked in the undocked ribozyme–substrate complex (Esteban et al. 1998).
It is distinctly visible from Figure 4 that variants that form docked complexes are the ones that undergo ligation. Bands with the docked complexes are clearly seen for the unmodified, P9 and P10 variants. 2AP10 and DAP10 variants also dock, although to a lesser extent. These data do not correlate with the cleavage performance of the variants. A similar observation was made previously with the abasic +2 substrate variant that cleaved faster than all other +2 nucleotide substitutions but did not show backbone protection or the formation of a docked complex (Walter et al. 2001). G9-, G10-, and I10-substituted ribozymes migrate in an anomalous fashion, which suggests the formation of an atypical tertiary structure possibly detrimental to ligation but not cleavage. These observations provide direct support for a model in which ligation requires a more stably docked structure than cleavage.
FIGURE 4.
Native gel electrophoresis of hairpin ribozyme variants. Lane 1 (+) for each variant is the LB-S (noncleavable substrate linked to 3′ Rz via a pentacytidine linker) that promotes folding. Lane 2 (−) for each variant is the negative control (G + 1A). First two lanes of each gel are the two individually labeled LB-S. Docked complex migrates faster than the coaxially stacked undocked complex.
DISCUSSION
The hairpin ribozyme catalyzes cleavage and ligation of its substrate, with its internal equilibrium shifted strongly toward ligation (Hegg and Fedor 1995; Esteban et al. 1997; Wilson et al. 2005). Under many conditions, reaction chemistry is rate limiting for cleavage, but tertiary structure formation can be rate limiting for ligation (Walter et al. 1998). Nucleotides essential for the cleavage reaction have been extensively studied, but most studies have focused on cleavage. In this study, we examine variants that selectively inhibit ligation and shift the reaction toward cleavage.
The hairpin ribozymes used in published studies have differences in their design, which can lead to significant differences in the reported activities of variants. These activity differences generally are a consequence of altered RNA folding behavior, which may be additionally influenced by choice of reaction conditions and folding protocols. Here, we have primarily used the modified minimal hairpin ribozyme SV5 (Butcher et al. 1995; Sargueil et al. 1995) instead of the “wild-type” ribozyme construct (Hampel and Tritz 1989; Anderson et al. 1994) because it shows superior folding and kinetic characteristics. The SV5 construct has permitted physical studies of the docked complex (Hampel et al. 1998; Walter et al. 1998), whereas similar efforts with the original “wild-type” hairpin ribozyme had not yielded evidence of stable tertiary structure with a lifetime amenable to biochemical detection (Butcher and Burke 1994).
The chemistry of ligation is assumed to be a microscopic reversal of cleavage. However, the ground state of the complex formed by the ribozyme and uncleaved substrate differs, particularly in constraint, from that formed by the ribozyme with the two short products, one containing a free 5′ hydroxyl group and the other containing a 2′, 3′- cyclic phosphate. The conformational changes required to form docked complexes and access the transition state from their respective ground states very likely differ from one another. Nucleotide substitutions in the ribozyme might have differential effects on cleavage versus ligation because (1) they might stabilize one ground state compared to the other or misfold one and not the other (for instance, by causing new interactions with the free ends of the products in the ground state); (2) they might interfere with transient interactions needed in conformational changes from one ground state and not the other in accessing the docked conformation; or (3) they could alter interactions specifically involved in alignment of the product termini for ligation.
Mutational and interference studies proposed the involvement of A9 and A10 in cleavage (Chowrira et al. 1993; Grasby et al. 1995; Shippy et al. 1998; Ryder and Strobel 1999) and are supported by the observation that they are located in the active site of the crystal structures (Rupert and Ferré-D'Amaré 2001; Salter et al. 2006; Torelli et al. 2007). Molecular dynamic studies based on the 1M5O crystal structure (Rupert and Ferré-D'Amaré 2001) with vanadate at the cleavage site suggest that A9, G8, and A38 function to stabilize the negatively charged oxygen atoms in the transition state (Park and Lee 2006).
Several studies carried out with the wild-type hairpin ribozyme or closely related constructs have suggested that A9 may play a role in the cleavage reaction (Chowrira et al. 1993; Grasby et al. 1995; Shippy et al. 1998). However, in the background of our construct, nucleobase substitutions at position 9 show minimal effects on cleavage. Similar findings have been reported for a hairpin ribozyme stabilized by a four-way junction, where an abasic substitution at position 9 decreases the cleavage rate only three- to fourfold (Kuzmin et al. 2004).
