Abstract
In yeast (Saccharoymces cerevisiae), the branchpoint binding protein (BBP) recognizes the conserved yeast branchpoint sequence (UACUAAC) with a high level of specificity and affinity, while the human branchpoint binding protein (SF1) binds the less-conserved consensus branchpoint sequence (CURAY) in human introns with a lower level of specificity and affinity. To determine which amino acids in BBP provide the additional specificity and affinity absent in SF1, a panel of chimeric SF1 proteins was tested in RNA binding assays with wild-type and mutant RNA substrates. This approach revealed that the QUA2 domain of BBP is responsible for the enhanced RNA binding affinity and specificity displayed by BBP compared with SF1. Within the QUA2 domain, a transposition of adjacent arginine and lysine residues is primarily responsible for the switch in RNA binding between BBP and SF1. Alignment of multiple branchpoint binding proteins and the related STAR/GSG proteins suggests that the identity of these two amino acids and the RNA target sequences of all of these proteins are correlated.
Keywords: KH domain, branchpoint binding protein, splicing factor 1, branchpoint sequence, pre-mRNA splicing
INTRODUCTION
There are three essential RNA elements for pre-mRNA splicing, the 5′ splice site, the 3′ splice site, and the branchpoint sequence. These three elements are necessary for the two transesterification reactions that remove an intron and ligate two exons together (Reed 1996). In the first reaction, the 2′ OH of the branchsite adenosine is used as the nucleophile to attack the phosphodiester bond between the 5′ exon and 5′ splice site, creating the free 5′ exon and lariat intermediate. In the second reaction, the 3′ OH of the free 5′ exon is the nucleophile that attacks the phosphodiester bond between the 3′ splice site and downstream exon, creating the ligated exons and excised intron. These steps are catalyzed by the spliceosome, which is composed of five snRNAs and ∼150 proteins (Staley and Guthrie 1998; Will and Lührmann 2006).
Although the branchpoint sequence, or more specifically, a branchsite nucleophile, is essential for splicing, this motif is less conserved in comparison with the 5′ and 3′ splice sites across eukaryotic organisms. In human introns the branchpoint sequence is degenerate with the consensus sequence of CURAY (R=purine, Y=pyrimidine) (Keller and Noon 1984). For some human introns it is difficult to identify a putative branchpoint sequence, and mutating a mapped branchsite often results in the splicing machinery using a cryptic branchsite (Ruskin et al. 1985). Multiple branchsites within a single human intron may be used to create different 3′ splice sites, resulting in different mRNAs (Wang et al. 2007). In contrast to the degenerate branchpoint sequence in humans, in yeast (Saccharoymces cerevisiae), the branchpoint sequence is almost always UACUAAC (Langford and Gallwitz 1983; Pikielny et al. 1983).
The branchpoint sequence is specifically recognized at least twice in splicing, first by the branchpoint binding protein and then subsequently by U2 snRNP through an RNA–RNA interaction in which the branchsite adenosine is unpaired and bulged out for use as a nucleophile in the first reaction of splicing (Query et al. 1994, 1996). The degeneracy of the branchpoint sequence in humans does not correlate with the U2 snRNA sequence, because the GUAGUA sequence of U2 snRNA that base pairs with the branchpoint sequence is highly conserved. Although natural branchpoint sequences in human introns are highly variable, the yeast UACUAAC sequence has been shown to be an optimal branchpoint sequence for splicing in the human system (Zhuang et al. 1989).
The branchpoint binding protein, referred to as branchpoint binding protein (BBP) in yeast (Berglund et al. 1997) and splicing factor 1 (SF1) in human (Kramer 1988) is a highly conserved protein that contains multiple RNA binding domains: a K homology (KH) domain, a QUA2 domain, and either one or two zinc knuckles (Berglund et al. 1998b; Rain et al. 1998). The KH domain is a common RNA binding domain that can be subdivided into two subclasses: types I and II. Type I domains, which include the KH domain of BBP/SF1, fold into a βααββα topology, while type II KH domains fold into an αββααβ topology (Grishin 2001; Auweter et al. 2006). KH domains bind RNA in a cleft between two loops: a small loop between the second and third α helices consisting of a conserved GXXG motif and a large “variable loop” of nonconserved sequence between β strands two and three.
A subclass of type I KH proteins called the signal transduction activator of RNA (STAR) family or the GRP33-Sam68-GLD-1 (GSG) family have KH domains that are extended at both ends (Vernet and Artzt 1997; Lukong and Richard 2003). The term STAR/GSG will be used below when referring to this subclass of KH domain containing proteins. The STAR/GSG proteins have extended KH domains with a QUA1 domain at the N-terminal side and a QUA2 domain at the C-terminal side. The QUA1 domain is responsible for homodimerization (Chen et al. 1997; Chen and Richard 1998). BBP/SF1 is unique among STAR/GSG proteins, in that it lacks a QUA1 domain. The NMR structure of SF1 in complex with the yeast branchpoint sequence UACUAAC RNA showed that the QUA2 domain is an α helix that extends the RNA binding site of these KH domains (Liu et al. 2001). Specifically, the QUA2 domain of SF1 interacts with the 5′ nucleotides of the branchpoint sequence RNA, the underlined nucleotides in the UACUAAC sequence (Liu et al. 2001).
Another notable feature of the STAR/GSG proteins is that they contain a single KH domain, whereas most other KH domain proteins contain multiple KH domains (Gibson et al. 1993; Burd and Dreyfuss 1994). Homodimerization significantly enhances the RNA binding affinity of the STAR/GSG proteins (Chen and Richard 1998; Chow et al. 2000; Ryder and Williamson 2004). Lacking the QUA1 domain, BBP/SF1 does not homodimerize; instead, it interacts with Mud2 in yeast or U2AF65 in human, and these proteins bind cooperatively to the branchpoint sequence and adjacent polypyrimidine tract at the 3′ end of the intron (Abovich and Rosbash 1997; Berglund et al. 1998a). Finally, both BBP and SF1 contain zinc knuckles, which are C-terminal of the QUA2 domains. BBP contains two zinc knuckles, the first of which enhances RNA binding, while the second zinc knuckle may slightly inhibit binding (Garrey et al. 2006). SF1 contains only one zinc knuckle and its presence enhances RNA binding (Berglund et al. 1998b). Zinc knuckles have been shown to be important for RNA binding in other proteins like the HIV-1 nucleocapsid protein (Dannull et al. 1994; De Guzman et al. 1998) and the human SR protein 9G8 (Cavaloc et al. 1999).
Although BBP and SF1 are orthologous proteins, these two proteins have different affinities for the branchpoint sequence and different specificities. These differences between BBP and SF1 are not surprising, because the yeast branchpoint sequence is the highly conserved UACUAAC (Langford and Gallwitz 1983; Pikielny et al. 1983), while human branchpoints are characterized by a degenerate consensus sequence, CURAY (Keller and Noon 1984). BBP binds RNAs containing the branchpoint sequence of UACUAAC with nanomolar affinity (Garrey et al. 2006). Mutational analysis of the branchpoint sequence along with SELEX has demonstrated the importance of all seven positions of the yeast branchpoint sequence for sequence-specific binding by BBP (Berglund et al. 1997; Garrey et al. 2006). Meanwhile, human SF1 binds the yeast BPS with more than 200-fold weaker affinity and much less specificity than that of BBP (Berglund et al. 1997; Garrey et al. 2006).
