Abstract
We have identified a Dictyostelium discoideum gene encoding a serine/threonine kinase, PAKa, a putative member of the Ste20/PAK family of p21-activated kinases, with a kinase domain and a long NH2-terminal regulatory domain containing an acidic segment, a polyproline domain, and a CRIB domain. PAKa colocalizes with myosin II to the cleavage furrow of dividing cells and the posterior of polarized, chemotaxing cells via its NH2-terminal domain. paka null cells are defective in completing cytokinesis in suspension. PAKa is also required for maintaining the direction of cell movement, suppressing lateral pseudopod extension, and proper retraction of the posterior of chemotaxing cells. paka null cells are defective in myosin II assembly, as the myosin II cap in the posterior of chemotaxing cells and myosin II assembly into cytoskeleton upon cAMP stimulation are absent in these cells, while constitutively active PAKa leads to an upregulation of myosin II assembly. PAKa kinase activity against histone 2B is transiently stimulated and PAKa incorporates into the cytoskeleton with kinetics similar to those of myosin II assembly in response to chemoattractant signaling. However, PAKa does not phosphorylate myosin II. We suggest that PAKa is a major regulator of myosin II assembly, but does so by negatively regulating myosin II heavy chain kinase.
Keywords: Dictyostelium discoideum, PAK, myosin II, chemotaxis, Rac/Cdc42
The actin cytoskeleton, composed of actin filaments and multiple actin-binding proteins, plays important roles in cell motility, cytokinesis, phagocytosis, and probably intracellular transport processes (Cooper 1991; Mabuchi 1994). Cell movement requires the protrusive and contractile forces generated via polymerization, depolymerization, and cross-linking of actin filaments and myosin motors. The Rho family of small G proteins are key regulators of changes in the actin cytoskeleton (Nobes et al. 1995; Tapon and Hall 1997; Hall 1998). In yeast, Cdc42 is required for cell polarization and bud formation (Ziman et al. 1993; Kron and Gow 1995; Li et al. 1995), whereas a mutation in a putative Rac exchange factor leads to cell polarization defects during Drosophila melanogaster embryogenesis (Häcker and Perrimon 1998). In mammalian cells, Cdc42 regulates the formation of filopodia, whereas Rac regulates lamellipodia formation and membrane ruffling, and RhoA regulates the formation of stress fibers. These pathways can be regulated through growth factor signaling and are important in controlling cellular movement and differentiation.
Conventional myosin (myosin II) is a key component in the regulation of cell motility, cytokinesis, and phagocytosis, functioning as an essential force-generating motor in these processes (Spudich et al. 1995). Myosin II localizes to the contractile ring (cleavage furrow) during cytokinesis of metazoan and Dictyostelium discoideum cells (Fukui 1990; Otto and Schroeder 1990; Moores et al. 1996). In Dictyostelium, myosin II (myoII) null cells show a conditional growth phenotype: the cells cannot undergo cytokinesis in suspension and form large, multinucleated cells (De Lozanne and Spudich 1987; Knecht and Loomis 1988). However, when grown on a substratum, the cells cleave by traction-mediated cytokinesis (Fukui et al. 1990). Myosin II is also involved in the regulation of chemotaxis. In polarized, chemotaxing cells, myosin II is predominantly localized to the posterior of the cell and forms a C-shaped cap that contracts during the last stage of cell movement, causing the posterior of the cell to detach from the substratum and move forward (Moores et al. 1996; Clow and McNally 1999). Wild-type cells predominantly form a single pseudopod at the leading edge; myoII null cells produce lateral pseudopodia, possibly due to reduced cortical tension (Pasternak et al. 1989) and have difficulty lifting the cell's posterior (Jay et al. 1995). This leads to a slower and less efficient chemotaxis.
Other key regulators of the cytoskeleton are PAKs, p21-activated Ser/Thr protein kinases, which are thought to lie downstream of Rac and Cdc42. The first members of this kinase family to be identified were Ste20 and Cla4 from Saccharomyces cerevisiae, which are regulated by Cdc42 (Herskowitz 1995). Members of the PAK family have an NH2-terminal regulatory domain that contains a CRIB (Cdc42/Rac binding) domain and a related COOH-terminal kinase domain. Mammalian p65PAK can be activated via autophosphorylation in vitro upon the binding of activated (GTP-bound) Rho family members, Cdc42 and Rac (Manser et al. 1994). The yeast PAK, Ste20, plays important roles in activating the MAP kinase cascade in the pheromone-response pathway (Herskowitz 1995), although mammalian PAKs have been implicated in the activation of the Rac/Cdc42-regulated MAP kinase cascades leading to the stimulation of stress-induced MAP kinases p38 and JNK (Bagrodia et al. 1995; Zhang et al. 1995).
There is increasing evidence that PAK family kinases regulate cytoskeletal changes mediated by Cdc42 and Rac. Ste20, along with its homologue Cla4, regulates polarized cell growth, presumably by controlling the actin cytoskeleton (Chant and Stowers 1995; Cvrckova et al. 1995). Microinjection of activated PAK1 protein into quiescent Swiss 3T3 cells induces the rapid formation of lamellipodia, filopodia, and membrane ruffles (Sells et al. 1997), which is very similar to the effect produced by microinjection of activated Rac and Cdc42 (Nobes and Hall 1995). Moreover, the Drosophila homologue of mammalian PAK1 is a key regulator of the cytoskeletal changes required for axonal guidance (Hing et al. 1999). Expression of various constitutively active forms of αPAK induces disassembly of stress fibers and focal adhesion complexes (Manser et al. 1997). A Dictyostelium myosin I heavy chain kinase (MIHCK)1 homologous to PAK and Ste20 has been cloned and proposed to provide a direct link between Cdc42/Rac signaling pathways and motile processes requiring myosin I molecules (Lee et al. 1996; Brzeska et al. 1997). PAKs also appear to regulate the function of myosin II, as the regulatory myosin light chains are phosphorylated by the Jurkat and placenta γPAK (Ramos et al. 1997), suggesting that different PAKs may regulate different cytoskeletal components.
We describe the properties of a putative Dictyostelium PAK (PAKa) that contains a potential polyproline SH3 binding domain, a highly acidic domain in its NH2 terminus, a CRIB domain, and a related kinase domain. paka null cells or wild-type cells expressing a kinase dead (putative dominant negative) PAKa produce many random, lateral pseudopodia and have a much higher frequency of making wrong turns than wild-type cells. Similarities in the distributions of PAKa and myosin II and in the phenotypes of paka and myoII null cells suggest that PAKa might be involved in the regulation of myosin II filament assembly in the posterior cortex. Furthermore, the increase of myosin II in the cytoskeleton in wild-type cells in response to cAMP was lacking in paka null cells, suggesting the assembly of myosin II into the cytoskeleton requires PAKa function. PAKa kinase is activated in response to stimulation with the chemoattractant cAMP, suggesting it controls chemotaxis by regulating myosin II function.
Materials and Methods
Materials
Sodium orthovanadate, β-glycerophosphate, aprotinin, and leupeptin were obtained from Sigma Chemical Co. Histone 2B was purchased from Boehringer Mannheim Corp. γ[32P]ATP was from ICN Biomedicals. Protein A–Sepharose CL-4B was obtained from Pharmacia Biotech Inc. Purified myosin II was a generous gift from Dr. Tom Egelhoff (Case Western Reserve University, Cleveland, OH).
Cloning of PAKa
To clone PAKa, a pair of degenerate primers was made based on the amino acid sequences conserved among the kinase domains of yeast Ste20 and human PAK65 (the two primers corresponded to the amino acid sequences VAIKK and DFGLAR). A full-length cDNA clone (3.6 kb) was obtained by screening a 12–16 h developmental λZAP library with a probe amplified by PCR. PAKa was disrupted in the thymidine auxotroph JH10 by inserting the Thy gene cassette into codon 791, and transformants were selected. For overexpression experiments, PAKa and PAKa mutants were subcloned into the DIP-J expression vector (Gaskins et al. 1996). PAKa was tagged at the COOH terminus with the FLAG epitope by using primer 5′-TTACTTGTCATCGTCGTCCTTGTATTGTGCAGTTAAATTTTGTAATTG-3′. The HA epitope tag was added to the NH2 terminus of PAKa by using primer 5′-AAAATGACCTACCCATA-CGATGTTCCAGATTACGCTGAAGAGAAACCAAAAAGTACA-ACTCCACCA-3′. myrPAKa was obtained by PCR using primer 5′-ATGGGTTCATCAAAATCAAAACCAAAAGATCCATCACAAC-GTCGTCGT-3′, a sequence coding for the first 16 amino acids of chicken c-Src, composed of the myristoylation signal and the basic amino acid cluster sufficient for stable membrane association.
Selection of Clones
To select for strains expressing a high level of PAKa, transformants were selected at 40 μg/ml of G418, which in our hands produces clones that express encoded proteins at high levels. To select clones that express at a low level, we selected and maintained cells at 15 μg/ml of G418. Clones were screened to identify those exhibiting a low level of exogenous PAKa expression and exhibiting wild-type phenotypes in actin staining.
Indirect Immunofluorescence Staining
For immunofluorescence staining, cells were starved in 12 mM sodium phosphate buffer (pH 6.2) for >5 h and fixed with 4% formaldehyde for 5 min. Cells were permeabilized with 0.5% Triton X-100, washed, and incubated with 1.4 μg/ml anti-FLAG mAb or rabbit anti-HA antibody (1/200 dilution) in PBS containing 0.5% BSA and 0.05% Tween-20 for 1 h. Cells were washed in 0.5% BSA containing PBS and incubated with FITC-labeled anti-mouse or anti-rabbit antibodies for 1 h. After washing, cells were observed with a 60× oil immersion lens on a Nikon Microphot-FX microscope. Images were captured with a Photometrics Sensys camera and IPLAB Spectrum software. Anti-HA antibody was purchased from Santa Cruz Biotechnology. Monoclonal anti-FLAG antibody was obtained from Kodak. F-actin was stained with FITC-labeled phalloidin (Sigma Chemical Co.). Rabbit antimyosin II antibody was a generous gift from Dr. James A. Spudich (Stanford University, Stanford, California).
PAKa Kinase Activity Assay
Log-phase vegetative cells were washed and resuspended at a density of 2–3 × 106 cells/ml in Na/K phosphate buffer and pulsed for 5 h with 30 nM cAMP every 10 min. The cells were collected by centrifugation and resuspended at a density of 2–3 × 107 cells/ml. The cAMP receptor activity was downregulated by bubbling air through the cell suspension for 10 min (Ma et al. 1997). The 200-μl samples were lysed by mixing with an equal volume of 2× lysis buffer (50 mM Tris, pH 7.6, at room temperature, 200 mM NaCl, 20 mM NaF, 2 mM vanadate, 50 mM β-glycerophosphate, 6 mM sodium pyrophosphate, 2 mM EDTA, 2 mM EGTA, 4 μg/ml leupeptin, 4 μg/ml aprotinin, 2% NP-40, 20% glycerol, 2 mM DTT). 1 μl of antibody was added to 200 μl supernatant and incubated on ice for 1 h. The formed immune complexes were collected with 50 μl of a 1:1 slurry of protein A beads in lysis buffer by incubation under agitation for 1 h at 4°C. The beads were washed twice with lysis buffer and twice with kinase buffer (25 mM MOPS, pH 7.4, at room temperature, 25 mM β-glycerophosphate, 20 mM magnesium chloride, 1 mM DTT, 30 mM KCl for myosin II assay). PAKa activity was measured in an immunocomplex kinase assay after immunoprecipitation with the anti-FLAG antibody. The beads were incubated with 75 μl kinase buffer containing 5 μCi γ[32P]ATP, 5 μM cold ATP, and 5 μg histone 2B or myosin II as a substrate. The reaction was stopped by the addition of 25 μl 4× sample buffer and boiling for 5 min. The samples were separated by SDS-PAGE (12.5%), blotted onto a PVDF membrane (Millipore Corp.), and exposed to film.
Chemotaxis Assay
Cells were pulsed with 30 nM cAMP at 6-min intervals for 5 h, and cells were washed and resuspended in Na/KPO4 buffer containing 200 μM CaCl2 and MgCl2. A small volume of cells was plated on glass-bottomed microwell plates (MatTek, Inc.) and allowed to adhere to the surface for ∼20 min. A micropipette filled with 150 μM cAMP was positioned to stimulate cells by using a micromanipulator (Eppendorf-Netheler-Hinz GmbH), and the response and movement of cells was recorded by using a time-lapse video recorder and NIH Image software (1 image every 6 s). The movement of cells and changes of cell shape were analyzed with the DIAS program (Wessels and Soll 1998).
Isolation of Cytoskeletal Proteins
Cytoskeletal proteins were isolated as proteins insoluble in detergent NP-40. Cells were harvested by centrifugation and resuspended at 108 cells/ml. cAMP (100 μM) was added to cells and 500 μl of cells were taken at each time point and immediately added to 500 μl of 2× lysis buffer as described for the kinase assay. After vortexing a few times, the tubes were placed on ice for 10 min, then allowed to warm to room temperature for 10 min. The samples were spun for 4 min at 11,000 g and the supernatant was discarded. After washing with 1× lysis buffer, the protein pellet was dissolved by boiling in 2× SDS-PAGE sample buffer. Samples were run in 8% gel, and protein bands were stained with Coomassie blue. Protein bands were scanned, and changes in myosin II content in cytoskeleton were measured by using IPLAB software.