The ligation reaction, in contrast to cleavage, shows a strong preference for the conserved A9 (Berzal-Herranz et al. 1993). While most variants at position 9 show moderate to extreme inhibition, the observed ligation rate for P9 is only 1.5-fold lower than that of A9. NAIM studies, also based on a ligation assay, showed a higher degree of interference from the substitution of a purine-α-phosphorothioate (PαS) nucleotide at position 9, leading to the conclusion that the 6-amino group of A9 was essential for activity (Ryder and Strobel 1999). The difference between the observations is most likely due to the difference in the constructs used (the NAIM analysis was performed with the wild-type hairpin ribozyme) and different reaction conditions (4°C for NAIM versus 25°C for these studies).
In the recent hairpin ribozyme crystal structure containing vanadate at the active site (Torelli et al. 2007), the N1 position of A9 is seen to coordinate with a water molecule that in turn interacts with one of the nonbridging oxygens of vanadate and the exocyclic amino group of G + 1. If this represents a meaningful stabilizing interaction, it might explain why the ribozyme with P9, whose N1 pKA is lower than those of the other variants, shows the presence of a docked ribozyme–substrate complex (Fig. 4), plus significant backbone protection from hydroxyl radicals (Fig. 3G) and a higher ligation rate (Fig. 2A).
Variants G9 and I9 do not decrease the cleavage activity of the ribozyme, but significantly inhibit ligation, suggesting that the keto group and/or the N1 proton might be making unfavorable interactions, although neither of the two positions is within hydrogen bonding distance to any other functional group as seen in the crystal structures. Both folding assays indicate that tertiary folding is inhibited—little or no docked complex is observed by native gel electrophoresis and low levels of protection are observed in the hydroxyl radical footprinting experiments. Together, these results suggest that these substitutions at position 9 act to inhibit formation of the active docked complex by (1) stabilizing ground state interactions or by (2) inhibiting essential interdomain contacts required for tertiary structure formation.
In accordance with prior results (Shippy et al. 1998), the 10 position of the hairpin ribozyme requires at least a purine ring to maintain minimal cleavage activity (Fig. 2A; Table 1). A single exocyclic amino group (2AP10, A10) seems to aid in cleavage; two (DAP10) or none (P10) (Grasby et al. 1995) hinder it while a 6-keto group (G10, I10) enhances it. Unlike the previous observation in the wild-type sequence ribozyme where a G10 variant had cleavage activity reduced by 18-fold (Shippy et al. 1998), data here suggest that the presence of the keto group in our construct increases the rate of the cleavage reaction about fivefold compared to the A10 ribozyme (Fig. 2A), although it does not stabilize the docked complex, as is evident from the native gel analysis (Fig. 4) and hydroxyl radical footprinting data (Fig. 3). The apparent destabilizing effect of the 6-keto group may disrupt docking enough in the wild-type ribozyme to inhibit cleavage while leaving the SV5 ribozyme still able to interact stably enough to cleave.
The same substitutions have very different effects on ligation. In spite of the high cleavage rates seen in G10 and I10 variants, their ligation in this background is strongly inhibited, implying that the same interactions that destabilize docking and enhance cleavage may concomitantly hinder ligation. Crystal structures suggest that A10 probably does not play a direct role in ligation chemistry; however, some of the mutations may interfere with the formation of the active site and/or with electrostatic stabilization of the transition state.
The 2AP10 substituted variant behaves similarly to the unmodified ribozyme, while P10 and DAP10 substitution reduces the rate nearly twofold. Similar substitutions with phosphorothioate analogs analyzed by NAIM gave slightly different results. High interference was observed with 2APαS and PurαS and none with DAPαS (Ryder and Strobel 1999; Ryder et al. 2001), emphasizing the importance of the Hoogsteen face of A10. Again, the observations may vary because of differences in the constructs and conditions used. The exocyclic amino group and N1 of A10 have been proposed to make some crucial water-mediated interactions to stabilize the reacting groups (Salter et al. 2006; Torelli et al. 2007) that would be disturbed in the presence of G10 and I10, possibly leading to a drastic shift in the reaction equilibrium favoring cleavage.