In this study, chimeras of BBP and SF1 were generated to determine which amino acids are responsible for their significant differences in branchpoint sequence RNA binding affinity and specificity. We find that the QUA2 domains of BBP and SF1 are primarily responsible for the differences in specificity and affinity between BBP and SF1. Within the QUA2 domain, a transposition of two amino acids confers yeast BBP-like RNA binding affinity and specificity to the human SF1 for the yeast branchpoint RNA sequence.
RESULTS
Difference in RNA binding affinity and specificity between BBP and SF1
Previously we had shown qualitatively that there are significant differences in RNA binding affinity and specificity between SF1 and BBP (Berglund et al. 1997). Here, we have quantified the differences between the two proteins. The wild-type protein constructs used for comparison contained only the RNA binding domains: KH, QUA2, and one zinc knuckle (Fig. 2, below). BBP contains a second C-terminal zinc knuckle, but this domain was excluded because we had previously shown it is not involved in RNA binding (Garrey et al. 2006). The BBP construct consisted of amino acids 147–296 and will be referred to as BBP-296. The SF1 construct consisted of amino acids 134–297 and will be referred to as SF1-297.
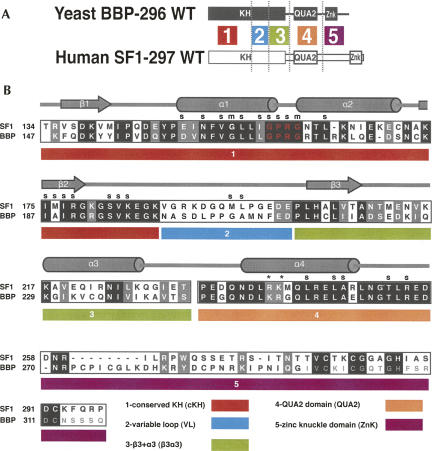
FIGURE 2.
A map of the five regions used to make the BBP and SF1 chimeras. (A) The domain structure of BBP-296 (black) and SF1-297 (white), and placement of the five different regions. (B) An alignment of BBP-296 and SF1-297 and detailed map of the five regions underneath. A key of the five regions and their names are listed at the bottom of the alignment. In the alignment, black boxes show identical amino acids between BBP and SF1, and gray boxes represent similar amino acids between the two proteins. The GXXG motif is shown in red letters. Secondary structure and contacts to the RNA are marked (s for side chain and m for main chain RNA contacts, cylinders for α-helices, and arrows for β-strands) based on the human SF1 NMR structure (Liu et al. 2001). An asterisk (*) above the alignment marks the important arginine (R) and lysine (K) discussed in Figure 6. The chimeras were named according to the region of the protein that was replaced with the yeast sequence and a y placed before the region name to represent yeast. For example, in the construct SF1-ycKH, the yeast-conserved KH (cKH) region was exchanged for the homologous human cKH region. The second zinc knuckle of BBP was not used in the protein constructs for binding studies, and those amino acids are shown in a smaller light gray in the alignment.
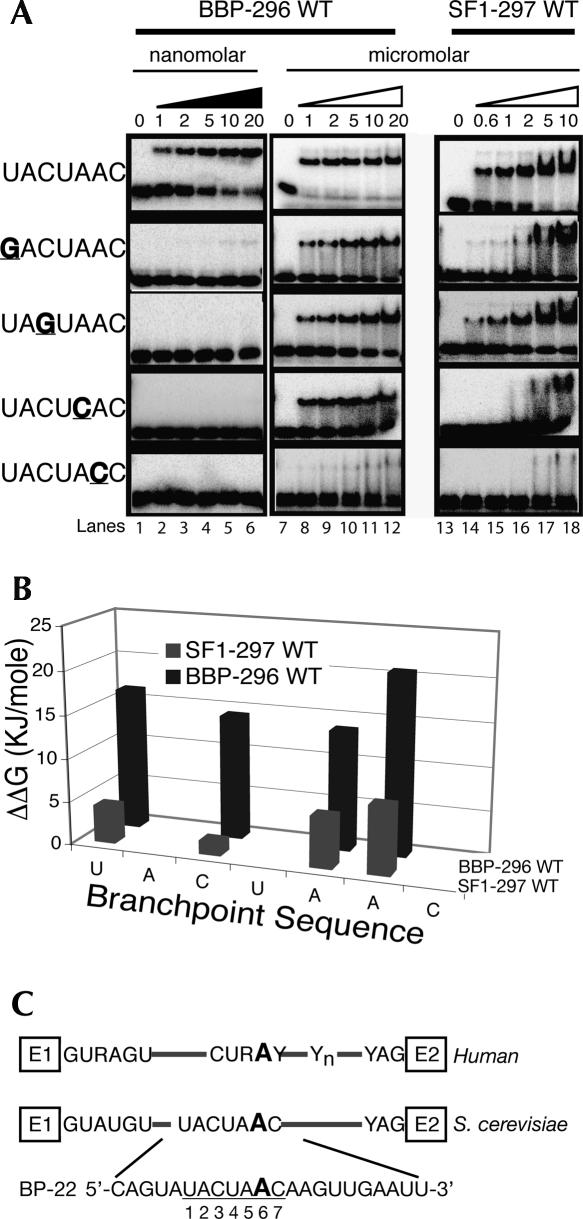
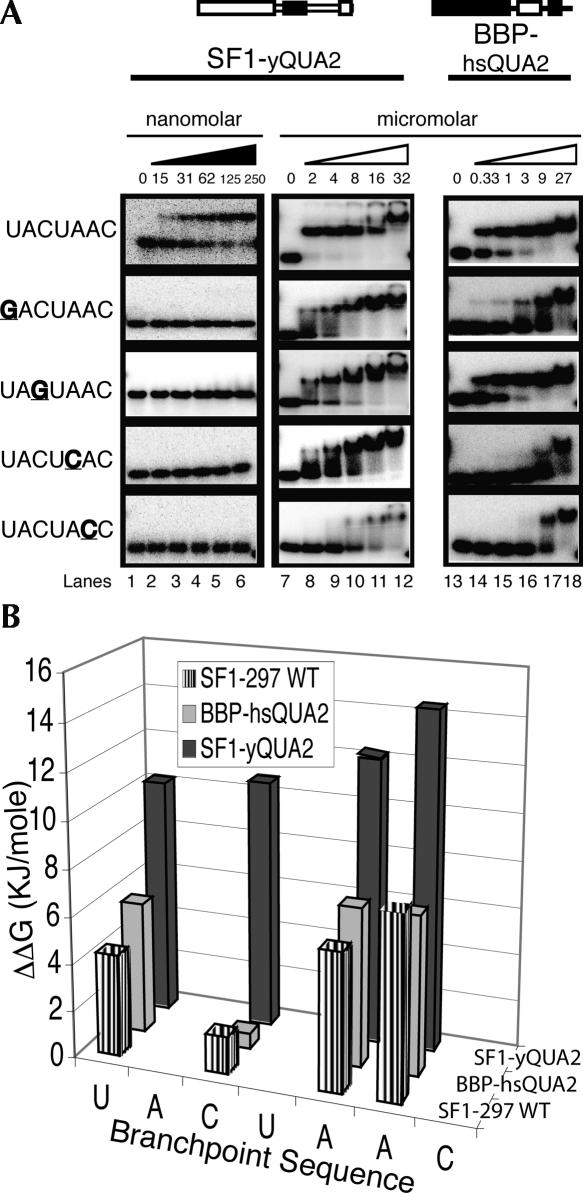
The affinities of BBP-296 and SF1-297 for a 22-mer RNA (BP-22, Fig. 1C) containing the yeast consensus branchpoint sequence UACUAAC were measured. BBP-296 bound the BP-22 RNA with a Kd of 3 nM (Fig. 1A, lanes 1–6; Table 1), while SF1-297 bound this RNA with a Kd of only 740 nM (Fig. 1, lanes 13–18; Table 1). Therefore, these proteins have an approximate 250-fold difference in their affinity for the optimal branchpoint sequence.