Two-Hybrid and Overlay Analysis
Rac1B, RacE, RasG, and human Cdc42 were amplified by PCR and cloned into yeast two-hybrid fish vector pG4-5 as described previously (Lee et al. 1997, Lee et al. 1999). The clones were sequenced to confirm the reading frame and the absence of mutations. Point mutations corresponding to Q61L in Ras were made with the Transformer Site Directed Mutagenesis Kit from CLONTECH Laboratories, Inc. Constructs containing site-directed mutants were sequenced to confirm the nucleotide substitutions and the absence of other mutations. Rac1B was a generous gift from Su Dharmawardhane (University of Texas, Austin, TX), RacE was from Arturo De Lozanne (Duke University, Durham, NC), RasG was from Robert Insall (University of Birmingham, UK), and HsCdc42 was from Michael Karin (University of California, San Diego, CA).
For overlay assays, Rac1B and HsCdc42 were cloned into GST fusion constructs. The constructs were sequenced to confirm the reading frame and the absence of errors in the cloning/amplification. GST fusion proteins were produced in Escherichia coli and overlay assays done according to the methods of Manser et al. 1994.
Results
Cloning of PAKa
We identified PAKa in a PCR screen for PAK/Ste20 family kinases using degenerate primers based on conserved sequences in the kinase domains of Ste20 and mammalian PAK. Sequenced PCR products carrying part of a potential PAK family member were used to screen a cDNA library and several family members were identified, one being PAKa. One ∼3.5-kb PAKa cDNA clone contained the complete PAKa open reading frame (Fig. 1 A). The domain structure of PAKa is similar to that of mammalian PAK1 (Fig. 1 B). PAKa has a predicted Cdc42/Rac, a small G protein binding domain (CRIB), a COOH-terminal kinase domain, and a long NH2-terminal putative regulatory domain containing a highly acidic region and a potential SH3-interacting polyproline stretch. In addition, the NH2-terminal domain contains an Akt/PKB and multiple cAMP-dependent kinase (PKA) consensus phosphorylation sites. Comparisons of the kinase and CRIB domains of PAKa and other members of this family are shown in Fig. 2A and Fig. B, respectively. The PAKa kinase domain has the highest sequence identity to the kinase domain of Dictyostelium MIHCK (59% identity) and human PAK1, mouse PAK3, and Saccharomyces pombe PAK1 (∼48% identity each).
Figure 1.

Deduced sequence of PAKa. A, The cDNA clone, PAKa 6-1, has an insert of 3670 nt and contains the entire PAKa open reading frame. The boxed region depicts the conserved kinase domain. The underlined region shows acidic domain. The bolded and underlined sequence represents a polyproline stretch. The shaded sequence indicates the CRIB domain. The GenBank/EMBL/DDBJ accession number for PAKa is AF131221. B, Schematic diagrams of PAKa and PAKa mutants. C, Developmental RNA blot of PAKa expression. 6 μg of total RNA was loaded in each lane and probed (Datta and Firtel 1987). Time in development is shown. 0 h, vegetative cells.
Figure 2.
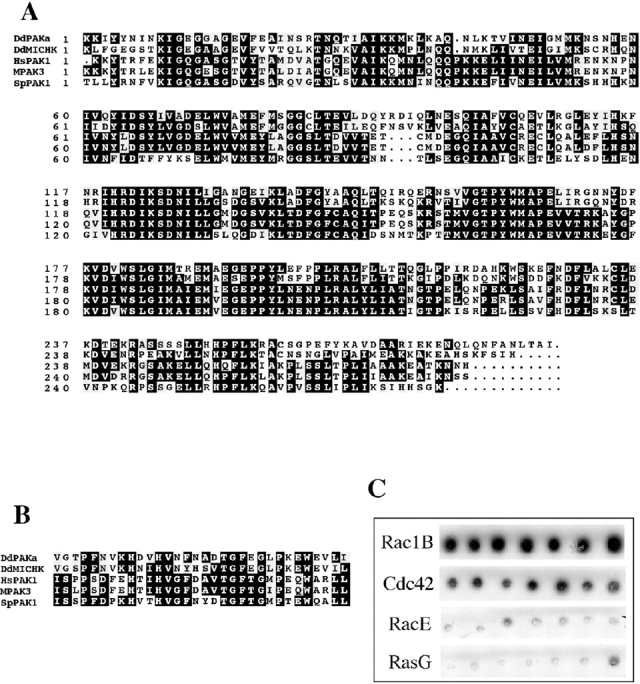
A, Amino acid sequence comparison of the kinase domain of PAKa with PAKs from a variety of organisms. DdP, Dictyostelium PAKa; DdM, Dictyostelium MIHCK; Hs, human PAK1; mo, mouse PAK3; Sp, Saccharomyces pombe PAK1. GenBank/EMBL/DDBJ accession numbers: human PAK1, Q13153; mouse mPAK-3, g1168176; Saccharomyces pombe PAK1, P50527; Dictyostelium discoideum MIHCK, U67716. B, Amino acid sequence comparison of the CRIB domain of PAKa with PAKs from other organisms. C, Interaction of CRIB domain of PAKa with small G proteins in the yeast two-hybrid system developed in the laboratory of Roger Brent (Gyuris et al. 1993), which was used to examine the interactions between PAKa and small G proteins as described previously (Lee et al. 1997). The CRIB domain of PAKa was cloned into bait vector pEG202. Constitutively active Rac1B, RacE, RasG, and human Cdc42 were cloned into fish vector pG4-5. EGY48. Yeast was transformed with PEG202, pG4-5, and a reporter construct. Several colonies were picked and plated on Gal/CM Ura−His−Trp− plates containing X-gal. The picture depicts color development 4 d after plating. DdRac1B and HsCdc42 show strong interaction with the CRIB domain of PAK, as colonies are dark blue on X-gal plates. In contrast, RacE and RasG have very weak interactions, as colonies do not show any color development. RasG interacts strongly with the Ras-interacting domains of RIP3 (Lee et al. 1999) and PI3K1 and PI3K2 in the same yeast two-hybrid system. Under these conditions, Rac1B and HsCdc42 do not interact with the RIP3 Ras interacting domain. There are no known proteins that interact with RacE.
The PAKa CRIB domain shares strong homologies with the CRIB domain of Dictyostelium MIHCK and PAKs from other species (Fig. 2 B). To examine the specificity of the PAKa CRIB domain for small G proteins, we used the yeast two-hybrid system (Gyuris et al. 1993) to assay the interactions of the PAKa CRIB domain with activated forms of Dictyostelium Rac1B, human Cdc42 (a Dictyostelium homologue has not yet been identified), Dictyostelium RacE, and Dictyostelium RasG, the latter two of which are required for cytokinesis in Dictyostelium (Vithalani et al. 1996; Tuxworth et al. 1997). As we show in Fig. 2 C, the PAKa CRIB domain preferentially interacts with DdRac1B and HsCdc42. We confirmed the interaction of DdRac1B and HsCdc42 with the PAKa CRIB using an overlay assay (Manser et al. 1994). We observed strong interaction of a GST-CRIB/PAK fusion protein with DdRac1BGTPγS and Cdc42GTPγS (data not shown). These results are consistent with, but do not prove, that PAKa is a member of the PAK family of Rac/Cdc42-regulated protein kinases.
Northern blot analysis indicates that PAKa encodes a single developmentally regulated transcript (3.7 kb) that is expressed in vegetative and early developing cells, and is upregulated during aggregation and multicellular stages (Fig. 1 C). No hybridization signal was detected when the probe was used on a blot carrying RNA isolated from paka null cells (data not shown).
paka Null Cells Have a Defect in Cytokinesis and Development
PAKa was disrupted by homologous recombination (see Materials and Methods). Random transformants were selected and screened for disruption of PAKa by Southern and RNA blot analysis. All clones that showed disruption of PAKa did not exhibit an mRNA signal on RNA blots, and had the same developmental and growth phenotype. As depicted in Fig. 3 A, paka null cells exhibit a defect in completing cytokinesis when grown in suspension. Under these conditions, the increase in cell number indicates paka null cells grow more slowly than wild-type cells because the cells become multinucleate, as revealed by an increase in number of nuclei in a cell (Fig. 3). When grown in suspension for five days, paka null cells are considerably larger than wild-type cells, and DAPI (4′, 6-diamidino-2-phenylindole) staining shows an average of eight nuclei per cell (Fig. 3 B). However, when paka null cells are grown on plastic, they grow predominantly as mono- and dinucleate cells (data not shown), dividing by traction-mediated cytofission, as previously described for myoII null cells (De Lozanne and Spudich 1987; Fukui et al. 1990). Our data suggest that paka null cells might have difficulties in regulating cytoskeletal organization required for cytokinesis, but karyokinesis is normal. The cytokinesis and developmental defects of paka null cells can be complemented by overexpression of PAKa from the Actin 15 (Act15) promoter (data not shown).
Figure 3.
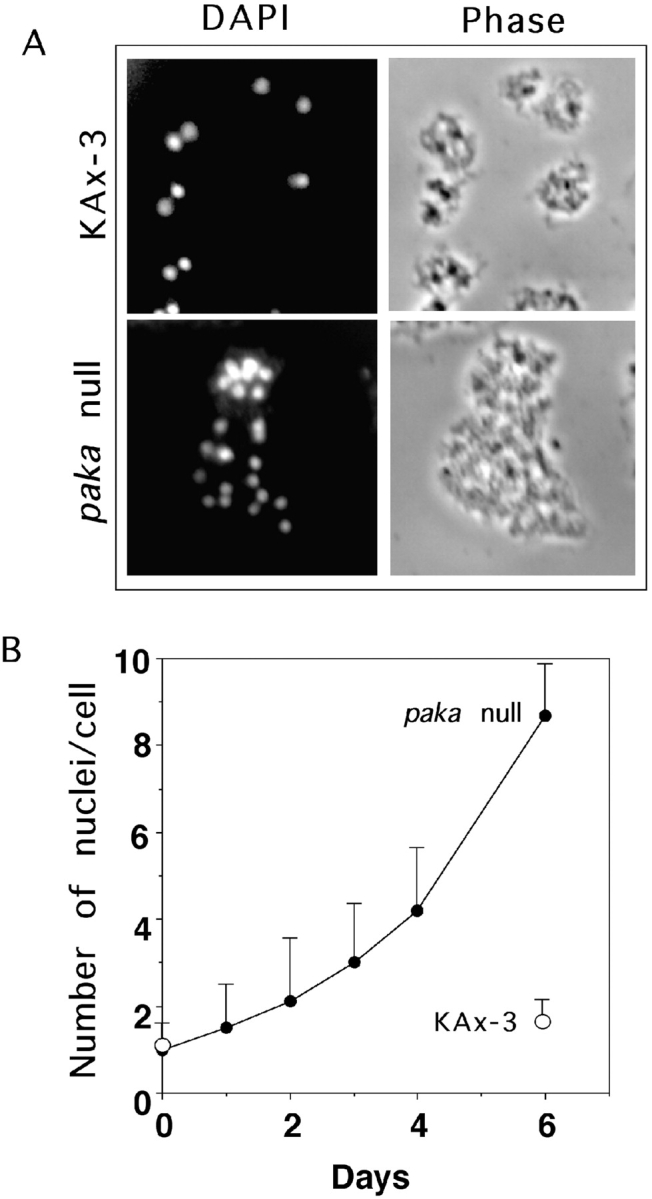
Multinucleated paka null cells due to defective cytokinesis. A, Defective cytokinesis in paka null cells. Phase-contrast and epifluorescent micrographs of multinucleated paka null cells grown in shaking culture for 5 d. Nuclei were visualized with DAPI. B, Increase of the number of nuclei in paka null cells. Wild-type KAx3 and paka null cells were transferred from plates into axenic medium in shaken culture at 0 h and the number of nuclei in a cell were counted at intervals thereafter. Cells were counted using a hematocytometer. When paka null cells are grown in suspension for 5 d, there are an average of eight nuclei per cell.
To examine the effect of disrupting PAKa on multicellular development, we plated paka null and wild-type cells on nonnutrient agar and followed development. Compared with wild-type cells, development of paka null cells is very delayed (Fig. 4). Tight aggregates do not form until 16–17 h, and some mounds arrest at this stage. For those that proceed further, the first fingers are not formed until ∼24 h. Mature fruiting bodies form at ∼36 h, containing sori that are smaller than those of wild-type cells (data not shown). To examine the role of PAKa further, we created a series of PAKa mutant constructs: a kinase dead, putative dominant negative mutant of PAKa made by changing the Lys residue in the ATP binding site in the kinase domain to Ala (PAKaK394A, designated PAKaDN); a putative constitutively active PAKa (PAKaCA) containing only the kinase domain (the NH2-terminal regulatory domain is thought to function as a negative regulatory module and its deletion in mammalian PAKs leads to a constitutively active kinase; Cairns et al. 1992); a PAKa mutant containing the CRIB and kinase domains (PAKaG+K); and a PAKa carrying the NH2-terminal myristoylation signal from Src (myrPAKa), which has been demonstrated to constitutively target mammalian and Dictyostelium proteins uniformly to the plasma membrane (Buser et al. 1994; Meili et al. 1999). The structures of these constructs are shown in Fig. 1 B.