Stability of docking is more important for ligation than it is for cleavage
Assembling all the strands of the ribozyme and the cleavage products into a conformation favorable for the ligation reaction to occur is more complicated than the analogous process for the cleavage reaction. The docking rate for assembling the cleaved products–ribozyme complex is slower than undocking a cleaved substrate–ribozyme complex (Liu et al. 2007), making it a limiting factor for the ligation reaction. The ligation rate is much faster than cleavage but it is more influenced by the stability of the docked complex. Many variants do not exhibit protection or the presence of a docked complex and yet they can cleave their substrates (i.e., I10, G10), sometimes faster than the variants that show protection and are docked. A similar observation was made in the past (Walter et al. 2001) in which an abasic +2 substrate variant that least affected the cleavage rate among other +2 substitutions did not show any detectable docking by FRET. This is similar to what is seen with some of the variants here—the protection data do not correlate well with the cleavage activity of the variant. These observations could be due either to extremely slow folding rates or dynamic docking during cleavage.
Ligation activity, on the other hand, is only observed for the variants that seem to form a stably docked structure and that show backbone protection. Apart from the unmodified SV5 ribozyme (A9, A10), only P9, 2AP10, and P10 variants show more than 20% protection in the hydroxyl radical footprinting assay and have some fraction of their molecules docked as measured by nondenaturing gel electrophoresis. The docked complex is stabilized by a network of weak interactions in such a way that disruption of any of a number of interactions could prevent docking (Walter et al. 1998), and variants with unstable or undocked complexes are unable to catalyze a transesterification reaction to ligate the cleavage products.
The SV5 construct can overcome many docking problems compared to the wild-type ribozyme construct (Butcher et al. 1995), but it is still sensitive to some mutants like G9 and G10, among others. The four-way junction construct stabilizes the docked tertiary structure such that it can remain active under conditions that are destabilizing and inhibit the two-way junction construct (Hampel and Tritz 1989; Fedor 1999; Walter et al. 1999; Zhao et al. 2000; Klostermeier and Millar 2001). Ligation was partially rescued in all the inhibited variants (including G9 and G10) by introducing the substitution into the four-way junction construct (data not shown). This indicates that the destabilizing effects of the substitutions can be overcome by structural elements that stabilize the global tertiary fold. Our results support the view that ligation activity is highly dependent on the stability of the tertiary structure.
Changing the nature of ribozymes from reversible to unidirectional
The base substitution studies at the A9 and A10 position of the minimal SV5 ribozyme have led to the identification of a ribozyme that cleaves faster than the unmodified SV5 and yet does not give detectable ligation. The G10 modification in the SV5 construct cleaves five times faster than A10 (unmodified) and does not ligate in the two-way junction construct. Previously, in the wild-type ribozyme construct, this same modification had reduced the cleavage rate 18-fold (Shippy et al. 1998). Unlike the base substitution mutant ribozymes in a previous study (Anderson et al. 1994), none of which gave activities significantly greater than the wild-type form, a number of variants were obtained in the SV5 background that cleaved slightly faster than the unmodified ribozyme, for example, 2AP9, I9, I10, and G10.
The G10 variant effectively changes a reversible cleavage reaction into an irreversible one under these conditions. Such substitutions might prove useful in the design of ribozymes for specific purposes where ligation is undesirable. This may be extremely valuable in ribozyme-based targeted RNA cleavage studies aimed at disease models and gene therapy (Zhang and Burke 2005).
MATERIALS AND METHODS
RNA preparation
All oligonucleotides were prepared by solid-phase synthesis using standard RNA phosphoramidite chemistry. Reagents were purchased from Glen Research. Following deprotection, RNA was purified by denaturing gel electrophoresis and reverse-phase HPLC, as previously described (Sargueil et al. 1995; Walter et al. 1998).
Ribozyme kinetic analysis
Cleavage assays were carried out in 50 mM HEPES/NaOH (pH 7.5) and 15 mM MgCl2 (reaction buffer) at 25°C. Unlabeled 5′ Rz and 3′ Rz (at 0.2 μM final concentration), shown in Figure 1A, were mixed with reaction buffer in one tube and ∼1 nM 5′-32P-labeled S (four-way substrate; Walter et al. 1999) with reaction buffer in another. Both the tubes were preincubated at 37°C for 10 min and then allowed to equilibrate to a reaction temperature of 25°C. Reactions were initiated by combining the two volumes. These concentrations were found to be saturating for all mutants examined. At least 16 time points were taken for each experiment by quenching 1.6-μL aliquots on ice into 18.4 μL of formamide loading solution (FLS, 15 mM EDTA, 0.02% [v/v] bromophenol blue and 0.02% [v/v] xylene cyanol in formamide). The reactions were run on 20% polyacrylamide gels, and bands were quantified using a Bio-Rad Molecular Imaging System GS-525 at the Vermont Cancer Center DNA Analysis Facility. The reaction rates and extents were determined by nonlinear regression using Microcal Origin software. Experiments were repeated at least three times for each variant, and the experimental variation did not exceed ∼15% of the reaction rates or amplitudes.