FIGURE 1.
A quantitative comparison of the RNA binding specificity of BBP and SF1. (A) Binding to the BP-22 RNA is shown in the top row of the gels. BP-22 RNA contains the wild-type branchpoint sequence and is labeled on the left as UACUAAC. Binding to the mutant RNAs are below the wild-type UACUAAC and labeled on the left. Point mutations are underlined and in bold. Concentrations are listed underneath the triangles. (Lanes 1–6) Binding to BBP-296 in the low nanomolar range, (black triangle). To determine Kds for the mutant RNAs, binding in the micromolar range (white triangle) was required (lanes 7–12). (B) A bar graph comparing the specificities measured in ΔΔG for the point mutations tested in A with BBP and SF1. (C) A schematic of yeast and human introns are shown with the BP-22 RNA sequence used in these studies shown at the bottom. The branchsite adenosine used as the nucleophile in the first step of chemistry is underlined and in a larger font.
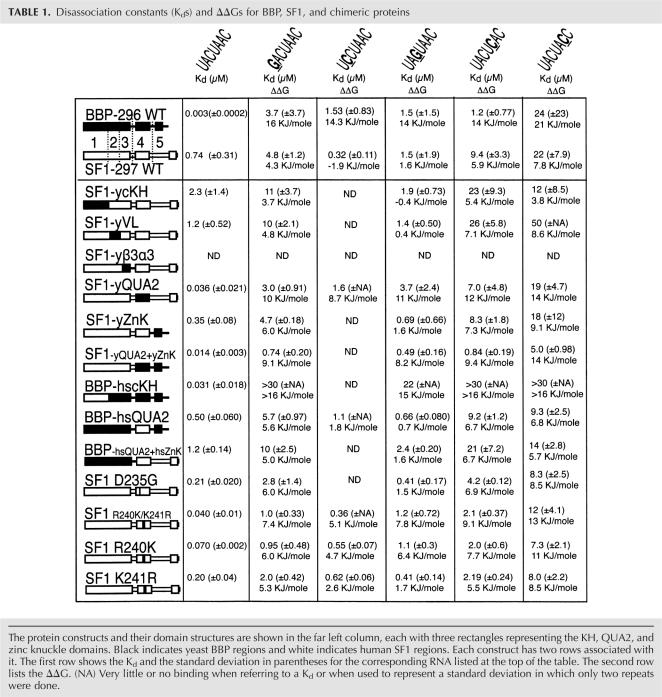
TABLE 1.
Disassociation constants (Kds) and ΔΔGs for BBP, SF1, and chimeric proteins
To compare the specificities of BBP-296 and SF1-297, we tested their binding to four mutant RNAs with different point mutations within the branchpoint sequence. The positions of the branchpoint sequence are numbers 1–7 and mutants will be referred to in the following format, wild-type nucleotide–position–mutant nucleotide (i.e., U1G). The binding of BBP to these mutant RNAs demonstrates the high-sequence specificity of the yeast protein (Fig. 1A). Whereas BBP-296 binds the wild-type branchpoint sequence with a Kd in the low nanomolar range, its apparent Kd for each of the mutant sequences is in the micromolar range (Fig. 1, lanes 7–12). Mutating any of these four nucleotides results in almost 400-fold to over 7000-fold weaker binding by BBP (Fig. 1A). This is shown graphically in Figure 1B, in which ΔΔG [ΔΔG=−RTln(Kd1/Kd2)] reflects the difference in affinity of a protein for two different RNA substrates and can be used as a measure of sequence specificity (Fig. 1B). For the human protein, SF1-297, these same RNA mutations reduce binding affinities between approximately twofold and 29-fold, or in terms of ΔΔG, from 1.6 KJ/mol to 7.8 KJ/mol (Fig. 1B; Table 1). These results show that BBP-296 is highly sensitive to changes in the branchpoint sequence, whereas SF1-297 has relaxed sequence specificity (Table 1).
Determining the regions of BBP and SF1 that are responsible for the differences in RNA binding affinity and specificity
To identify the region or regions of BBP-296 that give it higher affinity and specificity over SF1-297, we created chimeric proteins in which each of five segments of BBP-296 replaced the corresponding region of SF1-297 (Fig. 2A). The alignment of BBP and SF1, as well as the domain architecture, were used to delineate the regions that were exchanged (Fig. 2B). The first region, which we call the conserved KH (cKH), (SF1 amino acids 134–187) contains the first half of the KH domain. It contains the most conserved part of the KH domain, and includes the GXXG motif and the first two β-strands and α-helices. The second region (SF1 amino acids 188–201) contains the variable loop (VL), which is the least conserved region of the protein. Region three is on the “backside” of the protein, away from the RNA binding site, and contains the third α-helix and third β-sheet (SF1 amino acids 202–232). The last two regions are the QUA2 domain (SF1 amino acids 233–257) and the zinc knuckle domain (ZnK) (amino acids 258–297).
