Abstract
Since its discovery, qnrA has been found in most common Enterobacteriaceae. Ciprofloxacin MICs conferred by different qnrA-positive plasmids could range from 0.1 μg/ml to 2 μg/ml in Escherichia coli J53. The reasons for different ciprofloxacin MICs conferred by qnrA have not been fully clarified. Five hundred forty-one consecutive gram-negative clinical strains that were resistant or intermediate to ciprofloxacin and that were isolated in Shanghai in 2005 were screened for qnrA by PCR. For qnrA-positive isolates, the transferability of quinolone resistance was determined by conjugation and mutations within the quinolone resistance-determining region (QRDR) of gyrA and parC. aac(6′)-Ib-cr was detected and qnrA RNA expression was determined using real-time reverse transcription-PCR for transconjugants with different ciprofloxacin MICs. The qnrA gene was detected in 7 of the 541 clinical isolates. Quinolone resistance was transferred in four strains by conjugation. Mutations in the QRDR of gyrA and parC were detected in five qnrA-positive clinical strains with higher ciprofloxacin MICs. Of four qnrA-bearing plasmids in E. coli J53, pHS4 and pHS5 conferred ciprofloxacin MICs of 0.094 to 0.125 μg/ml; pHS3, which harbored the aac(6′)-Ib-cr gene as well, conferred a ciprofloxacin MIC of 0.25 μg/ml, and pHS6, which had both the aac(6′)-Ib-cr gene and a high expression level of qnrA, had a ciprofloxacin MIC of 1.0 μg/ml. The prevalence of qnrA appeared to be higher in Enterobacter cloacae than in other Enterobacteriaceae. The coexistence of qnrA and aac(6′)-Ib-cr in a single plasmid and increased qnrA expression can account for the different levels of ciprofloxacin resistance seen in transconjugants.
Plasmid-mediated quinolone resistance associated with the qnrA gene was discovered in 1998 (10). The originally described qnr gene is now referred to as qnrA because of recent findings of other related qnr genes, qnrB (7) and qnrS (5). Since the discovery of qnrA, qnrA was found in almost all populated continents and in most common Enterobacteriaceae including Escherichia coli, Klebsiella spp., Enterobacter spp., Citrobacter freundii, and Providencia stuartii (6, 16). The prevalence of qnrA varied from less than 1% to higher than 20%, depending on the population sampled. Plasmids harboring qnrA may also encode an extended-spectrum β-lactamase (ESBL) (6). qnrA was absent in nonfermenting gram-negative bacteria such as Pseudomonas aeruginosa and Acinetobacter spp. in small surveys, but whether the lack of detection reflects a true absence or the small number of strains tested is not clear (16). Most of these studies screened for qnrA from clinical isolates collected in the late 1990s or early 2000s, several years prior to each study. Usually, the strains screened had additional restriction conditions such as resistance to expanded-spectrum cephalosporins or carriage of genes encoding ESBLs simultaneously. Consecutive strains of Enterobacteriaceae were screened for qnrA in a few surveys (1).
qnrA conferred a low level of quinolone resistance, and the clinical strains that harbored qnrA usually had much higher MICs of ciprofloxacin than their respective transconjugants. The qnrA-bearing clinical strains might have an active efflux mechanism, decreased permeability, and/or additional mutations in the genes encoding the subunits of DNA gyrase and topoisomerase IV (22). Another bacterial defense mechanism is aac(6′)-Ib-cr, a variant of aminoglycoside acetyltransferase. In one study of these plasmids carrying qnrA, it was found that aac(6′)-Ib-cr had the ability to N-acetylate ciprofloxacin and norfloxacin at the amino nitrogen on its piperazinyl substituent. Other quinolones lacking unsubstituted piperazinyl nitrogen were unaffected. When the aac(6′)-Ib-cr gene was cloned into pBC SK and introduced into E. coli DH10B cells, the MIC of ciprofloxacin increased threefold to fourfold, and aac(6′)-Ib-cr increased the frequency of selection of chromosomal mutants upon exposure to ciprofloxacin substantially (19).