Ligation assays
5′-32P-labeled 5′ product (5′p) was generated by large-scale cleavage of 5′ labeled S. The 5′p was purified on a 20% polyacrylamide denaturing gel. 5′p had to be isolated by cleavage since the 2′, 3′-cyclic phosphate containing product cannot be synthetically prepared, and the long substrate (S) was used instead of the regular 14-nucleotide (nt) SV5 substrate to improve the recovery of the labeled 5′ cleavage product. All other strands were used at saturating concentrations of 15 μM. Ligation reactions were similar to cleavage except that reactions were initiated with the 3′ product (3′p). Briefly, 5′ Rz, 3′ Rz, and ∼1 nM 5′-32P labeled 5′p were mixed with reaction buffer and the reaction was initiated by combining it with the 3′p in reaction buffer subsequent to being separately incubated at 37°C for 10 min and then allowed to equilibrate to a reaction temperature of 25°C. Buffers used for the pH profile were MES buffers for pH 4.5–6.5 and HEPES/NaOH for pH 7–8.
Hydroxyl radical footprinting
Cleavage reactions were carried out as previously described (Hampel et al. 2001; Hampel and Burke 2003). All reactions were carried out in a final volume of 10 μL with noncleavable substrate or nonligatable 3′-phosphorylated 5′p (Walter et al. 1998). To allow the ribozyme to fold, trace amount of 5′-32P-end-labeled RNA and saturating concentrations (1 μM each) of the unlabeled RNAs were preincubated in 25 mM sodium cacodylate (pH 7) and 15 mM MgCl2 at room temperature for 20 min, as suggested by FRET experiments for docking kinetics of the two domains (Walter et al. 1998). For the cleavage reaction, 0.75 μL of each of the following components—H2O2 (0.375% v/v), Fe(II)-EDTA (5 mM Fe(NH4)2(SO4)2, 5 mM Na2-EDTA at pH 8.0, freshly mixed), and 60 mM sodium ascorbate—were added as individual drops in the cap of the reaction tube. The reaction was initiated by a quick microcentrifuge spin. The reactions were terminated by addition of 190 μL stopping cum precipitating buffer (0.125 μg/μL tRNA, 0.075 mM thiourea, 300 mM sodium acetate at pH 7.0). Each reaction was precipitated with ethanol and the pellet resuspended in 15 μL FLS. The products were analyzed on a 15% polyacrylamide 8 M urea gel and quantified using a Bio-Rad Molecular Imager FX system.
Fractional protection of a particular position was determined by subtracting from it the counts at the same nucleotide position but from the lane without hydroxyl radical treatment. The counts from “control” positions, whose protection does not change with docking, were also included in the calculations to account for gel loading differences. The control positions for the different bases were 9 for helix 2, 23 for nucleotides 25–27, and 45 for 3′ Rz.
Native gel electrophoresis
A noncleavable (deoxy A − 1) construct (Walter et al. 1998) was used, in which the 3′ Rz is attached to the substrate via a pentacytidine linker, RLS (Hampel and Burke 2001). RLS with sequence 5′-GGUCGUGGUACAUUACCUGGUACCCCCUCGCdAGUCCUAUUU-3′ promotes folding in the presence of 5′ Rz but inhibits cleavage. The negative control strand is of the same sequence except for a G + 1A modification instead of deoxy A − 1 (Hampel et al. 1998; Walter et al. 1998), which prevents docking of the two domains of the ribozyme. Each variant 5′ Rz (5 pmol in 10 μL) is incubated in nondenaturing buffer (0.4 M Tris-acetate at pH 7.5 and 0.25 M Mg-acetate), 5% glycerol, and a trace amount of 5′-32P-end-labeled RLS for 10 min at 25°C and then run on a 10% nondenaturing gel in the same buffer for 18 h at 4°C.
ACKNOWLEDGMENTS
We thank Dr. Ken Hampel and Dr. Iwona Buskiewicz for valuable scientific discussions, Mike Fay for RNA synthesis, and Anne MacLeod for assistance in manuscript preparation. This work was supported by the U.S. National Institutes of Health, award no. AI044186.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.716108.