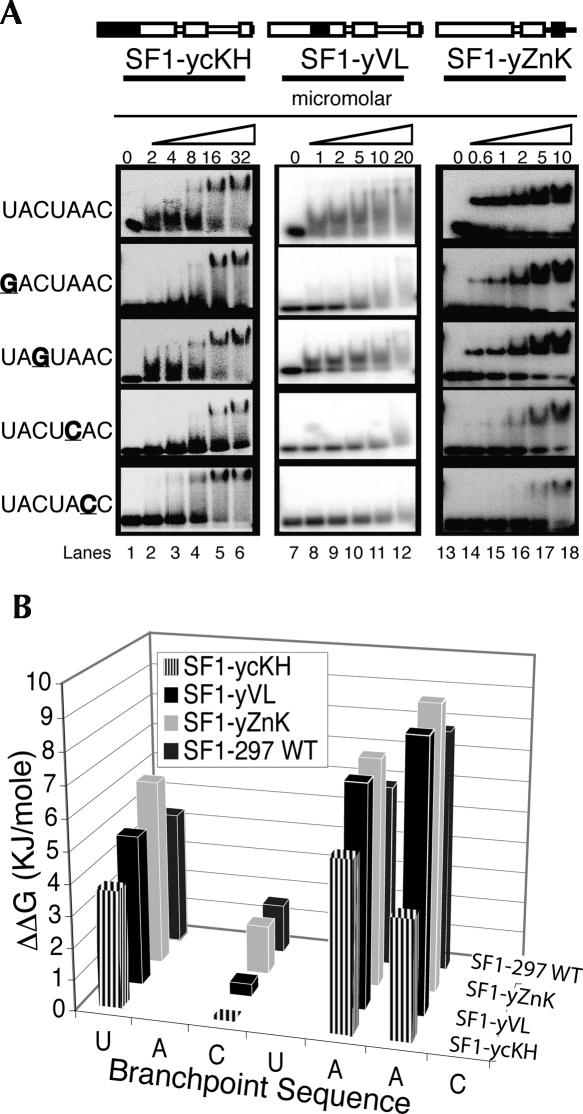
The human SF1-297 construct was exchanged with one of each of the five different regions from yeast BBP-296: creating five different yeast–human chimeras; SF1-ycKH, SF1-yVL, SF1-yβ3α3, SF1-yQUA2, and SF1-yZnK (see Table 1). These five chimeric proteins were made and assayed for binding to the wild-type BP-22 RNA (UACUAAC) and the four mutant RNAs as shown in Figure 1. Four of the five chimeric proteins bind RNA, but SF1-yβ3α3 did not (data not shown), suggesting that this protein was not properly folded. This result is not surprising, because amino acids from this region are part of the core of the protein (Liu et al. 2001). The RNA binding data for SF1-ycKH, SF1-yVL, and SF1-yZnK are shown in Figure 3. The RNA binding specificity and affinity for these three chimeras are similar to SF1-297 (Table 1). The subtle differences between SF1-297 and the chimeras included modestly reduced sequence specificity of one chimera and a general enhancement of RNA binding affinity for another (Fig. 3B). The sequence specificity of SF1-ycKH is reduced compared with SF1-297, specifically at C3 (1.6 KJ/mole for SF1-297 versus −0.4 KJ/mole for SF1-ycKH) and A6 (7.8 KJ/mole for SF1-297 versus 3.8 KJ/mole for SF1-ycKH) of the branchpoint sequence (Fig. 3; Table 1). Meanwhile, SF1-yZnK binds the wild-type RNA with a twofold lower Kd in comparison with the human SF1-297 (Fig. 3; Table 1). The general RNA binding affinity was enhanced for SF1-yZnK, and only minimal changes in specificity were observed when the zinc knuckles were exchanged between the yeast and human proteins (Table 1, cf. ΔΔGs for SF1-297 and SF1-yZnK for mutant RNAs), consistent with previous research (Berglund et al. 1998b; Garrey et al. 2006).
FIGURE 3.
RNA binding specificities for three BBP-SF1 chimeras. (A) All three have a different yeast BBP region exchanged into the SF1-297 protein, the conserved KH (cKH) (lanes 1–6), the variable loop (VL) (lanes 7–12), and the yeast first zinc knuckle (ZnK) (lanes 13–18). RNA binding is in the micromolar range with concentrations listed above the gels. (B) A bar graph comparing the binding specificities (ΔΔG) of SF1–297 wild-type protein and the three chimeras analyzed in A.
The QUA2 domain is primarily responsible for the differences in specificity and affinity between BBP and SF1
In contrast to the other chimeras, SF1-yQUA2 binds the wild-type RNA and the mutant RNAs with dramatically altered affinity and specificity compared with SF1-297. The RNA binding properties of SF1-yQUA2 are very similar to those of BBP-296. Figure 4 (lanes 1–6) shows that SF1-yQUA2 binds BP-22 with a Kd of 36 nM, which is more than 20-fold tighter than wild-type SF1-297 and only 12-fold weaker than BBP-296 (Fig. 1, lanes 1–6). Again, similar to BBP-296, affinity of this protein for the mutant RNAs is drastically reduced (Fig. 4A, lanes 7–12). Comparing the ΔΔGs for SF1-yQUA2 and BBP-296 (Fig. 4B; Table 1) shows that much of the enhanced binding by SF1-yQUA2 can be attributed to interactions with the C3 of the branchpoint sequence (11 KJ/mole for SF1-yQUA2 versus 1.6 KJ/mole for SF1-297) and U1, but other positions are affected as well (Table 1). These results clearly show that the QUA2 domain is important in modulating the specificity of these two proteins. The remaining differences in ΔΔG between SF1-yQUA2 and BBP-296 for the different RNAs are likely the result of subtle effects caused by amino acid differences in other regions of the protein.
FIGURE 4.
Exchanging the QUA2 domains of SF1 and BBP result in transformed RNA binding specificities and affinities. (A) Lanes 1–6 show gel-shift assays for SF1-yQUA2 in a low nanomolar concentration range (black triangle). For this construct, the yeast QUA2 domain replaces the human QUA2. Lanes 7–12 show SF1-QUA2 binding the RNAs with protein in the micromolar range. In lanes 13–18, BBP-hsQUA2 protein binds the RNAs with protein concentrations in the micromolar range. BBP-hsQUA2 is a BBP-296 protein in which the human QUA2 replaces the yeast QUA2. (B) A bar graph comparing the binding specificity (ΔΔG) of SF1-297 wild-type protein and the two chimeras, SF1-yQUA2 and BBP-hsQUA2.
To determine whether or not the addition of the yeast zinc knuckle (region 5) to SF1-yQUA2 would enhance affinity but not alter the specificity, the human zinc knuckle was replaced by the yeast zinc knuckle domain (SF1-yQUA2+yZnK). This change does indeed enhance binding and the Kd for BP-22 drops from 36 to 14 nM (data not shown; Table 1). The specificity of SF1-yQUA2 and SF1-yQUA2+yZnK for BP-22 RNA and the mutant RNAs remain approximately the same as shown by the similar ΔΔGs for these chimeras (Table 1), indicating that the zinc knuckle only affects the general affinity of the protein–RNA interaction.
Given the observations above, we expected that replacing the QUA2 domain in BBP with the QUA2 domain from SF1 should drastically reduce the affinity and specificity of this chimera (BBP-hsQUA2) for RNA. As expected, the affinity of BBP-hsQUA2 changed to a Kd of 496 nM for the BP-22 RNA, a 165-fold decrease in affinity, and the specificity of this protein was significantly reduced as well (Fig. 4A, lanes 13–18). Figure 4B shows that BBP-hsQUA2 displays the same relatively small differences in affinities for the variant branchpoint sequence RNAs as that of SF1-297 (Table 1), indicating similarly low-sequence specificity. These two chimeric proteins (SF1-yQUA2 and BBP-hsQUA2) demonstrate that the QUA2 domain plays a significant role in modulating the RNA binding affinity and specificity of both SF1 and BBP.