Figure 4.
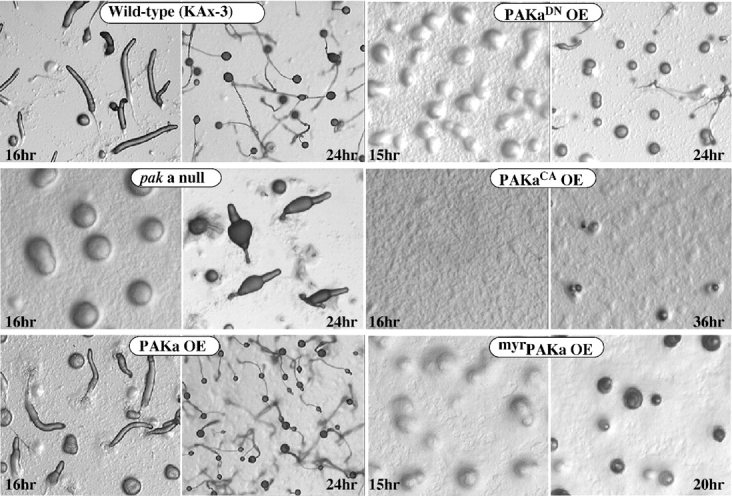
Development of paka null cells and cells expressing PAKa mutants. Cells grown in axenic medium were washed and plated on nonnutrient agar plates. Photographs were taken at various developmental stages thereafter. Development of paka null cells is delayed in aggregation and late development. Cells expressing dominant negative PAKa (PAKaDN) and myristoylated PAKa (myrPAKa) exhibit delayed aggregation similar to that of paka null cells. The phenotype of myrPAKa is more severe than either paka null or PAKaDN cells. Cells expressing constitutively active PAKa (PAKaCA) have a severe defect in aggregation and never form mounds.
We overexpressed the mutant PAKs and wild-type PAKa from the constitutive Act15 promoter. For these experiments, the overexpression mutants were selected at high G418 concentrations to yield clones expressing a high level of the specific PAKa protein (see Materials and Methods). Cells expressing PAKaDN have a phenotype similar to, but more severe than, that of paka null cells (Fig. 4). Aggregation is very delayed and most mounds arrest and do not develop further. The more severe phenotype of PAKaDN-expressing cells compared with paka null cells may be due to the inhibition of another PAK-like kinase by PAKaDN. Cells overexpressing PAKaCA exhibit severe aggregation defects and never form mounds.
We targeted PAKa to the plasma membrane by expressing myrPAKa. Indirect immunofluorescence indicates that myrPAKa, also tagged at the COOH terminus with the FLAG epitope, localizes uniformly to the plasma membrane around cells (see Fig. 8B, Fig. f). These cells exhibit very delayed aggregation and development, suggesting that proper subcellular localization of PAKa might be important for regulating the in vivo function of PAKa during aggregation (Fig. 4). Cells overexpressing wild-type PAKa or PAKaG+K exhibit a wild-type pattern of aggregation and multicellular development, and form normal-looking fruiting bodies (data not shown).
Figure 8.
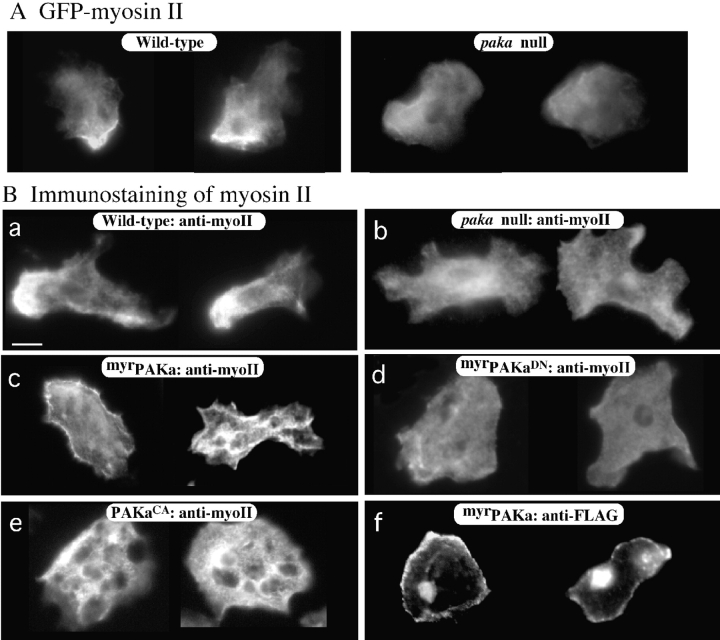
A, Localization of GFP–myosin II in aggregation stage wild-type or paka null cells. Myosin II is localized at the posterior cortex of wild-type cells, but not in paka null cells. GFP–myosin II is very diffuse throughout the null cells. B, The localization of myosin II was examined by indirect immunofluorescence staining with antimyosin II heavy chain antibody (a–e). Similar to GFP–myosin II, the staining of myosin II is concentrated at the posterior cortex. In paka null cells, the staining of myosin II is very diffuse over the cell. In cells expressing myrPAKa, myosin II staining is localized at the membrane cortex throughout the cells. However, this localized myosin II assembly at membrane cortex is not observed in cells expressing myrPAKaDN. Cells expressing PAKCA show regions of very strong myosin II staining, presumably due to the high level of assembled myosin II. myrPAKa appears to be targeted to the membrane as determined by localization of myrPAKa–FLAG by immunofluorescence staining (f). The intracellular staining is nuclear, which was sometimes observed in Dictyostelium cells using the FLAG antibody. Bar, 5 μm.
Aberrant F-actin Organization in paka Null Cells and Cells Expressing PAKa Mutants
To examine whether overexpression of PAKa mutants has an impact on the actin cytoskeleton, we performed phalloidin staining of aggregation-competent cells. Wild-type cells are usually polarized and show localized F-actin assembly at lamellipodia of the leading edge and sometimes, to a lesser degree, at the posterior cortical region of the retracting cell body (Fig. 5). paka null cells also appear elongated, but are not as polarized in the distribution of the actin cytoskeleton as wild-type cells. paka null cells have multiple, randomly localized F-actin–rich pseudopodia-like protrusions (Fig. 5, arrows). Cells overexpressing wild-type PAKa (PAKaOE), although lacking a developmental phenotype, exhibit changes in F-actin organization. PAKaOE cells have multiple F-actin–enriched crowns along the periphery of cells (Fig. 5). This pattern is very similar to that of racgap1 null cells and cells overexpressing constitutively active DdRac1B (DdRac1BQ61L; Chung, C.Y., S. Lee, C. Briscoe, and R.A. Firtel, manuscript submitted for publication), suggesting that the activity of PAKa might be regulated via Rac1B. Despite this aberrant F-actin organization, PAKaOE cells are elongated.
Figure 5.
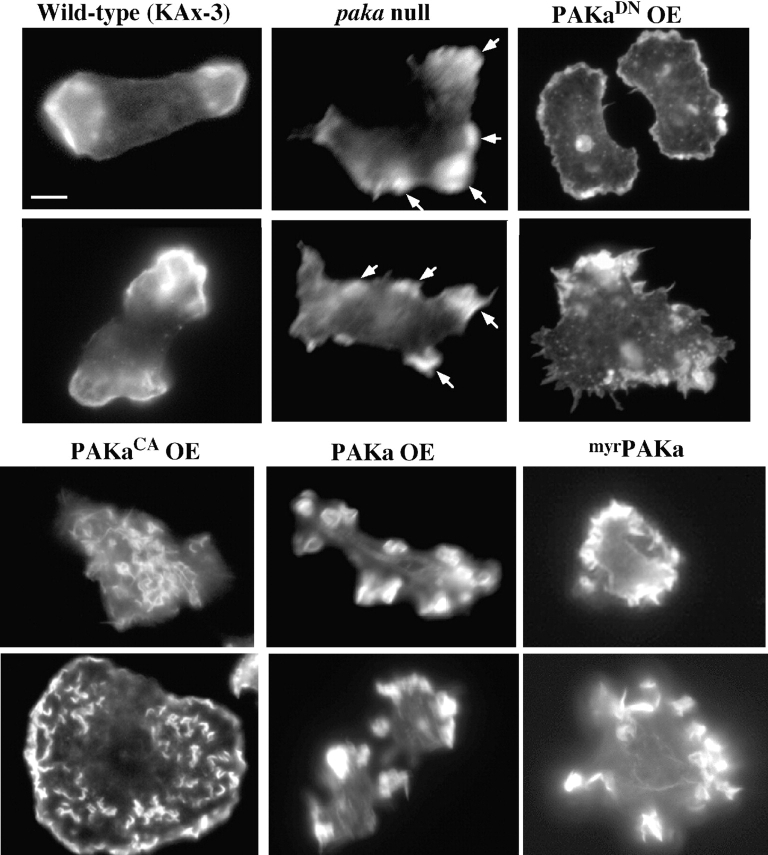
Organization of the actin cytoskeleton in paka null cells and cells expressing PAKa mutants. Cells were plated on glass coverslips and starved for 5–6 h before fixation with 3.7% formaldehyde in 12 mM Na/KPO4 (pH 6.1) for 5 min. After washing, cells were treated with blocking solution (PBS, 0.1% Triton X-100, 0.1% BSA) and 10 ng of FITC or rhodamine-labeled phalloidin was added to each glass coverslip. Note that wild-type cells have F-actin–rich lamellipodia at the leading edge that are absent in paka null cells and cells expressing PAKaDN. Cells expressing PAKaCA accumulate F-actin all over the cells. Cells overexpressing wild-type PAKa are elongated and have discrete actin-enriched domains along the periphery of the cell. Cells expressing myrPAKa exhibit domains of assembled F-actin along the entire membrane cortex. paka null cells exhibit multiple pseudopod-like, actin-enriched blebs. Some of these are marked with an arrow. Bar, 5 μm.
Cells expressing PAKaDN lack prominent actin-rich lamellipodia and actin staining is very diffuse and scattered around the cell periphery (Fig. 5). Some of the cells show microspikes (see lower panel for PAKaDN, Fig. 5), but these microspikes do not appear to be enriched in F-actin. In contrast, cells expressing PAKaCA accumulate assembled F-actin in multiple membrane ruffles over the entire cell. Some of these cells are multinucleate, although not to the extent of paka null cells, suggesting that a putative nonregulated form of PAKa, as well as a lack of PAKa, can result in cytoskeletal defects that prevent proper cytokinesis. Moreover, although these cells elongate, they fail to show a polarization of the actin cytoskeleton, consistent with their inability to aggregate properly. These results suggest that PAKa can mediate rearrangements in the actin cytoskeleton.
Cells expressing PAKaG+K do not exhibit a prominent change in F-actin distribution (data not shown) compared with wild-type cells and in contrast to cells expressing PAKaCA, suggesting that the CRIB domain might negatively regulate kinase function, consistent with work on human PAK1 (Sells et al. 1997). As wild-type PAKa has the CRIB domain, it is possible that the upstream, NH2-terminal sequences deleted in PAKaG+K are involved in regulating the function of the CRIB domain in controlling PAKa activation. Cells expressing myrPAKa exhibit domains of F-actin along the entire membrane cortex, consistent with a uniform distribution of myrPAKa along the cortex of the cells (see Fig. 8 f). These cells fail to elongate or polarize (Fig. 5). We expect that the upregulated assembly of F-actin results from the activity of the membrane-localized PAKa.
Abnormal Chemotactic Movement of paka Null Cells and Cells Expressing PAKa Mutants
To examine whether the changes in the cytoskeletal organization described above alter chemoattractant-induced cell migration, we compared the chemotactic movement of paka null cells to that of wild-type cells by examining their movement toward the chemoattractant using the DIAS image analysis software (Wessels and Soll 1998). Cells competent to chemotax toward cAMP (aggregation-competent cells) were obtained by pulsing cells in suspension for 5 h with 30 nM cAMP, conditions that mimic the cAMP signaling during the phase leading up to aggregation in vivo (Devreotes et al. 1987; Mann and Firtel 1987; Saxe et al. 1991; Insall et al. 1994). This protocol maximally induces the expression of aggregation stage genes required for aggregation, including the cAMP receptor, cAR1, and the coupled G protein α subunit, Gα2. Wild-type cells are usually well-polarized and move quickly and linearly toward the cAMP source. As we show in Fig. 6 A, wild-type cells produce pseudopodia almost exclusively at the edge in the direction of the micropipette, and produce very few random lateral or rear pseudopodia (an average of 1.5 lateral pseudopodia per 10 min). Wild-type cells make few changes in the direction of movement, as shown in Fig. 6 A. In sharp contrast, paka null cells produce more random pseudopodia (an average of 6 lateral pseudopodia per 10 min, some of which are marked with open arrowheads in Fig. 6 B) in various directions. In addition, paka null cells exhibit random changes in the direction of movement, which might result from the protrusion of lateral pseudopodia, leading to an inefficient chemotaxis toward the cAMP source. The movement of the four paka null cells shown in Fig. 6 B illustrates these phenotypes. Cells protrude multiple pseudopodia and, in cell c, two pseudopodia in the same cell become dominant (rather than one), resulting in the formation of two independent leading edges and the independent movement of the two halves that remain connected.
Figure 6.