REFERENCES
- Anderson, P., Monforte, J., Tritz, R., Nesbitt, S., Hearst, J., Hampel, A. Mutagenesis of the hairpin ribozyme. Nucleic Acids Res. 1994;22:1096–1100. doi: 10.1093/nar/22.6.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzal-Herranz, A., Joseph, S., Chowrira, B.M., Butcher, S.E., Burke, J.M. Essential nucleotide sequences and secondary structure elements of the hairpin ribozyme. EMBO J. 1993;12:2567–2573. doi: 10.1002/j.1460-2075.1993.tb05912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, S.E., Burke, J.M. Structure-mapping of the hairpin ribozyme. Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme–substrate complex. J. Mol. Biol. 1994;244:52–63. doi: 10.1006/jmbi.1994.1703. [DOI] [PubMed] [Google Scholar]
- Butcher, S.E., Heckman, J.E., Burke, J.M. Reconstitution of hairpin ribozyme activity following separation of functional domains. J. Biol. Chem. 1995;270:29648–29651. doi: 10.1074/jbc.270.50.29648. [DOI] [PubMed] [Google Scholar]
- Buzayan, J.M., Gerlach, W.L., Bruening, G. Satellite tobacco ringspot virus RNA: A subset of the RNA sequence is sufficient for autolytic processing. Proc. Natl. Acad. Sci. 1986a;83:8859–8862. doi: 10.1073/pnas.83.23.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan, J.M., Hampel, A., Bruening, G. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res. 1986b;14:9729–9743. doi: 10.1093/nar/14.24.9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan, J.M., Feldstein, P.A., Bruening, G., Eckstein, F. RNA mediated formation of a phosphorothioate diester bond. Biochem. Biophys. Res. Commun. 1988a;156:340–347. doi: 10.1016/s0006-291x(88)80846-5. [DOI] [PubMed] [Google Scholar]
- Buzayan, J.M., Feldstein, P.A., Segrelles, C., Bruening, G. Autolytic processing of a phosphorothioate diester bond. Nucleic Acids Res. 1988b;16:4009–4023. doi: 10.1093/nar/16.9.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowrira, B.M., Berzal-Herranz, A., Keller, C.F., Burke, J.M. Four ribose 2′-hydroxyl groups essential for catalytic function of the hairpin ribozyme. J. Biol. Chem. 1993;268:19458–19462. [PubMed] [Google Scholar]
- Esteban, J.A., Banerjee, A.R., Burke, J.M. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem. 1997;272:13629–13639. doi: 10.1074/jbc.272.21.13629. [DOI] [PubMed] [Google Scholar]
- Esteban, J.A., Walter, N.G., Kotzorek, G., Heckman, J.E., Burke, J.M. Structural basis for heterogeneous kinetics: Reengineering the hairpin ribozyme. Proc. Natl. Acad. Sci. 1998;95:6091–6096. doi: 10.1073/pnas.95.11.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor, M.J. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- Grasby, J.A., Mersmann, K., Singh, M., Gait, M.J. Purine functional groups in essential residues of the hairpin ribozyme required for catalytic cleavage of RNA. Biochemistry. 1995;34:4068–4076. doi: 10.1021/bi00012a025. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Burke, J.M. A conformational change in the “loop E-like” motif of the hairpin ribozyme is coincidental with domain docking and is essential for catalysis. Biochemistry. 2001;40:3723–3729. doi: 10.1021/bi0028385. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Burke, J.M. Solvent protection of the hammerhead ribozyme in the ground state: Evidence for a cation-assisted conformational change leading to catalysis. Biochemistry. 2003;42:4421–4429. doi: 10.1021/bi020659c. [DOI] [PubMed] [Google Scholar]
- Hampel, A., Tritz, R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Hampel, A., Tritz, R., Hicks, M., Cruz, P. “Hairpin” catalytic RNA model: Evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990;18:299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel, K.J., Walter, N.G., Burke, J.M. The solvent-protected core of the hairpin ribozyme–substrate complex. Biochemistry. 1998;37:14672–14682. doi: 10.1021/bi981083n. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Pinard, R., Burke, J.M. Catalytic and structural assays for the hairpin ribozyme. Methods Enzymol. 2001;341:566–580. doi: 10.1016/s0076-6879(01)41177-3. [DOI] [PubMed] [Google Scholar]