To again test the modular nature of the zinc knuckles, we replaced the first yeast zinc knuckle from BBP-hsQUA2 protein with the human zinc knuckle (BBP-hsQUA2+hsZnK). This change further reduced the binding affinity of the protein to all five RNAs from 1.5-fold to threefold (Table 1). The specificity (ΔΔGs) of this protein is no different compared with BBP-hsQUA2, indicating that the general affinity has been weakened but specificity has not been affected.
Transposing an arginine and lysine in the QUA2 domain endows SF1 with similar RNA binding characteristics to that of BBP
There are only five amino acid differences between the yeast and human QUA2 domains (see Fig. 2). To determine which of these amino acids are modulating the RNA binding we made individual (D235G, this numbering is based on SF1 sequence) and double mutations (D235G/M242G and R240K/K241R) in human SF1. These mutations covered four of the five amino acid differences between the SF1 and BBP QUA2 domains (Fig. 2). The D235G mutation removed a negative charge. We hypothesized that this would enhance the ability of the QUA2 domain to fully engage in binding due to the possible removal of charge repulsion with the phosphate backbone of the RNA. However, our results showed that the SF1-D235G mutation bound RNA with only threefold greater affinity than SF1-297 (Table 1). Specificity changes for this mutant were very modest, with only the U1G mutation exhibiting an effect compared with wild-type SF1 (ΔΔGs in Table 1). The double mutation D235G/M242G behaved the same as the single mutation D235G, indicating that M242 does not play a role in RNA affinity or specificity (data not shown).
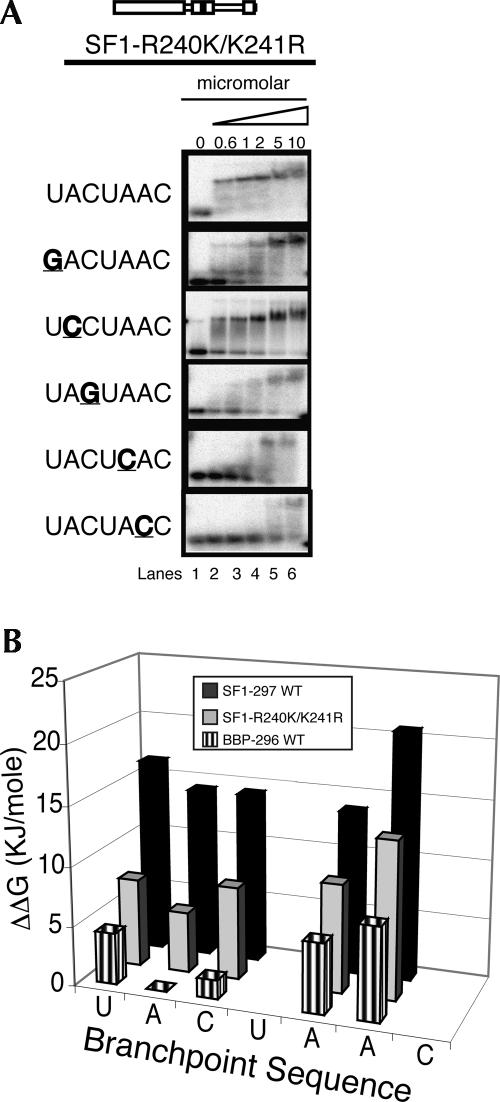
The R240K/K241R double mutation in human SF1-297 transposes adjacent arginine and lysine residues. Surprisingly, the exchange of these two similar amino acids has a significant effect on RNA binding specificity and affinity (Fig. 5). Similar to BBP-296 and SF1-yQUA2, the SF1-R240K/K241R protein binds with a Kd in the low nanomolar range to wild-type branchpoint RNA, and binds with drastically reduced affinity to the mutant RNAs (Fig. 5A, lanes 1–6). This indicates that R240 and K241 in human SF1 (or K252 and R253 in yeast) QUA2 domain are responsible for much of the differences in specificity and affinity between BBP and SF1.
FIGURE 5.
The transposition chimera, SF1-R240K/K241R binds the branchpoint sequence RNA like BBP. (A) Lanes 1–6 show binding in the micromolar concentration range for the mutant SF1 protein (triangle) to BP-22 RNA and five mutant RNAs. (B) A bar graph of the binding specificities (ΔΔG) of BBP-296, SF1-297, and SF1-R240K/K241R.
Because the QUA2 domain makes significant contacts with A2 (Liu et al. 2001), this position was mutated to a cytosine to determine its effect on SF1-R240K/K241R binding. As shown in Figure 5, A and B, the A2C mutation does affect SF1-R240K/K241R binding (Kd changes from 40 to 360 nM), but not nearly as significantly as this mutation disrupts BBP-296 binding (Fig. 5B, cf. ΔΔGs). While for SF1-297, the A2C mutation enhances binding slightly (Kd changes from 740 to 320 nM).
Single mutations in the QUA2 domain alter affinity and specificity of SF1
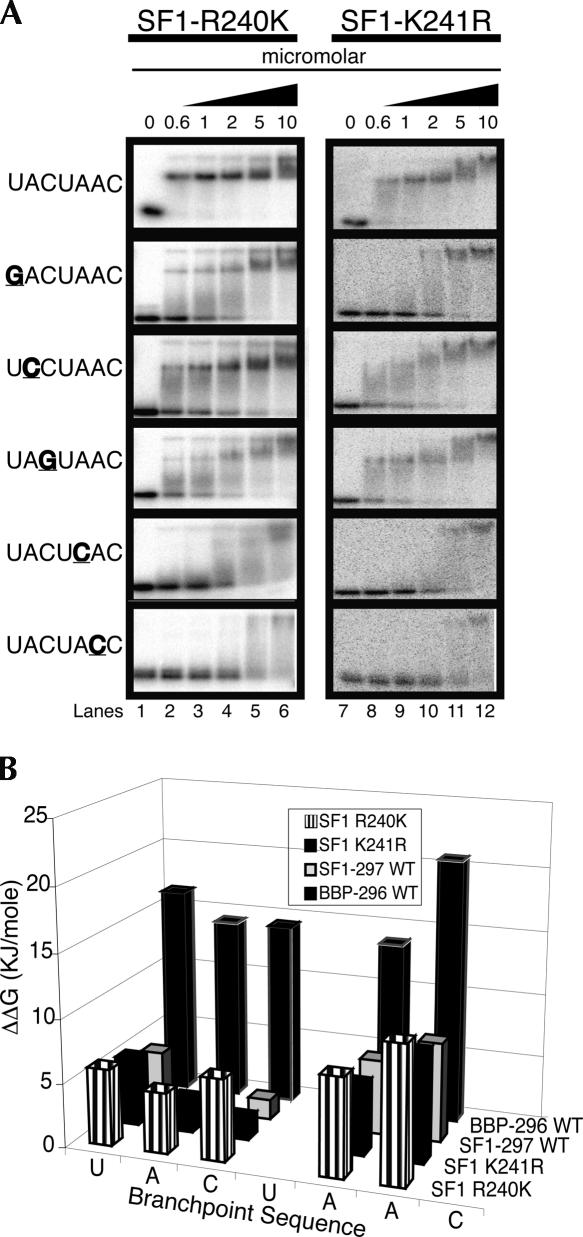
To determine the individual roles of R240 and K241 in the RNA binding of SF1 and BBP, these amino acids in SF1 were mutated individually to their yeast counterparts. Changing R240 to a lysine enhances binding 10-fold (Kd changes from 740 to 70 nM) (Fig. 6A), while changing K241 to an arginine enhances binding a little more than threefold (Kd changes from 740 to 200 nM) (Fig. 6A). With regard to specificity, Figure 6B shows that mutating A2 and C3 have a significant impact on the binding of SF1-R240K, while these RNA mutations have much less of an effect on the binding of SF1-K241R and SF1-297. These results suggest that both amino acids play a role in RNA binding and that the presence of an arginine at position 240 inhibits binding.