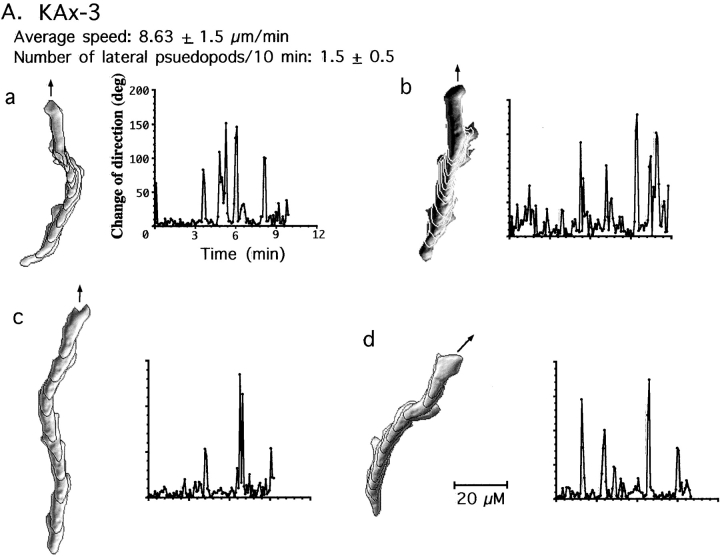
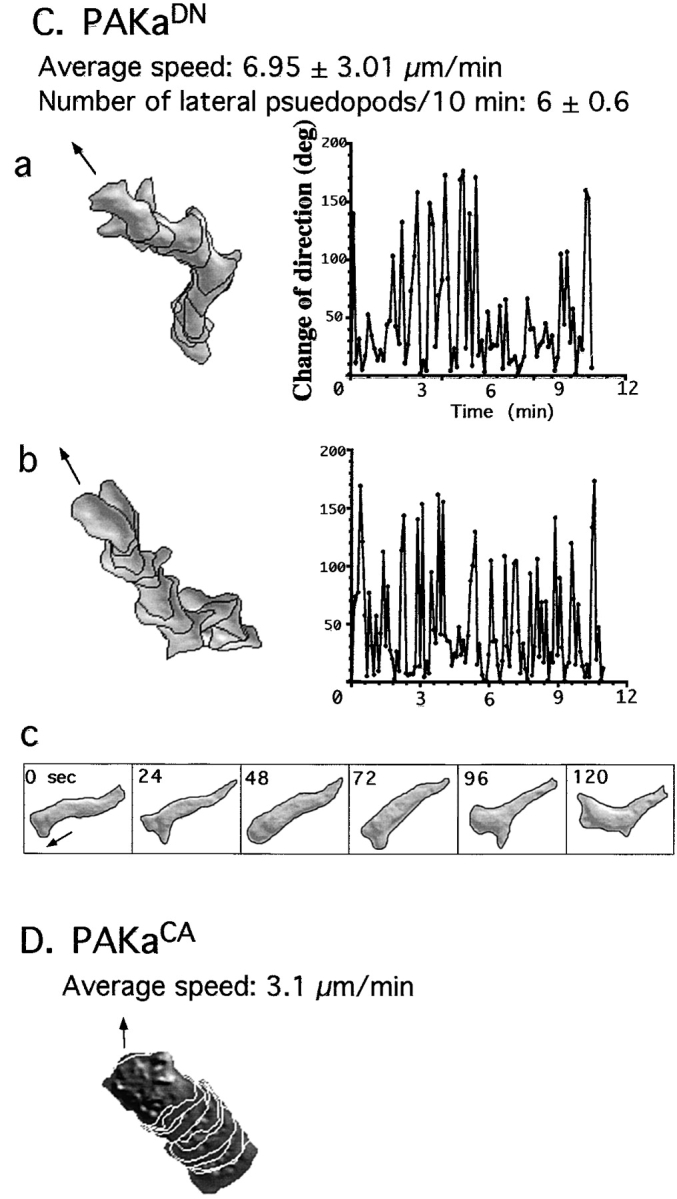
Abnormal chemotactic movement of paka null cells and cells expressing PAKa mutants. Wild-type and paka null cells were washed and pulsed for 4.5 h with 30 nM cAMP every 6 min (see Materials and Methods). Cells were plated on a plastic petri dish containing a hole, over which a glass microscope slide was glued. The tip of the micropipette containing 100 μM cAMP was placed near the cell as described in detail previously (Meili et al. 1999). The movement of cells toward the tip was recorded with NIH Image software at 6-s intervals and the movement of cells and shape changes were analyzed with the DIAS program, a newly developed image analysis system. A, Wild-type cells are well-polarized and move toward the cAMP gradient without making lateral pseudopodia. Left panel shows superimposed images representing cell shape at 1-min intervals. Right panel shows the change of direction in which the leading edge of the migrating cell headed. Solid arrows point out the direction of the micropipette filled with 100 μM cAMP. Note that wild-type cells do not make lateral pseudopodia and do not change their direction of movement very often. Bar, 20 μm. The number of newly formed pseudopodia was counted in each image for the 10-min time period of the experiment. B, paka null cells protrude many random lateral pseudopodia (average ∼6 lateral pseudopodia in 10 min, some of which are marked with open arrows) and the posterior cell bodies (marked with asterisks, a and c) of null cells are elongated, due to the difficulty in retracting. Due to frequent changes of the direction of movement, the speed of chemotactic movement of paka null cells is slower than that of wild-type cells. C, Cells expressing PAKaDN make many random lateral pseudopodia and show elongated posterior cell bodies due to the difficulty of retraction (c). D, Cells expressing PAKaCA are very flattened and stationary. They are not polarized and move very slowly in random directions.
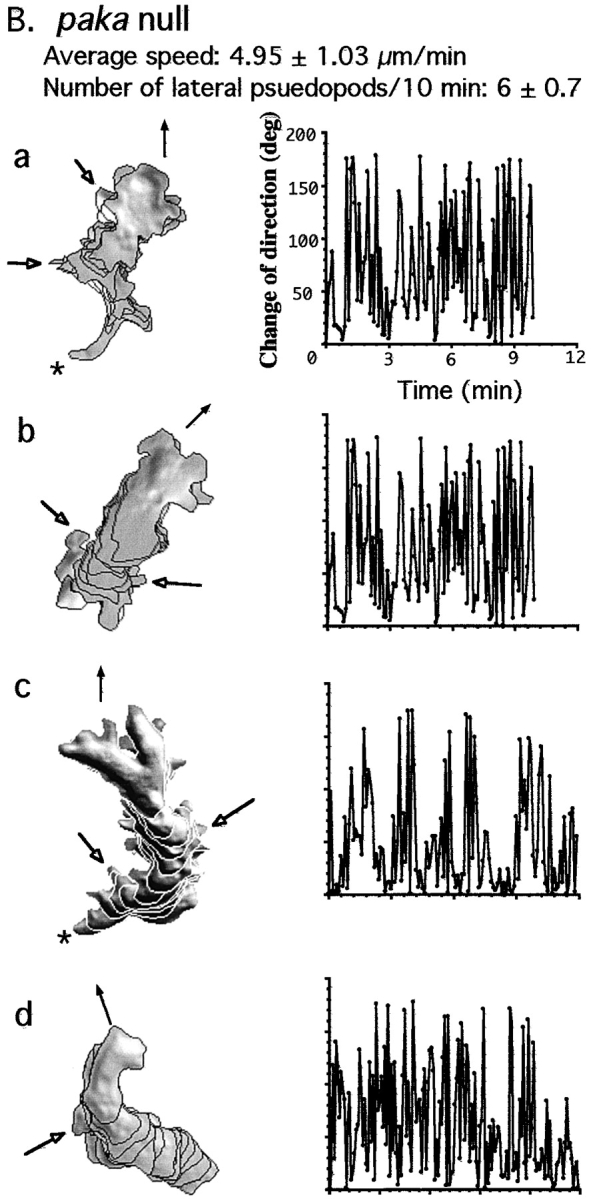
In addition, paka null cells do not retract their rear cell body as efficiently as wild-type cells. The posterior of wild-type cells contracts and lifts from the substratum after the leading-edge pseudopod has been extended (Jay et al. 1995); paka null cells appear defective in this process (Fig. 6 B: asterisk in a and c marks the rear cell body of the paka null cells, which remain tightly associated with the substratum). As illustrated by Fig. 6 B, cells a and c, the anterior of the paka null cell continues to extend its cell body toward the cAMP source until the cell reaches a certain length and then the posterior retracts very rapidly. Probably due to the combination of random changes of the direction of movement and the difficulty in retracting the rear cell body, the speed of movement of paka null cells (4.95 μm/min) is about half that of wild-type cells (8.63 μm/min).
PAKaDN cells produce more random pseudopodia than wild-type cells and have an increased frequency of turning, similar to paka null cells (Fig. 6 C). PAKaDN cells exhibit difficulty in retracting the rear cell body as depicted in Fig. 6C, Fig. c. As the leading edge of a cell moves forward, the rear cell body elongates, due to the lack of retraction, and then retracts very rapidly. PAKaCA cells, as expected from the upregulated F-actin assembly over the entire cell, are flattened and appear adherent and stationary. The cell–substratum contact area of these cells is significantly larger than that of wild-type cells (Fig. 6 D). Upon stimulation by cAMP, PAKaCA cells do not polarize and produce few dominant pseudopodia. These cells are not capable of changing their shape; hence, their migration is very slow. Our results suggest that PAKa plays an important role in the regulation of cytoskeletal organization, which is presumably required for maintaining cellular polarity and preventing the formation of random, lateral pseudopodia. The inhibition of the formation of random pseudopodia might prevent cells from making random changes of direction when moving up a chemoattractant gradient.
Subcellular Localization of PAKa
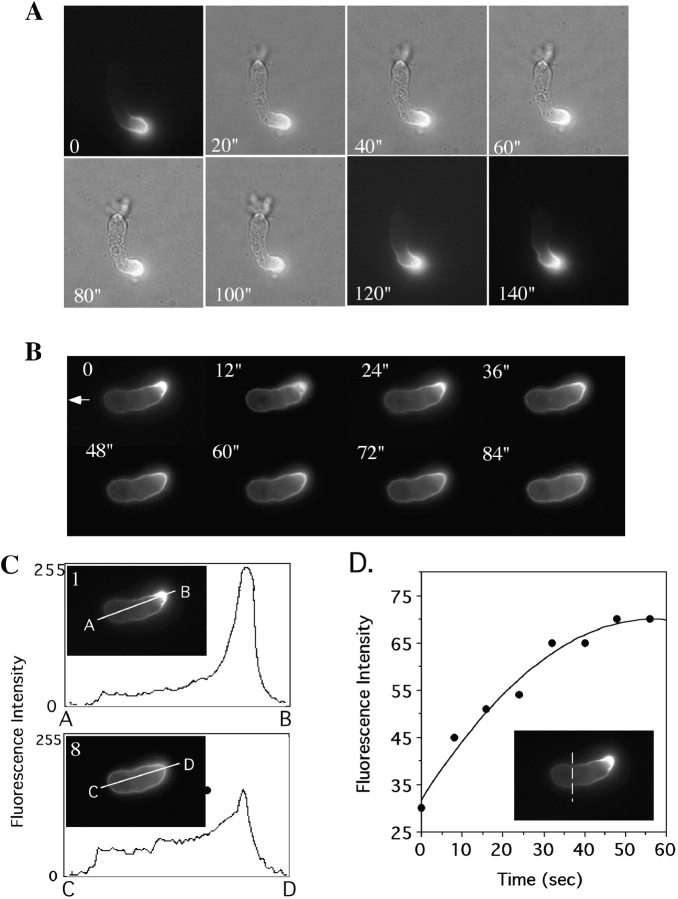
To examine where PAKa is localized in polarized, aggregation-competent cells, PAKa was tagged with FLAG at the COOH terminus or HA at the NH2 terminus, and expressed in wild-type cells. The transformants carrying a construct of tagged PAKa were selected and maintained at a low concentration of G418 to select strains that expressed PAKa at a low level. Strains were selected for those that did not exhibit the altered actin cytoskeletal defects shown in the high overexpression strains used in Fig. 4 and Fig. 5. The localization of tagged PAKa was determined in aggregation-competent cells by indirect immunofluorescence. FLAG-tagged PAKa mostly localizes to the posterior cortical region of the cell body (Fig. 7 B) and is not found in the leading edge where F-actin is concentrated (Fig. 7 A). The staining pattern of FLAG-PAKa in the posterior cortex overlaps with the phalloidin staining of F-actin in that region. Similarly, HA-tagged PAKa localizes to the posterior cortex (Fig. 7D and Fig. E). Knowing that paka null cells have a cytokinesis defect, we examined the localization of HA-tagged PAKa in wild-type cells undergoing cytokinesis. As depicted in Fig. 7F and Fig. G, HA-PAKa localizes to the cortex of the cleavage furrows of dividing cells. These results are consistent with involvement of PAKa in cytokinesis.
Figure 7.