- Hegg, L.A., Fedor, M.J. Kinetics and thermodynamics of intermolecular catalysis by hairpin ribozymes. Biochemistry. 1995;34:15813–15828. doi: 10.1021/bi00048a027. [DOI] [PubMed] [Google Scholar]
- Klostermeier, D., Millar, D.P. Tertiary structure stability of the hairpin ribozyme in its natural and minimal forms: Different energetic contributions from a ribose zipper motif. Biochemistry. 2001;40:11211–11218. doi: 10.1021/bi010773f. [DOI] [PubMed] [Google Scholar]
- Kuzmin, Y.I., Da Costa, C.P., Fedor, M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Liu, S., Bokinsky, G., Walter, N.G., Zhuang, X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting.”. Proc. Natl. Acad. Sci. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt, S., Hegg, L.A., Fedor, M.J. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- Park, H., Lee, S. Role of solvent dynamics in stabilizing the transition state of RNA hydrolysis by hairpin ribozyme. J Chem. Theory Comput. 2006;2:858–862. doi: 10.1021/ct0503015. [DOI] [PubMed] [Google Scholar]
- Pinard, R., Lambert, D., Walter, N.G., Heckman, J.E., Major, F., Burke, J.M. Structural basis for the guanosine requirement of the hairpin ribozyme. Biochemistry. 1999;38:16035–16039. doi: 10.1021/bi992024s. [DOI] [PubMed] [Google Scholar]
- Pinard, R., Hampel, K.J., Heckman, J.E., Lambert, D., Chan, P.A., Major, F., Burke, J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert, P.B., Ferré-D'Amaré, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B., Massey, A.P., Sigurdsson, S.T., Ferré-D'Amaré, A.R. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Strobel, S.A. Nucleotide analog interference mapping of the hairpin ribozyme: Implications for secondary and tertiary structure formation. J. Mol. Biol. 1999;291:295–311. doi: 10.1006/jmbi.1999.2959. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Oyelere, A.K., Padilla, J.L., Klostermeier, D., Millar, D.P., Strobel, S.A. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- Salter, J., Krucinska, J., Alam, S., Grum-Tokars, V., Wedekind, J.E. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargueil, B., Pecchia, D.B., Burke, J.M. An improved version of the hairpin ribozyme functions as a ribonucleoprotein complex. Biochemistry. 1995;34:7739–7748. doi: 10.1021/bi00023a021. [DOI] [PubMed] [Google Scholar]
- Sargueil, B., Hampel, K.J., Lambert, D., Burke, J.M. In vitro selection of second site revertants analysis of the hairpin ribozyme active site. J. Biol. Chem. 2003;278:52783–52791. doi: 10.1074/jbc.M306703200. [DOI] [PubMed] [Google Scholar]
- Shippy, R., Siwkowski, A., Hampel, A. Mutational analysis of loops 1 and 5 of the hairpin ribozyme. Biochemistry. 1998;37:564–570. doi: 10.1021/bi9721288. [DOI] [PubMed] [Google Scholar]
- Torelli, A.T., Krucinska, J., Wedekind, J.E. A comparison of vanadate to a 2′-5′ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, N.G., Hampel, K.J., Brown, K.M., Burke, J.M. Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J. 1998;17:2378–2391. doi: 10.1093/emboj/17.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, N.G., Burke, J.M., Millar, D.P. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- Walter, N.G., Chan, P.A., Hampel, K.J., Millar, D.P., Burke, J.M. A base change in the catalytic core of the hairpin ribozyme perturbs function but not domain docking. Biochemistry. 2001;40:2580–2587. doi: 10.1021/bi001609f. [DOI] [PubMed] [Google Scholar]
- Wilson, T.J., Nahas, M., Ha, T., Lilley, D.M. Folding and catalysis of the hairpin ribozyme. Biochem. Soc. Trans. 2005;33:461–465. doi: 10.1042/BST0330461. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Burke, J.M. Inhibition of viral replication by ribozyme: Mutational analysis of the site and mechanism of antiviral activity. J. Virol. 2005;79:3728–3736. doi: 10.1128/JVI.79.6.3728-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z.Y., Wilson, T.J., Maxwell, K., Lilley, D.M. The folding of the hairpin ribozyme: Dependence on the loops and the junction. RNA. 2000;6:1833–1846. doi: 10.1017/s1355838200001230. [DOI] [PMC free article] [PubMed] [Google Scholar]