FIGURE 6.
Binding of arginine-lysine 240 (R240K) and lysine-arginine 241 (K241R) mutant SF1 proteins to BP-22 RNA and RNA mutants. (A) Lanes 1–6 show binding in the micromolar concentration range for the mutant SF1 proteins (black triangle) to BP-22 RNA and five mutant RNAs. (B) A bar graph of the binding specificities (ΔΔG) of BBP-296, SF1-297, SF1-R240K, and SF1-K241R.
Fluorescence anisotropy
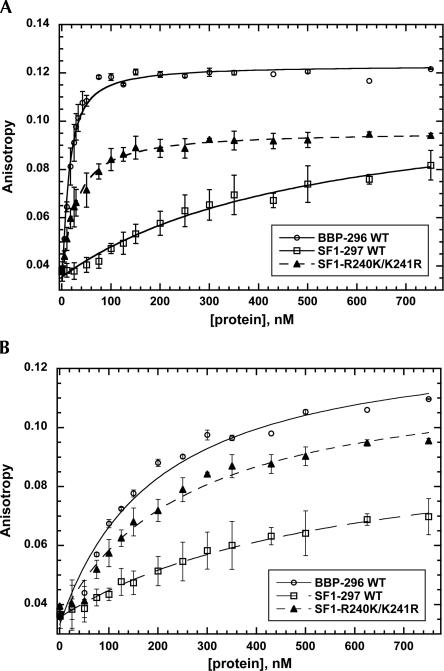
Fluorescent anisotropy measurements were used to confirm our gel-shift assay measurements, the BP-22 RNA and the A2C mutant RNAs were fluorescein labeled and used to measure the binding affinities of the SF1-297, BBP-296, and SF1-R240K/K241R proteins. Figure 7A shows that the binding affinity of the SF1-R240K/K241R protein is shifted closely to that of BBP-296, confirming that this transposed protein is binding BP-22 RNA much more like BBP-296 than SF1-297. The Kds derived from these curves are 11 nM for BBP-296, 30 nM for SF1-R240K/K241R, and 600 nM for SF1-297, while the Kds derived from the gel-shift experiments are 3 nM for BBP-296, 40 nM for SF1-R240K/K241R, and 740 nM for SF1-297. The measurements with the mutant RNA (A2C) show that all three proteins have reduced levels of binding; BBP-296 binds with a Kd of 250 nM, SF1-297 binds with a Kd of 620 nM, and SF1-R240K/K241R binds with a Kd of 270 nM (Fig. 7B). The measurements determined using the gel-shift assay and fluorescence anisotropy are in good agreement except for BBP-296, for which the Kds are shifted over threefold for BP-22 RNA and fivefold for the A2C mutant RNA. The use of two different methods to measure the RNA binding affinities of the various proteins and the demonstration that all of the proteins are between 40% and 100% active (Supplemental Fig. 1) shows that R240 and K241 play an important role in determining the RNA binding affinity and specificities of BBP and SF1.
FIGURE 7.
Fluorescence anisotropy of BP-22 RNA (A) and a mutant RNA (B) upon addition of BBP-296, SF1-297, or SF1-R240K/K241R. (A) As BP-22 RNA is titrated with BBP-296 (○), SF1-297 (□), or SF1-R240K/K241R (▴), increased anisotropy is observed, indicating protein–RNA binding. (B) Titration of BBP-296, SF1-297, or SF1-R240K/K241R with mutant RNA required higher concentrations of protein to achieve binding. No error bars are shown for points on the titration curve where a single measurement was taken.
DISCUSSION
Our results show that the QUA2 and zinc knuckle domains in the yeast and human branchpoint binding proteins can be used as modular domains to achieve different levels of RNA binding affinities and specificities. Replacing the zinc knuckle of human SF1 with that of yeast BBP mildly increases the overall RNA binding affinity, but does not significantly alter RNA binding specificity, indicating that these zinc knuckles are general RNA binding modules. Exchanging the QUA2 domains of SF1 and BBP significantly alters the RNA binding affinity and specificity of these proteins so that they mimic those of the protein from which the QUA2 domain was derived. This shows that the QUA2 domain plays an important role in modulating the RNA binding of these two proteins. Furthermore, a transposition of two adjacent basic amino acids (R240 and K241 to K240 and R241) within the QUA2 domain is sufficient to transform a RNA binding protein with low affinity and specificity into a protein that binds RNA with high affinity and specificity.
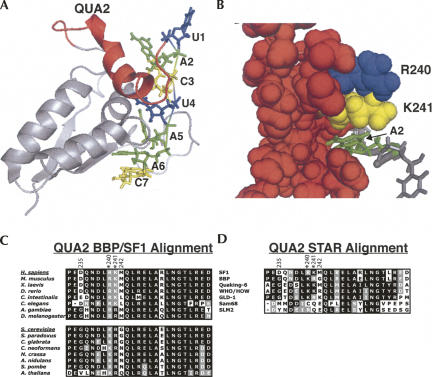
The NMR structure of SF1 in complex with the yeast branchpoint sequence shows that the QUA2 domain interacts with the 5′ end of the RNA (Fig. 8A), specifically A2, C3, and U4 (Liu et al. 2001), and that K241 sits on top of A2 (Fig. 8B). As one might predict, changing the lysine at position 241 to an arginine enhances binding, because favorable stacking interactions likely occur between the more planar side chain of arginine and the adenosine base, while lysine's conformations do not allow stacking interactions to occur (Flocco and Mowbray 1994). In the STAR/GSG family protein, GLD-1, it was found that the equivalent amino acid to K241 (K314) is important for RNA binding (Lehmann-Blount and Williamson 2005). Interestingly, it is R240 that has the most significant affect on binding, but R240 is distant from A2 and any other nucleotides based on the NMR structure (Fig. 8B). One possible interpretation of our data is that when R240 is mutated to a lysine, the QUA2 adopts a conformation facilitating additional interactions between the QUA2 and the RNA, thus, the observed enhanced binding affinity and specificity.