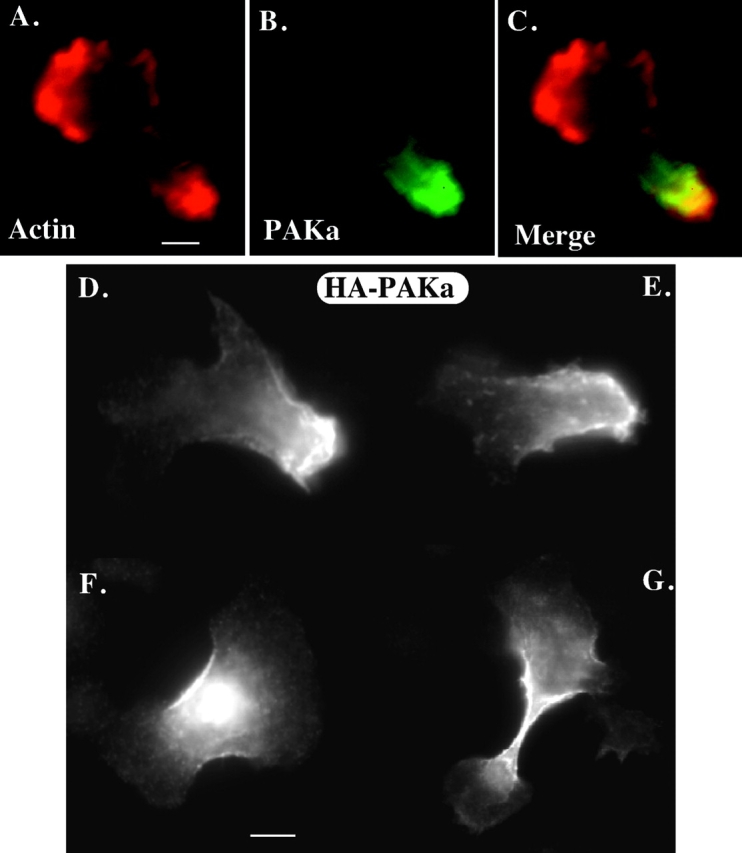
Localization of PAKa in the cell. The distribution of PAKa was examined in cells expressing FLAG- (at the COOH terminus) or HA- (at the NH2 terminus) tagged PAKa by indirect immunofluorescence staining as described in the Materials and Methods. A, Distribution of F-actin stained with FITC-phalloidin. F-actin is mainly concentrated at the leading edge of a migrating cell, but is found in the posterior cell body. B, Localization of FLAG-tagged PAKa in a cell at the aggregation stage. PAKa localizes in the posterior cell body. C, Merge of A and B. Note that F-actin staining and PAKa staining are overlapped in the posterior cell body. D and E, Distribution of HA-tagged PAKa in cells at the aggregation stage. Note that PAKa is concentrated at the posterior cortex. F and G, Localization of HA–PAKa in the cleavage furrow. Bar, 5 μM.
The subcellular localization of PAKa during chemotaxis and cytokinesis is very similar to that of myosin II. This raises the possibility that PAKa might regulate myosin II and, in turn, control the cytoskeletal organization in the posterior cell body. To test this possibility, we examined the distribution of a myosin II–green fluorescent protein (GFP) fusion protein (Moores et al. 1996) in wild-type cells and paka null aggregation stage cells. Myosin II–GFP mainly localizes at the cortex of the rear cell body in wild-type cells (Fig. 8 A) as has been described previously (Yumura et al. 1984; Yumura and Fukui 1985; Moores et al. 1996). However, in paka null cells, we observe a much more diffuse staining pattern of myosin II–GFP throughout the cells and no posterior cortical cap. We examined the localization of myosin II by indirect immunofluorescence staining using the antimyosin II antibody (Egelhoff et al. 1991). Myosin II is concentrated in the posterior cell body in aggregation-competent wild-type cells (Fig. 8 B) as shown by the myosin II–GFP fusion, whereas the staining of myosin II in paka null cells does not exhibit a strong subcellular localization. In contrast, we found strong myosin II staining along the membrane cortex of cells expressing myrPAKa, suggesting that anchoring PAKa to the plasma membrane promotes assembly of myosin II filaments at the membrane cortex (Fig. 8 B). This increased myosin II assembly at the membrane cortex is not found in cells expressing myrPAKaDN (Fig. 8B, Fig. d), reflecting that myosin II assembly at the membrane cortex is due to the PAKa activity. However, cells expressing myrPAKaDN do not have a posterior that is enriched in myosin II. We expect this is because myrPAKaDN functions (like non–myr-tagged PAKaDN) as a dominant negative protein and inhibits intrinsic PAKa activity. Like myrPAKa, myrPAKaDN predominantly localizes to the membrane cortex as determined by immunostaining (Fig. 8B, Fig. f; data not shown for myrPAKaDN).
Cells expressing PAKaCA have regions highly enriched in myosin II throughout the cell. These highly enriched myosin II-containing regions probably contain highly assembled myosin II. This result strongly suggests that PAKa might be involved in the regulation of myosin II assembly into the cytoskeleton. To test this idea, we determined the amount of myosin II in cytoskeleton fraction in wild-type cells, paka null cells, and cells expressing PAKCA in both vegetative growth and aggregation stages. In vegetatively growing cells, the level of myosin II assembled into the cytoskeleton in both wild-type and paka null cells is low, compared with cells at the aggregation stage (Fig. 9 A). However, cells expressing PAKCA display a higher level (approximately four times higher than wild-type cells) of myosin II assembly in the cytoskeleton. This result is consistent with PAKa activity being involved in regulating myosin II assembly.
Figure 9.
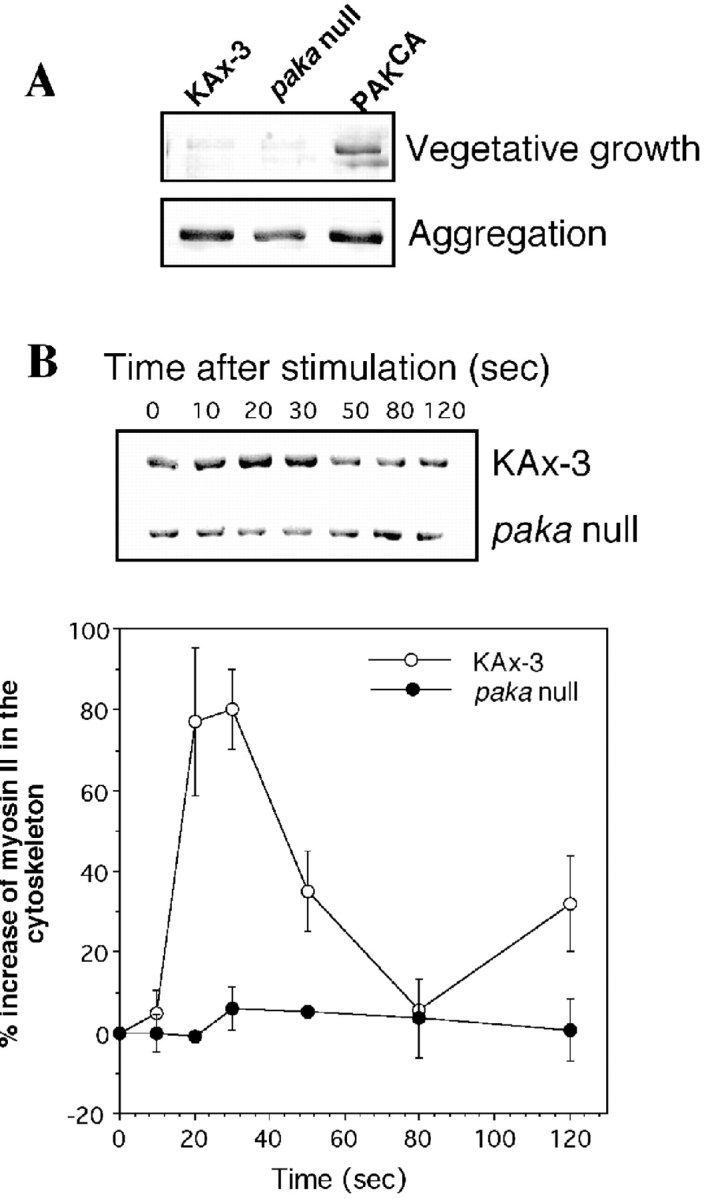
A, Level of myosin II content in cytoskeleton fractions of wild-type cells, paka null cells, and cells expressing PAKaCA. Cells (2 × 107) grown in axenic medium in shaken culture were collected by centrifugation and lysed with lysis buffer (see Materials and Methods). For aggregation-competent cells, the same number of cells was plated on a 100-mm petri dish, washed twice with phosphate buffer, and starved for 5 h. After 5 h, cells were lysed by adding lysis buffer and lysates were collected by using a rubber policeman. Lysates (containing 500 μg of protein) were spun at 11,000 g for 4 min to collect the pellet containing the cytoskeletal fraction, which was washed once with lysis buffer. The pellet was dissolved in 2× SDS-PAGE sample buffer and boiled for 5 min. Samples were run on 8% SDS gels and proteins were stained with Coomassie blue. Gel bands were scanned and changes in myosin II content were quantified with IPLAB software. In the vegetative, growing stage in wild-type and paka null cells, little of the myosin II is assembled into the cytoskeleton fraction, but the cytoskeleton from cells expressing PAKaCA has a much higher myosin II content, resulting from higher PAKa activity. In the aggregation stage, the myosin II level is elevated in all three cell types, but paka null cells exhibit a lower level of myosin II content than wild-type cells. B, Changes of myosin II content in the cytoskeleton upon cAMP stimulation. Cells were pulsed (see Materials and Methods), washed with phosphate buffer, and incubated in 1 mM caffeine for 10 min. After washing the cells, 100 μM cAMP was added and an equal number of cells was collected at specific time points. Cell lysis was immediate. Determination of the myosin II content in the cytoskeleton fraction was done as described above and graphed in the lower part of the figure. The data are from four separate experiments. Myosin II content peaks at 30 s after cAMP stimulation in wild-type KAx3 cells, but this peak is missing in paka null cells.
The level of myosin II in the cytoskeleton in aggregation-competent, wild-type cells is significantly higher (seven to eight times higher) than that in growth-stage, vegetative cells (Fig. 9 A). This difference is probably due to myosin II assembly in the rear cell body upon cell polarization in aggregation-competent cells. In paka null cells, the level of assembled myosin II is ∼65% of that of wild-type cells, suggesting that PAKa is required to control the level of myosin II assembly. Interestingly, there is no striking difference of myosin II level between wild-type cells and cells expressing PAKCA at the aggregation stage (probably because the assembly of myosin II is already activated in aggregation-competent cells).
We examined whether PAKa function is important in controlling changes in myosin II assembly into the cytoskeleton upon cAMP stimulation (Liu and Newell 1988). As shown in Fig. 9 B, in response to cAMP stimulation, the level of myosin II heavy chain in the cytoskeletal fraction in wild-type cells rises to a peak at 20–30 s and returns to the basal level by 60–80 s, consistent with previous observations (Liu and Newell 1988). However, this peak of myosin II assembly upon cAMP stimulation into the cytoskeleton is absent in paka null cells. These results suggest that PAKa might be involved in regulating myosin II assembly via either direct phosphorylation of myosin II or regulation of kinases controlling myosin II assembly.
Regulation of PAKa in Kinase Activity and its Localization upon Chemoattractant Stimulation
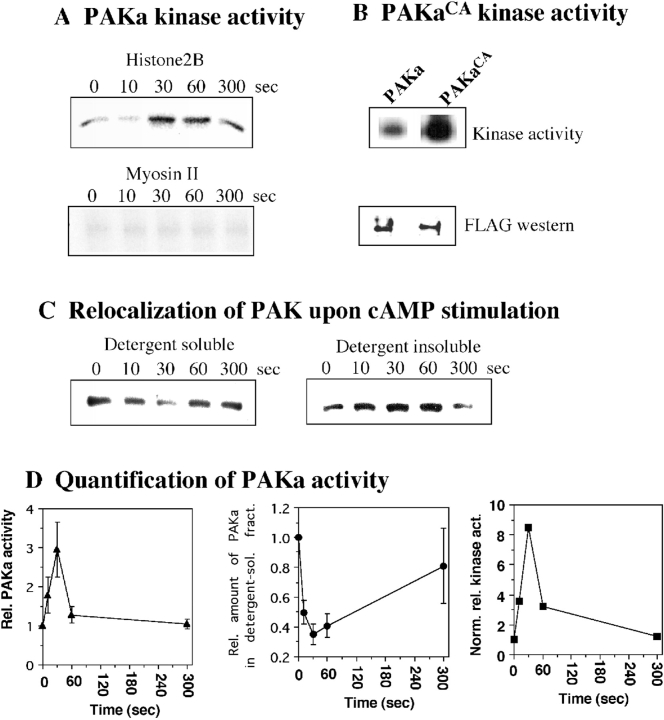
To determine whether Dictyostelium PAKa activity is stimulated in response to the chemoattractant, aggregation stage cells expressing FLAG-tagged PAKa were stimulated by cAMP, lysed at various times after stimulation, and PAKa–FLAG was immunoprecipitated from the detergent-soluble fraction (see Materials and Methods). Fig. 10 A shows that the kinase activity of immunoprecipitated PAKa towards a nonspecific substrate, H2B, rapidly increases three- to fourfold in response to cAMP. Maximal H2B phosphorylation is obtained within 30 s, after which the activity decreases slowly with kinetics similar to those of the adaptation of cAMP receptor-stimulated ERK2 kinase activity (Maeda and Firtel 1997). The kinetics of PAKa activation are very similar to those of PAK activation in human neutrophil stimulated by fMet-Leu-Phe (Knaus et al. 1995).