FIGURE 8.
Analyzing the QUA2 domain structure from the NMR structure of SF1 in complex with the branchpoint sequence RNA (PDB #1K1G) (Liu et al. 2001) and alignments of branchpoint binding proteins from multiple organisms and the related STAR/GSG proteins. (A) The QUA2 domain (red) relative to the KH domain and branchpoint sequence. The nucleotides of the branchpoint sequence are numbered 1 through 7. (B) A spaced-filled view of R240 (blue) and K241 (yellow) and the rest of the QUA2 domain (red) binding RNA. The A2 nucleotide (green) fits in a pocket formed by the QUA2 domain. (C) An alignment of QUA2 domains of SF1, BBP, and branchpoint binding proteins from different organisms. (D) A QUA2 domain alignment of SF1, BBP, and other STAR/GSG proteins. Accession numbers for these proteins can be found in Supplemental Table 1.
The U2 snRNA sequence used to base pair with the branchpoint sequence is highly invariant, yet the branchpoint sequence varies amongst organisms as do the RNA binding affinities and specificities of the branchpoint binding proteins, raising the intriguing possibility that the evolution of this protein and the branchpoint sequence are linked. Specifically, we propose that the identity of amino acids 240 and 241 coevolved with the branchpoint sequences. In general, fungal introns have branchpoint sequences that are more similar to the highly conserved UACUAAC branchpoint sequence of Saccharomyces cerevisiae introns (Bon et al. 2003), and the fungal branchpoint binding proteins all have lysine at position 240 and arginine at 241 (Fig. 8C). In contrast, the branchpoint sequences in vertebrate introns are less conserved (Keller and Noon 1984), and the vertebrate branchpoint binding proteins have arginine at position 240 and lysine at 241 (Fig. 8C). This analysis suggests that having K240 and R241 in the branchpoint binding protein correlates to the UACUAAC branchpoint sequence, while the transposition (R240 and K241) correlates with the CURAY degenerate branchpoint sequence. The consensus branchpoint sequence in Drosophila is CUAAU (Mount et al. 1992), which might be considered an intermediate consensus sequence compared with the yeast and human branchpoint consensus sequences, suggesting that the presence of arginines at positions 240 and 241 in the insect branchpoint binding proteins might modulate the binding to match this CUAAU consensus sequence.
SF1 and BBP are members of the STAR/GSG family of RNA binding proteins, and analysis of the QUA2 domains from the other members of this group supports the model that one or both of these amino acids play an important role in RNA binding specificity. In the alignment of the QUA2 domains of STAR/GSG proteins (Fig. 8D), except for Sam68 and Slm2, these two amino acids are lysines and arginines. GLD-1 and BBP have the lysine and arginine in the same positions, and GLD-1, like BBP, binds the UACUAAC sequence with high affinity and specificity (Ryder and Williamson 2004; Lehmann-Blount and Williamson 2005). The quaking (QKI) protein binds YACUAAY (Ryder and Williamson 2004; Galarneau and Richard 2005), which is very similar to the UACUAAC site for BBP and has two lysines at positions 240 and 241. Sam68 binds to the consensus sequence of UAAA, which lacks the 5′ nucleotides recognized by the other members of the STAR/GSG family of proteins (Lin et al. 1997). Sam68 does not contain a basic amino acid at either position 240 or 241 (Fig. 8D). These correlations between RNA binding sites and the identity of the amino acids at positions 240 and 241 suggest that these two amino acids may play an important role in tuning the affinity and specificity of the STAR/GSG proteins for their specific RNA targets.
MATERIALS AND METHODS
Plasmid construction
Our previously described SF1, here known as SF1-297, was used as a template to make all point mutants. Primers and templates used to make each of the BBP/SF1 chimeras are shown in Supplemental Table 1. Those chimeras that took multiple PCR steps are divided into A, B, and C. For some chimeras with multiple PCR steps, the product of the first PCR reaction (A) was used as a primer for the second PCR reaction (B), or the product of the second PCR reaction (B) was used as a primer for the third PCR reaction (C). These are indicated under the primer sequence column as Product of A or Product of B. All final PCR fragments, once the last PCR step was completed, were cut with BamHI at the N terminus and EcoRI at the C terminus and cloned into pGEX-6P-1 (Pharmacia).
Protein purification and expression
BBP, SF1, and all other chimeras and mutant constructs were transformed into BL21(STAR) cells (Novagen). Cells were resuspended in 25 mM Tris (pH 7.5), 300 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.1% Triton X-100 (Sigma). Cells were sonicated and spun to pellet insoluble debris at 19,800g for 30 min.
The supernatant was added to glutathione-agarose beads (Sigma product# G 4510) for 15 min and washed three times with resuspension buffer (above). Beads were washed once with 1X cleavage buffer (50 mM Tris at pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.1% Triton X-100). The protein of interest was cleaved from GST using Precision protease (Pharmacia). All constructs were purified over an ion exchange column (Mono S HR 10/10) and dialyzed into protein storage buffer (25 mM Tris at pH 7.5, 150 mM NaCl, 1 mM DTT, and 15% glycerol). A concentration step was not needed.
The activities of all of the proteins were determined by titrating RNA against a constant amount of protein. We found that all of the proteins except SF1-yβ3α3, which did not bind RNA to begin with, had activities in the 40%–100% range (Supplemental Fig. 1). Also, to compare the overall secondary structures of SF1-297 and BBP-296, as well as three of the mutants (SF1-R240K/K241R, SF1-R240K, and SF1-K241R), we used circular dichroism and found that all five of these proteins contained approximately the same percentage of secondary structure (data not shown).
Gel-shift assays
RNAs were labeled with [γ-32P]ATP using T4 polynucleotide kinase, purified on denaturing polyacrylamide gels, and cleaned up using Bio-Rad P6 spin columns. Binding buffer consisted of 25 mM Tris (pH 7.5), 50 mM NaCl, 2 mM DTT, 0.1% Triton X-100, and 0.07 mg/mL tRNA (Bakers yeast from Sigma). The radiolabeled oligonucleotide was at an approximate concentration of 0.1 nM. Free RNA and protein/RNA complexes were separated on 0.5X TBE 6% polyacrylamide gels run at 150 V for 1 h at 4°C. The gel-shift assay was repeated three times for each RNA substrate except for those indicated in Table 1. Gels were dried and exposed on a Storm 860 PhosphorImager (Amersham). Quantification of the fraction of RNA bound was done using ImageQuant. Data was plotted in Kaleidagraph and data points fit to the following equation to calculate an approximate Kd: Fraction RNA bound=((Kd+[RNATotal]+[SF1Total])-((-Kd-[RNATotal]-[SF1Total])2-(4([SF1Total][RNATotal]))1/2)/(2[RNATotal]). To determine ΔG and ΔΔG, the following formulas were used: ΔG=-RTln(1/Kd) and ΔΔG=-RTln(Kd1/Kd2), Kd1 is protein binding to the wild-type BP-22 RNA, and Kd2 is protein binding to the mutant RNA in that column. For example, the ΔΔG in Table 1 for BBP-296 WT and the UACUACC mutant was calculated using the Kd for BBP-296 with UACUAAC and the Kd for BBP-296 with the mutant RNA, UACUACC.