Figure 10.
A, Activation of PAKa upon stimulation of cells with cAMP. PAKa activity of immunoprecipitates towards histone 2B and myosin II as substrates is shown. Aggregation stage cells expressing FLAG-tagged PAKa (see Materials and Methods) were activated by cAMP. Aliquots were taken at various time points after stimulation. Cells were lysed, PAKa was precipitated using an anti-FLAG mAb, and kinase activity was measured as described in Materials and Methods. Phosphorylation of histone 2B (H2B), but not myosin II, is stimulated by cAMP. B, Activity of PAKaCA compared with PAKa. Cells expressing FLAG-tagged PAKa and PAKaCA were pulsed and PAKa and PAKaCA were immunoprecipitated with anti-FLAG antibody. Kinase activity was measured as described above. The same amount of PAKaCA (determined by Western analysis with anti-FLAG antibody) showed a much higher basal activity than PAKa. C, Incorporation of PAKa–FLAG into the cytoskeleton upon the stimulation of cAMP. Cells were prepared as described in the kinase assay and cell lysate was spun at 11,000 g for 5 min to collect the detergent-insoluble cytoskeleton fraction. The amount of PAKa–FLAG in the detergent-soluble cytosol and insoluble cytoskeleton fraction was determined by immunoblotting with the anti-FLAG antibody. The level of PAKa–FLAG in the detergent-soluble fraction was rapidly decreased upon the cAMP stimulation and the level of PAKa–FLAG in the detergent-insoluble cytoskeleton fraction was reciprocally increased. This relocalization of PAKa–FLAG was reversible and the level of PAKa–FLAG in the detergent-soluble fraction returned to the level of unstimulated cells after 5 min. The kinetics of the incorporation of PAK–FLAG are similar to those of PAKa activation by cAMP. D, Quantification of PAKa activity. The PAKa activity normalized by the amount of PAKa in the detergent-soluble fraction was plotted. The data are from four separate experiments.
In a process of verifying that there is an equal amount of PAKa–FLAG in each immunoprecipitate, we observed that the amount of PAKa–FLAG in the detergent-soluble fraction is rapidly decreased in response to cAMP. This decrease of PAKa–FLAG in the detergent-soluble fraction is due to the incorporation of PAKa–FLAG into a detergent-insoluble cytoskeleton fraction (Fig. 10 C). The kinetics of PAKa relocalization into the cytoskeleton are very similar to the kinetics of myosin II assembly into the cytoskeleton upon cAMP stimulation. This result suggests that PAKa is involved in the regulation of myosin II assembly. When PAKa kinase activity is normalized for the amount of PAKa protein, activity increases approximately ninefold.
We tested whether PAKa might directly regulate myosin II through its phosphorylation in the COOH-terminal tail fragment (Egelhoff et al. 1991; Yumura and Kitanishi-Yumura 1992). As shown in Fig. 10 A, we do not observe myosin II phosphorylation by PAKa, suggesting that PAKa indirectly regulates the assembly of myosin II, probably via the regulation of MHCKs. As expected, PAKCA immunoprecipitated from aggregation-competent cells has three to four times higher basal activity against H2B (Fig. 10 B), but the activity is not regulated in response to cAMP stimulation, probably because the activity is already maximal.
Translocation of PAKa upon Change of Cell Polarity
To access the localization of PAKa in live, migrating cells, we fused the NH2-terminal regulatory domain of PAKa (excluding the CRIB and kinase domains) to GFP. Cells expressing N-PAKa–GFP chemotax normally (data not shown). As we demonstrate in Fig. 11 A, N-PAKa–GFP fusion protein localizes to the cortex at the cell's posterior, as we observed with FLAG- or HA-tagged PAKa, suggesting the NH2-terminal domain used in this fusion is sufficient for proper subcellular localization of PAKa. As cells migrate, the N-PAKa–GFP fusion protein remains at the rear cortical region (Fig. 11 A), suggesting that PAKa activity is localized in the posterior and thus, may be important to maintain the polarity of cells during chemotaxis and retract the posterior cell body. To determine whether the localization of PAKa can be altered by disrupting cell polarity, we examined the distribution of N-PAKa–GFP by overlaying the cells with a receptor-saturating cAMP solution (Fig. 11 B). As cells lose their polarity and round up, N-PAKa–GFP remains associated with the cortex, but diffuses away from its previous location to be distributed along the membrane cortex over the entire cell. Fig. 11 C shows the quantification of the stimulus-induced relocalization of N-PAK–GFP at the plasma membrane. The differences in fluorescence intensities before and after the addition of cAMP over the cell were measured along a thin line through the central portion of cells. In the polarized cell (frame 1), the fluorescence intensity in the rear cell body is tenfold higher than the intensity in the leading edge. However, the fluorescence intensity in the rear cell body is remarkably reduced by 50 s after cAMP stimulation. The intensity in the leading edge is increased ∼2.5 times. The kinetics of the translocation of N-PAK–GFP from the rear cell body to leading edge are presented in Fig. 11 D as a measure of the fluorescence intensity along the thin line through the anterior part of the cell. The translocation of N-PAK–GFP reached maximum ∼30–40 s after stimulation, which is similar to the kinetics of PAK kinase activity upon the cAMP stimulation.
Figure 11.
Localization of N-PAKa–GFP protein in a migrating Dictyostelium cell. A, N-PAKa–GFP fusion protein was expressed in a wild-type cell and the localization of fusion protein was examined in a live, migrating cell. Cells expressing GFP fusion protein were pulsed with 30 nM cAMP for 5 h and plated on a coverslip. The fusion protein stays at the posterior cortex while cell is moving. A single cell was viewed at 20-s intervals. A low level of visible light was used to view the cell body in combination with fluorescent GFP fusion protein. B, Localization of fusion protein in a cell was examined when the cell lost polarity by overlaying 150 μM cAMP solution on the cell. GFP fusion protein is originally localized at the posterior cortex and diffuses along the membrane cortex as the cell loses polarity and starts rounding up. A single cell was viewed at 12-s intervals. A solid white arrow points to the leading edge. C, Quantification of the stimulus-induced relocalization of N-PAKa–GFP at the plasma membrane. The difference in fluorescence intensities before and after the addition of cAMP over the cell was measured along a thin line through the central portion of cells (Fig. 9 B, frames 1 and 8). D, Kinetic analysis of the relocalization of N-PAKa–GFP, along with plasma membrane. The kinetics of the translocation of N-PAKa–GFP from the rear cell body to the membrane cortex over the entire cell are presented as a measure of the fluorescence intensity along the thin line through the anterior part of the cell. The translocation of N-PAKa–GFP reached maximum ∼30–40 s after stimulation. The data shown are representative of multiple experiments.
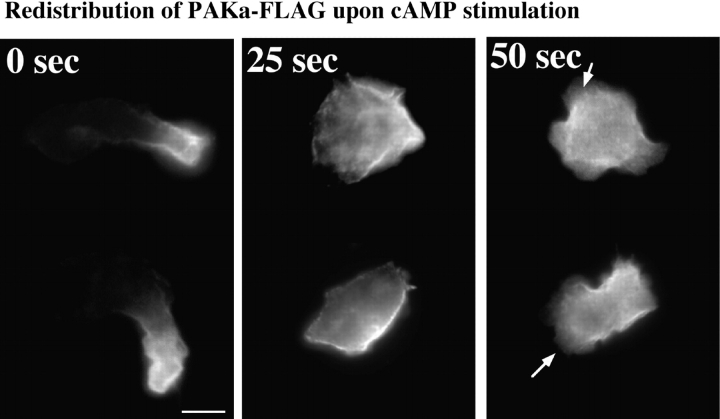
To examine whether the localization of holo-PAKa shows similar changes upon cAMP stimulation, cells expressing PAKa–FLAG were pulsed and plated on coverslips. Cells were bathed with 100 μM cAMP and fixed at 0, 25, and 50 s after cAMP stimulation and the localization of PAKa–FLAG was determined by indirect immunofluorescence staining. PAKa was localized in the rear cell body of polarized, unstimulated cells (Fig. 12) and relocalized along the membrane cortex 25 s after cAMP stimulation, which is very similar to the relocalization of N-PAK–GFP. In contrast to the distribution of N-PAK–GFP, PAKa staining in cells fixed at 50 s was absent in the cortex where new pseudopodia were formed, suggesting PAKa might be excluded in newly formed pseudopodia. The difference in the behavior between PAKa and N-PAK–GFP might result from the lack of a CRIB domain in N-PAK–GFP.
Figure 12.
Redistribution of PAKa upon cAMP stimulation. Cells expressing PAKa–FLAG were pulsed with 30 nM cAMP for 5 h and plated on a coverslip. Cells were bathed with 100 μM cAMP and fixed at 0, 25, and 50 s after stimulation. The localization of PAKa–FLAG was examined by indirect immunofluorescence staining with anti-FLAG antibody. Arrows indicate newly formed pseudopodia. Bar, 5 μm.
Discussion
PAKa Exhibits a Dynamic Subcellular Localization
We have identified a gene encoding a serine/threonine kinase, PAKa, that is a putative member of the PAK family of protein kinases. We find that PAKa has a dynamic subcellular localization. PAKa localizes to the cleavage furrow of cells undergoing cytokinesis and the posterior cortex of polarized cells and colocalizes in the same region of the cell with myosin II. Using a GFP fusion, we demonstrate that the NH2-terminal region lacking the CRIB domain is sufficient for the proper subcellular localization of PAKa in live, chemotaxing cells. If cells are overlaid with a receptor-saturating concentration of cAMP, the N-PAKa–GFP fusion proteins remain associated with the cell cortex, but delocalize along the cortex of the cell as it loses its polarized shape. Our data suggest that the NH2-terminal domain associates with a dynamically localized component of the cell cortex that is found in the posterior of polarized cells. Moreover, we suggest that the establishment of polarity in the cortical region responsible for this localization and the subsequent rearrangements of the cortical region are controlled through the chemoattractant receptors. We suggest that the localization of PAKa to a specific subdomain of the cell is a mechanism for restricting its activity to a specific site in the cell. This might be particularly important if its substrate is more uniformly distributed. Expression of PAKaCA, which is randomly localized in the cells, or myrPAKa, which is uniformly membrane-localized, upregulates the actin and myosin cytoskeletons and the cells' lack of cell polarity. These results are in agreement with our view that PAKa's tethering to the posterior of the cell is an important aspect of regulating PAKa function and may be important in maintaining the polarity of chemotaxing cells.
PAKa is a Regulator of the Myosin Component of the Cytoskeleton: Regulation of Cytokinesis
paka null cells exhibit two major phenotypes associated with an inability to properly regulate the cytoskeleton: the cells fail to complete cytokinesis when grown in suspension and the cells exhibit defects in chemotaxis. Our results provide the first identification of an essential role of a putative PAK in cytokinesis. Dictyostelium cells and mammalian cells undergo cytokinesis via the formation and constriction of a myosin II-rich contractile ring in the cleavage furrow. myoII null cells do not divide in suspension due to the inability to perform cytokinesis, but undergo cytokinesis on an adhesive surface by traction-mediated cytofission in which dividing daughter cells are pulled apart (Yumura et al. 1984; Yumura and Fukui 1985; De Lozanne and Spudich 1987; Knecht and Loomis 1988; Manstein et al. 1989; Fukui et al. 1990; Moores et al. 1996). Several small G proteins and GAPs are required for proper cytokinesis in Dictyostelium. These genes include: RasG (Tuxworth et al. 1997); two IQGAP-related genes, Ddrasgap1 and GapA (Adachi et al. 1997; Lee et al. 1997); and a novel Rac protein, RacE (Larochelle et al. 1997). RasG might function via the activation of the PI3 kinase because deletions in two PI3 kinase genes (PI3K1 and PI3K2) lose the ability to grow in suspension (Zhou et al. 1995) and RasG interacts with the Ras binding domain of PI3K1 and PI3K2 (Reddy, T.B.K., and R.A. Firtel, unpublished observations). One of the IQGAP-related proteins, Ddrasgap1, interacts with activated DdRac1B and is thus a potential effector of Rac1B (Faix et al. 1998; Lee, S., and R.A. Firtel, unpublished observations), which interacts with PAKa. It is not known if Rac1B interacts with Ddrasgap1 in vivo and regulates its role in cytokinesis. racE null cells fail to divide in suspension and become multinucleated (Larochelle et al. 1997). RacE could be an upstream regulator of PAKa. However, we only observe a strong interaction with activated Rac1B in yeast two-hybrid system and no interactions with activated RacE.
PAKa Functions during Chemotaxis as a Potential Regulator of the Myosin II Component of the Cytoskeleton
paka null cells exhibit defects in chemotaxis and morphogenesis during multicellular development. The chemotaxis phenotype is exemplified by inability of paka null cells or wild-type cells expressing PAKaDN to maintain polarity, regulate the formation of pseudopodia, and control the contraction of the rear cell body during cell migration. In chemotaxing cells, myosin II aligns along the posterior of the cell, forming a C-shaped cap (Yumura et al. 1984; Clow and McNally 1999). This myosin cap contracts, causing the posterior of the cell to lift and allowing it to retract towards the leading edge (Jay et al. 1995). In myoII and paka null cells, the retraction of the posterior of the cell during chemotaxis is defective (Jay et al. 1995; Clow and McNally 1999; this manuscript). We find that this C-shaped cap of myosin II in wild-type cells is much less visible or absent in paka null cells. Moreover, we found that paka null cells are defective in the assembly of myosin II into the cytoskeleton upon cAMP stimulation, suggesting that PAKa is involved in the regulation of myosin II assembly. In addition, cells expressing PAKaCA have a much higher level of myosin II assembly in the cytoskeleton in vegetatively growing cells. These data are consistent with PAKa playing an important role in the regulation of myosin II assembly into the cytoskeleton.
Consistent with this, PAKa, as previously demonstrated for myosin II (Varnum-Finney et al. 1987), is important for suppressing lateral pseudopod formation. Both myoII and paka null cells form more lateral pseudopodia than do wild-type cells (Wessels et al. 1988; Wessels and Soll 1990; this manuscript). The inability to inhibit lateral pseudopod formation in myoII null cells may be associated with a reduction in cortical tension of these cells (Pasternak et al. 1989). The higher frequency of protrusion of lateral pseudopodia in paka null cells probably results from the reduction in cortical tension due to the lack of myosin II filament assembly at the posterior cortex. These cytoskeletal defects might also be a major cause of the morphological defects during multicellular development of paka null cells and myoII null cells (Springer et al. 1994; Chen et al. 1998).
Possible Model for PAKa Function in Controlling Myosin II Assembly
We demonstrate that PAKa kinase activity against H2B is stimulated in response to cAMP; however, our data indicate that myosin II is not phosphorylated by PAKa. This observation is consistent with a requirement of PAKa for the maintenance, but not the disassembly, of myosin II fibers. The regulation of myosin II filament assembly occurs through receptor-mediated phosphorylation of myosin II heavy chain tail and myosin II light chain (Berlot et al. 1985; Gerisch et al. 1993). Phosphorylations at threonine residues in the myosin II tail by two kinases, MHCKA and MHC-PKC, lead to the disassembly of myosin filaments (Berlot et al. 1985; Côté and McCrea 1987; Vaillancourt et al. 1988; Nachmias et al. 1989; Ravid and Spudich 1989; Egelhoff et al. 1991). MHCKA is expressed during growth and development (Futey et al. 1995). The mhckA null cells are viable, but display partial defects in myosin localization (Kolman et al. 1996). Cells overexpressing MHCKA display reduced myosin II assembly in the cytoskeleton and become large and multinucleated in suspension, which is very similar to phenotypes of myoII null and paka null cells. In this context, PAKa is likely to regulate myosin II assembly via inhibition of MHCKA. MHC-PKC is particularly interesting, since it translocates from the cytosol to the plasma membrane/cortex and is activated upon cAMP stimulation, leading to the disassembly of myosin II fibers (Dembinsky et al. 1996). Disruption of the gene encoding MHC-PKC leads to an upregulation of the myosin II cytoskeleton and chemotaxis defects (Abu-Elneel et al. 1996). Our data are consistent with a model in which PAKa phosphorylates and thereby inhibits the activity of MHC-PKC and possibly MHCKA. According to such a model, this posterior localization of PAKa would allow MHCK-mediated disassembly of myosin II fibers at the leading edge while inhibiting the disassembly of myosin II fibers at the posterior. Myosin II light chain kinases are not likely to be substrates of PAKa, since light chain phosphorylation controls myosin II motor activity and not its assembly (Griffith et al. 1987; Chen et al. 1995; Smith et al. 1996). Our results are consistent with the finding that cells expressing a myosin II mutant in which the three mapped MHC-PKC phosphorylation sites are converted to Ala do not maintain their shape and are unable to suppress the formation of lateral pseudopodia (Stites et al. 1998). Thus, the ability to sequentially assemble and disassemble myosin II, possibly in response to the activation and adaptation of PAKa kinase activity, appears to be critical for cell movement.
Dictyostelium PAKa has high homology to Dictyostelium MIHCK. In Acanthamoeba and Dictyostelium, PAK homologues phosphorylate myosin I heavy chain and stimulate the Mg2+-dependent ATPase activity (Lee et al. 1996; Brzeska et al. 1997). Immunohistochemical studies demonstrate that three classic myosin Is, myoI-B, myoI-C, and myoI-D, are concentrated at the leading edge of chemotaxing cells (Jung et al. 1993; Morita et al. 1996), and are not found in the cleavage furrow, suggesting that myosin Is are not PAKa substrates.
PAKs have been implicated in the morphological changes resulting from changes in the actin cytoskeleton associated with Rac and Cdc42 (Knaus and Bokoch 1998). Microinjection of activated PAK1 protein into quiescent Swiss 3T3 cells induces the rapid formation of lamellipodia, filopodia, and membrane ruffles (Sells et al. 1997), which is very similar to the effect of microinjection of activated Rac and Cdc42. The interaction between the proline-rich domain of PAK1 and a protein containing an SH3 domain appears to be important for regulating PAK1 activity. Nck, an adapter protein containing SH2 and SH3 domains, binds PAK1 and induces PAK1 relocalization to the membrane, leading to its activation (Lu et al. 1997). Other studies demonstrate that localization of PAK to membranes stimulates the kinase activity of PAK (Bokoch et al. 1996; Daniels et al. 1998) and our data with myrPAKa are consistent with this model. Although we did not examine whether the proline-rich domain of PAKa interacts with proteins containing an SH3 domain such as Nck, PAKa probably associates with an SH3 domain via the PPxP sequence, resulting in the localization of PAKa to the cortex. We note that PAKCA leads to a hyperpolymerization of actin. We do not know if this is a direct effect of PAKa on regulators of the actin cytoskeleton or is indirect through the control of myosin II. It is also possible that the PAKa kinase domain, when expressed by itself, may not show the same level of substrate specificity as the whole protein. Our observation that the expression of myrPAKa results in a hyperpolymerization of myosin and actin along the periphery of the cell, reinforces the model that the subcellular localization of PAKa to the posterior of the cell may be essential for its proper function. We suggest that this overexpression along the entire membrane or the localization by the myristoylation leads to a constitutive activation of the kinase.
Although we have not demonstrated that PAKa is directly regulated by Rac1 and/or Cdc42, the GTP-bound, but not GDP-bound, forms of Rac1B and HsCdc42 bind to the PAKa CRIB domain. These data are consistent with, but do not prove, that PAKa is a bone fide PAK. In contrast to the kinase domain alone, overexpression of the entire PAKa protein or the kinase and CRIB domains (PAKmG+K) does not cause a major change in the actin or myosin cytoskeleton. This finding is consistent with observations of mammalian PAKs in which the CRIB domain and an adjacent domain negatively regulate PAK kinase activity (Sells et al. 1997). We expect that binding of activated Rac or Cdc42 to the CRIB domain of PAK regulates the activation of the kinase, whereas the more NH2-terminal domain regulates PAKa's localization.
Cells overexpressing PAKa exhibit an upregulated assembly of F-actin, resulting in multiple actin-enriched crowns, which is very similar to the F-actin organization of cells expressing Rac1BQ61L or of null cells of the Rac1 GAP, DdRacGAP1 (Chung, C.Y., S. Lee, C. Briscoe, and R.A. Firtel, manuscript submitted for publication). These similarities of F-actin organization suggest PAKa may be important in regulating F-actin organization (Manser et al. 1997; Sells et al. 1997). A Dictyostelium Ste20 family kinase that phosphorylates severin, a Ca2+-dependent F-actin fragmenting protein, was purified and cloned recently (Eichinger et al. 1998), indicating a direct signal transduction from the plasma membrane to the cytoskeleton by phosphorylating actin-binding proteins. PAKa might control F-actin organization by phosphorylation of actin binding or actin-bundling proteins. The identification of the downstream substrates of PAKa and the determination of the role of phosphorylation by PAKa in regulating the function of the substrate should resolve these issues.
Acknowledgments
We would like to thank Hans Warrick and Jim Spudich (Stanford University) for the antimyosin II antibody, and Paul Steimle and Tom Egelhoff (Case Western Reserve University) for purified myosin II. We would also like to thank members of the Firtel lab for helpful suggestions throughout the period of this work.
This work was supported by National Institutes of Health grants to R.A. Firtel.
Footnotes
1.used in this paper: GFP, green fluorescent protein; H2B, histone 2B; MIHCK, myosin I heavy chain kinase; myrPAKa, a PAKa carrying the NH2-terminal myristoylation signal from Src; PAKaCA, a putative constitutively active PAKa containing only the kinase domain; PAKaDN, kinase dead, putative dominant, negative mutant of PAKa; PAKaG+K, a PAKa mutant containing the CRIB and kinase domains
References
- Abu-Elneel K., Karchi M., Ravid S. Dictyostelium myosin II is regulated during chemotaxis by a novel protein kinase C. J. Biol. Chem. 1996;271:977–984. doi: 10.1074/jbc.271.2.977. [DOI] [PubMed] [Google Scholar]
- Adachi H., Takahashi Y., Hasebe T., Shirouzu M., Yokoyama S., Sutoh K. Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J. Cell Biol. 1997;137:891–898. doi: 10.1083/jcb.137.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagrodia S., Dérijard B., Davis R.J., Cerione R.A. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J. Biol. Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- Berlot C.H., Spudich J.A., Devreotes P.N. Chemoattractant-elicited increases in myosin phosphorylation in Dictyostelium . Cell. 1985;43:307–314. doi: 10.1016/0092-8674(85)90036-4. [DOI] [PubMed] [Google Scholar]
- Bokoch G.M., Wang Y., Bohl B.P., Sells M.A., Quilliam L.A., Knaus U.G. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J. Biol. Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- Brzeska H., Knaus U.G., Wang Z.Y., Bokoch G.M., Korn E.D. p21-activated kinase has substrate specificity similar to Acanthamoeba myosin I heavy chain kinase and activates Acanthamoeba myosin I. Proc. Natl. Acad. Sci. USA. 1997;94:1092–1095. doi: 10.1073/pnas.94.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser C., Sigal C., Resh M., McLaughlin S. Membrane binding of myristylated peptides corresponding to the NH2 terminus of Src. Biochemistry. 1994;33:13093–13101. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- Cairns B.R., Ramer S.W., Kornberg R.D. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 1992;6:1305–1318. doi: 10.1101/gad.6.7.1305. [DOI] [PubMed] [Google Scholar]
- Chant J., Stowers L. GTPase cascades choreographing cellular behaviormovement, morphogenesis, and more. Cell. 1995;81:1–4. doi: 10.1016/0092-8674(95)90363-1. [DOI] [PubMed] [Google Scholar]
- Chen T.L., Kowalczyk P.A., Ho G., Chisholm R.L. Targeted disruption of the Dictyostelium myosin essential light chain gene produces cells defective in cytokinesis and morphogenesis. J. Cell Sci. 1995;108:3207–3218. doi: 10.1242/jcs.108.10.3207. [DOI] [PubMed] [Google Scholar]
- Chen T.L., Wolf W.A., Chisholm R.L. Cell-type–specific rescue of myosin function during Dictyostelium development defines two distinct cell movements required for culmination. Development. 1998;125:3895–3903. doi: 10.1242/dev.125.19.3895. [DOI] [PubMed] [Google Scholar]
- Clow P., McNally J.G. In vivo observations of myosin II dynamics support a role in rear retraction. Mol. Biol. Cell. 1999;10:1309–1323. doi: 10.1091/mbc.10.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.A. The role of actin polymerization in cell motility. Ann. Rev. Physiol. 1991;53:585–605. doi: 10.1146/annurev.ph.53.030191.003101. [DOI] [PubMed] [Google Scholar]
- Côté G.P., McCrea S.M. Selective removal of the carboxyl-terminal tail end of the Dictyostelium myosin II heavy chain by chymotrypsin. J. Biol. Chem. 1987;262:13033–13038. [PubMed] [Google Scholar]
- Cvrckova F., De Virgilio C., Manser E., Pringle J.R., Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Daniels R.H., Hall P.S., Bokoch G.M. Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Firtel R.A. Identification of the sequences controlling cyclic AMP regulation and cell-type–specific expression of a prestalk-specific gene in Dictyostelium discoideum . Mol. Cell. Biol. 1987;7:149–159. doi: 10.1128/mcb.7.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J.A. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Dembinsky A., Rubin H., Ravid S. Chemoattractant-mediated increases in cGMP induce changes in Dictyostelium myosin II heavy chain-specific protein kinase C activities. J. Cell Biol. 1996;134:911–921. doi: 10.1083/jcb.134.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., Fontana D., Klein P., Sherring J., Theibert A. Transmembrane signaling in Dictyostelium . Meth. Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- Egelhoff T.T., Brown S.S., Spudich J.A. Spatial and temporal control of nonmuscle myosin localizationidentification of a domain that is necessary for myosin filament disassembly in vivo. J. Cell Biol. 1991;112:677–688. doi: 10.1083/jcb.112.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L., Bähler M., Dietz M., Eckerskorn C., Schleicher M. Characterization and cloning of a Dictyostelium Ste20-like protein kinase that phosphorylates the actin-binding protein severin. J. Biol. Chem. 1998;273:12952–12959. doi: 10.1074/jbc.273.21.12952. [DOI] [PubMed] [Google Scholar]