Fluorescence anisotropy assays
Titrations were performed using a Jobin Yvon Horiba FluoroMax-3 fluorescence spectrometer. 5′-Fluorescently labeled RNA oligonucleotides were obtained from IDT. The sequence of the wt RNA was 5′-fluorescein-CAGUAUACUAACAAGUUGAAUU-3′. The sequence of the mutant RNA was 5′-fluorescein-CAGUAUAGUAACAA GUUGAAUU-3′. Excitation and emission wavelengths were 495 and 525 nm, respectively. The excitation and emission bandpasses were set to 3.5 nm, and the instrument was set at 1 nm resolution. Samples were maintained at 25°C with a recirculating water bath. Proteins were dialyzed into storage buffer (25 mM Tris at pH 7.5, 150 mM NaCl, 1 mM TCEP [Tris(2-carboxyethyl) phosphine hydrochloride, Fluka], 15% glycerol) containing TCEP instead of DTT, as DTT caused significant light scattering. RNA samples were prepared in a buffer containing 25 mM Tris (pH 7.5), 50 mM NaCl, and 0.07 mg/mL tRNA (Sigma). The fluorescent RNA oligonucleotide concentration was 10 nM. Protein was added to a final protein concentration of 750 nM. Data was plotted in Kaleidagraph and fit to the following equation to calculate an approximate Kd: Anisostropy=A0+(A-A0)*(([ProteinTotal]+[RNATotal]+Kd)-((-[ProteinTotal]-[RNATotal]-Kd)2-(4*[RNATotal]*[ProteinTotal]))1/2)/(2*[RNA])). For each combination of BBP-296 WT or SF1–297 WT and RNA, three independent experiments were performed. The approximate Kd values were determined for each experiment, and the Kd values were averaged. The error reported is the standard deviation of the three measurements. For SF1-R240K/K241R, two experiments were performed and the approximate Kd values were averaged. The error reported is the standard deviation of the two measurements.
SUPPLEMENTAL DATA
Supplemental Materials can be obtained via email: aberglund@molbio.uoregon.edu.
ACKNOWLEDGMENTS
We thank Alice Barkan, Vickie DeRose, Rodger Voelker, and members of the Berglund laboratory for experimental advice and comments on the manuscript. This work was supported by a grant from NSF (0616264-MCB) to J.A.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.633808.
REFERENCES
- Abovich, N., Rosbash, M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Auweter, S.D., Oberstrass, F.C., Allain, F.H. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, J.A., Chua, K., Abovich, N., Reed, R., Rosbash, M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berglund, J.A., Abovich, N., Rosbash, M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes & Dev. 1998a;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, J.A., Fleming, M.L., Rosbash, M. The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA. 1998b;4:998–1006. doi: 10.1017/s1355838298980499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon, E., Casaregola, S., Blandin, G., Llorente, B., Neuveglise, C., Munsterkotter, M., Guldener, U., Mewes, H.W., Van Helden, J., Dujon, B., et al. Molecular evolution of eukaryotic genomes: Hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G., Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Cavaloc, Y., Bourgeois, C.F., Kister, L., Stevenin, J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Richard, S. Structure-function analysis of Qk1: A lethal point mutation in mouse quaking prevents homodimerization. Mol. Cell. Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T., Damaj, B.B., Herrera, C., Lasko, P., Richard, S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: Role of the KH domain. Mol. Cell. Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, L.C., Gelinas, R.E., Broker, T.R., Roberts, R.J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. 1977. Rev. Med. Virol. 2000;10:362–371. discussion 355–366. [PubMed] [Google Scholar]
- Dannull, J., Surovoy, A., Jung, G., Moelling, K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman, R.N., Wu, Z.R., Stalling, C.C., Pappalardo, L., Borer, P.N., Summers, M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- Flocco, M.M., Mowbray, S.L. Planar stacking interactions of arginine and aromatic side-chains in proteins. J. Mol. Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- Galarneau, A., Richard, S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- Garrey, S.M., Voelker, R., Berglund, J.A. An extended RNA binding site for the yeast branch point-binding protein and the role of its zinc knuckle domains in RNA binding. J. Biol. Chem. 2006;281:27443–27453. doi: 10.1074/jbc.M603137200. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., Thompson, J.D., Heringa, J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Grishin, N.V. KH domain: One motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, E.B., Noon, W.A. Intron splicing: A conserved internal signal in introns of animal pre-mRNAs. Proc. Natl. Acad. Sci. 1984;81:7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, A. Presplicing complex formation requires two proteins and U2 snRNP. Genes & Dev. 1988;2:1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- Langford, C.J., Gallwitz, D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983;33:519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Lehmann-Blount, K.A., Williamson, J.R. Shape-specific nucleotide binding of single-stranded RNA by the GLD-1 STAR domain. J. Mol. Biol. 2005;346:91–104. doi: 10.1016/j.jmb.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Lin, Q., Taylor, S.J., Shalloway, D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J. Biol. Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Luyten, I., Bottomley, M.J., Messias, A.C., Houngninou-Molango, S., Sprangers, R., Zanier, K., Kramer, A., Sattler, M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- Lukong, K.E., Richard, S. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Mount, S.M., Burks, C., Hertz, G., Stormo, G.D., White, O., Fields, C. Splicing signals in Drosophila: Intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny, C.W., Teem, J.L., Rosbash, M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: Implications for U1 RNA and metazoan mRNA splicing. Cell. 1983;34:395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Moore, M.J., Sharp, P.A. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes & Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Strobel, S.A., Sharp, P.A. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Rain, J.C., Rafi, Z., Rhani, Z., Legrain, P., Kramer, A. Conservation of functional domains involved in RNA binding and protein–protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- Ruskin, B., Greene, J.M., Green, M.R. Cryptic branch point activation allows accurate in vitro splicing of human β-globin intron mutants. Cell. 1985;41:833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Williamson, J.R. Specificity of the STAR/GSG domain protein Qk1: Implications for the regulation of myelination. RNA. 2004;10:1449–1458. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P., Guthrie, C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Vernet, C., Artzt, K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- Wang, J., Tse, S.W., Andreadis, A. Tau exon 6 is regulated by an intricate interplay of trans factors and cis elements, including multiple branch points. J. Neurochem. 2007;100:437–445. doi: 10.1111/j.1471-4159.2006.04252.x. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Lührmann, R. Spliceosome structure and function. In: Gesteland R., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Zhuang, Y.A., Goldstein, A.M., Weiner, A.M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl. Acad. Sci. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]