- Faix J., Clougherty C., Konzok A., Mintert U., Murphy J., Albrecht R., Mühlbauer B., Kuhlmann J. The IQGAP-related protein DGAP1 interacts with Rac and is involved in the modulation of the F-actin cytoskeleton and control of cell motility. J. Cell Sci. 1998;111:3059–3071. doi: 10.1242/jcs.111.20.3059. [DOI] [PubMed] [Google Scholar]
- Fukui Y. Actomyosin organization in mitotic Dictyostelium amoebae. Ann. NY. Acad. Sci. 1990;582:156–165. doi: 10.1111/j.1749-6632.1990.tb21676.x. [DOI] [PubMed] [Google Scholar]
- Fukui Y., De Lozanne A., Spudich J.A. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J. Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futey L.M., Medley Q.G., Côté G.P., Egelhoff T.T. Structural analysis of myosin heavy chain kinase A from Dictyostelium. Evidence for a highly divergent protein kinase domain, an amino-terminal coiled-coil domain, and a domain homologous to the beta-subunit of heterotrimeric G proteins. J. Biol. Chem. 1995;270:523–529. doi: 10.1074/jbc.270.2.523. [DOI] [PubMed] [Google Scholar]
- Gaskins C., Clark A.M., Aubry L., Segall J.E., Firtel R.A. The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes Dev. 1996;10:118–128. doi: 10.1101/gad.10.1.118. [DOI] [PubMed] [Google Scholar]
- Gerisch G., Albrecht R., De Hostos E., Wallraff E., Heizer C., Kreitmeier M., Müller-Taubenberger A. Actin-associated proteins in motility and chemotaxis of Dictyostelium cells. Symp. Soc. Exp. Biol. 1993;47:297–315. [PubMed] [Google Scholar]
- Griffith L.M., Downs S.M., Spudich J.A. Myosin light chain kinase and myosin light chain phosphatase from Dictyosteliumeffects of reversible phosphorylation on myosin structure and function. J. Cell Biol. 1987;104:1309–1323. doi: 10.1083/jcb.104.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis E., Chertkov H., Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Häcker U., Perrimon N. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila . Genes Dev. 1998;12:274–284. doi: 10.1101/gad.12.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeastfor mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hing H., Xiao J., Harden N., Lim L., Zipursky S. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila . Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Insall R.H., Soede R.D.M., Schaap P., Devreotes P.N. Two cAMP receptors activate common signaling pathways in Dictyostelium . Mol. Biol. Cell. 1994;5:703–711. doi: 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P.Y., Pham P.A., Wong S.A., Elson E.L. A mechanical function of myosin II in cell motility. J. Cell Sci. 1995;108:387–393. doi: 10.1242/jcs.108.1.387. [DOI] [PubMed] [Google Scholar]
- Jung G., Fukui Y., Martin B., Hammer J.A., III. Sequence, expression pattern, intracellular localization, and targeted disruption of the Dictyostelium myosin ID heavy chain isoform. J. Biol. Chem. 1993;268:14981–14990. [PubMed] [Google Scholar]
- Knaus U.G., Bokoch G.M. The p21Rac/Cdc42-activated kinases (PAKs) Int. J. Biochem. Cell Biol. 1998;30:857–862. doi: 10.1016/s1357-2725(98)00059-4. [DOI] [PubMed] [Google Scholar]
- Knaus U.G., Morris S., Dong H.-J., Chernoff J., Bokoch G.M. Regulation of human leukocyte p21-activated kinase through G protein-coupled receptors. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- Knecht D.A., Loomis W.F. Developmental consequences of the lack of myosin heavy chain in Dictyostelium discoideum . Dev. Biol. 1988;128:178–184. doi: 10.1016/0012-1606(88)90280-1. [DOI] [PubMed] [Google Scholar]
- Kolman M.F., Futey L.M., Egelhoff T.T. Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J. Cell Biol. 1996;132:101–109. doi: 10.1083/jcb.132.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron S.J., Gow N.A. Budding yeast morphogenesissignaling, cytoskeleton, and cell cycle. Curr. Opin. Cell Biol. 1995;7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- Larochelle D.A., Vithalani K.K., De Lozanne A. Role of Dictyostelium racE in cytokinesismutational analysis and localization studies by use of green fluorescent protein. Mol. Biol. Cell. 1997;8:935–944. doi: 10.1091/mbc.8.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Escalante R., Firtel R.A. A Ras GAP is essential for cytokinesis and spatial patterning in Dictyostelium . Development. 1997;124:983–996. doi: 10.1242/dev.124.5.983. [DOI] [PubMed] [Google Scholar]
- Lee S., Parent C.A., Insall R., Firtel R.A. A novel Ras interacting protein required for chemotaxis and cAMP signal relay in Dictyostelium . Mol. Biol. Cell. 1999;In press doi: 10.1091/mbc.10.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.F., Egelhoff T.T., Mahasneh A., Côté G.P. Cloning and characterization of a Dictyostelium myosin I heavy chain kinase activated by Cdc42 and Rac. J. Biol. Chem. 1996;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]
- Li R., Zheng Y., Drubin D.G. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J. Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Newell P.C. Evidence that cyclic GMP regulates myosin interaction with the cytoskeleton during chemotaxis of Dictyostelium . J. Cell Sci. 1988;90:123–129. doi: 10.1242/jcs.90.1.123. [DOI] [PubMed] [Google Scholar]
- Lu W., Katz S., Gupta R., Mayer B.J. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr. Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- Ma H., Gamper M., Parent C., Firtel R.A. The Dictyostelium MAP kinase kinase DdMEK1 regulates chemotaxis and is essential for chemoattractant-mediated activation of guanylyl cyclase. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:4317–4332. doi: 10.1093/emboj/16.14.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I. Cleavage furrowtiming of emergence of contractile ring actin filaments and establishment of the contractile ring by filament bundling in sea urchin eggs. J. Cell Sci. 1994;107:1853–1862. doi: 10.1242/jcs.107.7.1853. [DOI] [PubMed] [Google Scholar]
- Maeda M., Firtel R.A. Activation of the mitogen-activated protein kinase ERK2 by the chemoattractant folic acid in Dictyostelium . J. Biol. Chem. 1997;272:23690–23695. doi: 10.1074/jbc.272.38.23690. [DOI] [PubMed] [Google Scholar]
- Mann S.K.O., Firtel R.A. Cyclic AMP regulation of early gene expression in Dictyostelium discoideummediation via the cell surface cyclic AMP receptor. Mol. Cell. Biol. 1987;7:458–469. doi: 10.1128/mcb.7.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E., Leung T., Salihuddin H., Zhao Z.S., Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- Manser E., Huang H.Y., Loo T.H., Chen X.Q., Dong J.M., Leung T., Lim L. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein D.J., Titus M.A., De Lozanne A., Spudich J.A. Gene replacement in Dictyosteliumgeneration of myosin null mutants. EMBO (Eur. Mol. Biol. Organ.) J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R., Ellsworth C., Lee S., Reddy T.B.K., Ma H., Firtel R.A. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium . EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores S.L., Sabry J.H., Spudich J.A. Myosin dynamics in live cells. Proc. Natl. Acad. Sci. USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y.S., Jung G., Hammer J.A., 3rd, Fukui Y. Localization of Dictyostelium myoB and myoD to filopodia and cell–cell contact sites using isoform-specific antibodies. Eur. J. Cell Biol. 1996;71:371–379. [PubMed] [Google Scholar]
- Nachmias V.T., Fukui Y., Spudich J.A. Chemoattractant-elicited translocation of myosin in motile Dictyostelium . Cell Motil. Cytoskel. 1989;13:158–169. doi: 10.1002/cm.970130304. [DOI] [PubMed] [Google Scholar]
- Nobes C.D., Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Nobes C.D., Hawkins P., Stephens L., Hall A. Activation of the small GTP-binding proteins rho and rac by growth factor receptors. J. Cell Sci. 1995;108:225–233. doi: 10.1242/jcs.108.1.225. [DOI] [PubMed] [Google Scholar]
- Otto J.J., Schroeder T.E. Association of actin and myosin in the contractile ring. Ann. NY. Acad. Sci. 1990;582:179–184. doi: 10.1111/j.1749-6632.1990.tb21678.x. [DOI] [PubMed] [Google Scholar]
- Pasternak C., Spudich J.A., Elson E.L. Capping of surface receptors and concomitant cortical tension are generated by conventional myosin. Nature. 1989;341:549–551. doi: 10.1038/341549a0. [DOI] [PubMed] [Google Scholar]
- Ramos E., Wysolmerski R.B., Masaracchia R.A. Myosin phosphorylation by human cdc42-dependent S6/H4 kinase/gammaPAK from placenta and lymphoid cells. Recept. Signal Transduct. 1997;7:99–110. [PubMed] [Google Scholar]
- Ravid S., Spudich J.A. Myosin heavy chain kinase from developed Dictyostelium cells. Purification and characterization. J. Biol. Chem. 1989;264:15144–15150. [PubMed] [Google Scholar]
- Saxe C.L., III, Johnson R.L., Devreotes P.N., Kimmel A.R. Expression of a cAMP receptor gene of Dictyostelium and evidence for a multigene family. Genes Dev. 1991;5:1–8. doi: 10.1101/gad.5.1.1. [DOI] [PubMed] [Google Scholar]
- Sells M.A., Knaus U.G., Bagrodia S., Ambrose D.M., Bokoch G.M., Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- Smith J.L., Silveira L.A., Spudich J.A. Myosin light chain kinase (MLCK) gene disruption in Dictyosteliuma role for MLCK-A in cytokinesis and evidence for multiple MLCKs. Proc. Nat. Acad. Sci. USA. 1996;93:12321–12326. doi: 10.1073/pnas.93.22.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M.L., Patterson B., Spudich J.A. Stage-specific requirement for myosin II during Dictyostelium development. Development. 1994;120:2651–2660. doi: 10.1242/dev.120.9.2651. [DOI] [PubMed] [Google Scholar]
- Spudich J.A., Finer J., Simmons B., Ruppel K., Patterson B., Uyeda T. Myosin structure and function. Cold Spring Harbor Symp. Quant. Biol. 1995;60:783–791. doi: 10.1101/sqb.1995.060.01.084. [DOI] [PubMed] [Google Scholar]
- Stites J., Wessels D., Uhl A., Egelhoff T., Shutt D., Soll D.R. Phosphorylation of the Dictyostelium myosin II heavy chain is necessary for maintaining cellular polarity and suppressing turning during chemotaxis. Cell Motil. Cytoskel. 1998;39:31–51. doi: 10.1002/(SICI)1097-0169(1998)39:1<31::AID-CM4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Tapon N., Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- Tuxworth R.I., Cheetham J.L., Machesky L.M., Spiegelmann G.B., Weeks G., Insall R.H. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt J.P., Lyons C., Côté G.P. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J. Biol. Chem. 1988;263:10082–10087. [PubMed] [Google Scholar]
- Varnum-Finney B., Edwards K.B., Voss E., Soll D.R. Amebae of Dictyostelium discoideum respond to an increasing temporal gradient of the chemoattractant cAMP with a reduced frequency of turningevidence for a temporal mechanism in ameboid chemotaxis. Cell Motil. Cytoskel. 1987;8:7–17. doi: 10.1002/cm.970080103. [DOI] [PubMed] [Google Scholar]
- Vithalani K.K., Shoffner J.D., De Lozanne A. Isolation and characterization of a novel cytokinesis-deficient mutant in Dictyostelium discoideum . J. Cell Biol. 1996;62:290–301. doi: 10.1002/(sici)1097-4644(199608)62:2<290::aid-jcb16>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Wessels D., Soll D.R. Computer assisted analysis of cytoskeletal mutants of Dictyostelium discoideum . In: Soll D.R., Wessels D., editors. Motion Analysis of Living Cells. John Wiley & Sons; New York: 1998. pp. 101–140. [Google Scholar]
- Wessels D., Soll D.R. Myosin II heavy chain null mutant of Dictyostelium exhibits defective intracellular particle movement. J. Cell Biol. 1990;111:1137–1148. doi: 10.1083/jcb.111.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D., Soll D.R., Knecht D., Loomis W.F., De Lozanne A., Spudich J. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev. Biol. 1988;128:164–177. doi: 10.1016/0012-1606(88)90279-5. [DOI] [PubMed] [Google Scholar]
- Yumura S., Fukui Y. Reversible cyclic AMP-dependent change in distribution of myosin thick filaments in Dictyostelium . Nature. 1985;314:194–196. doi: 10.1038/314194a0. [DOI] [PubMed] [Google Scholar]
- Yumura S., Kitanishi-Yumura T. Release of myosin II from the membrane-cytoskeleton of Dictyostelium discoideum mediated by heavy-chain phosphorylation at the foci within the cortical actin network. J. Cell Biol. 1992;117:1231–1239. doi: 10.1083/jcb.117.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S., Mori H., Fukui Y. Localization of actin and myosin for the study of amoeboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol. 1984;99:894–899. doi: 10.1083/jcb.99.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Han J., Sells M.A., Chernoff J., Knaus U.G., Ulevitch R.J., Bokoch G.M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J. Biol. Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- Zhou K., Takegawa K., Emr S.D., Firtel R.A. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideumbiological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologs during growth and development. Mol. Cell. Biol. 1995;15:5645–5656. doi: 10.1128/mcb.15.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Preuss D., Mulholland J., O'Brien J.M., Botstein D., Johnson D.I. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]