In this study, we examined the presence of the plasmid-mediated quinolone resistance determinant qnrA in consecutive gram-negative clinical strains that were resistant or intermediate to ciprofloxacin, regardless of the susceptibility to other antimicrobials including cephalosporins, isolated in a teaching hospital of Fudan University in Shanghai from 15 February to 15 July 2005. We have found that the transcriptional level of the qnrA gene had substantial effects on the qnrA expression level and hence the resistance of the strain to ciprofloxacin. Furthermore, aac(6′)-Ib-cr was identified as coexisting with qnrA in two plasmids. Finally, we identified the mutations within the quinolone resistance-determining region (QRDR) of gyrA and parC associated with resistance to quinolones.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Five hundred forty-one consecutive gram-negative clinical strains that were resistant or intermediate to ciprofloxacin were collected from a teaching hospital of Fudan University in Shanghai, China, between 15 February and 15 July 2005, including 169 strains of K. pneumoniae, 128 of P. aeruginosa, 98 of E. coli, 77 of A. baumannii, 13 of Enterobacter spp. (11 Enterobacter cloacae and 2 Enterobacter aerogenes), 9 of Citrobacter spp., 20 of other Enterobacteriaceae (6 of Proteus mirabilis, 6 of Klebsiella oxytoca, 5 of Serratia marcescens, and 3 of Morganella morganii), and 27 of other nonfermenting bacilli (11 strains of Stenotrophomonas maltophilia, 9 of Flavobacterium spp., 4 of Alcaligenes spp., and 3 of other Acinetobacter spp.). Inhibition zone diameter interpretive standards and MIC breakpoints for ciprofloxacin followed CLSI criteria: susceptible, zone diameter of ≥21 mm or MIC of ≤1 μg/ml; intermediate, zone diameter of 16 to 20 mm or MIC of 2 μg/ml; resistant, zone diameter of ≤15 mm or MIC of ≥4 μg/ml (2). Each isolate was from a separate patient, and most were from hospitalized patients. Additional strains and plasmids used in the study are given in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli J53AzR | Resistant to azide, met pro | 10 |
| E. coli HB101 | F′ Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr) xyl-5 mtl-1 recA13 rpsL20 (Strr) | TaKaRa |
| E. coli V517 | Multiple-plasmid-containing strain used as the molecular marker for plasmid | 22 |
| E. coli R1 | E. coli J53 containing plasmid R1 (92 kb) | 22 |
| E. coli Plac | E. coli J53 containing plasmid Plac (152 kb) | 22 |
| E. coli R27 | E. coli J53 containing plasmid R27 (182 kb) | 22 |
| UAB1 | K. pneumoniae containing pMG252, a qnrA-bearing plasmid | 10 |
| Plasmids | ||
| pKK232-8 | Promoter selection vector, replicon pMB1, Apr | Pharmacia |
| pKK232-8-HS3 | pKK232-8 containing the promoter for qnrA from plasmid pHS3 | This study |
| pKK232-8-HS4 | pKK232-8 containing the promoter for qnrA from plasmid pHS4 | This study |
| pKK232-8-HS5 | pKK232-8 containing the promoter for qnrA from plasmid pHS5 | This study |
| pKK232-8-HS6 | pKK232-8 containing the promoter for qnrA from plasmid pHS6 | This study |
Screening for the qnrA gene and conjugation.
The strains were screened for the presence of the qnrA gene by PCR with primers qnrA F and qnrA R (Table 2) to produce a 627-bp amplification product as previously described (21). Both strands of purified PCR products of qnrA were sequenced using dye terminators with an ABI 3730 automated sequencer.
TABLE 2.
Primers used in this study
| Gene or plasmid and primer | Sequence | Source or reference |
|---|---|---|
| qnrA | ||
| qnrA Fa | 5′-TCAGCAAGAGGATTTCTCA-3′ | 21 |
| qnrA Ra | 5′-GGCAGCA CTATTACTCCCA-3′ | 21 |
| RTqnrA Fb | 5′-TGCCAACTGCTTTGGCATAGA-3′ | This study |
| RTqnrA Rb | 5′-TGGCCACTCAAGTTGGTATAGG-3′ | This study |
| 16S rRNA | ||
| RT16S Fb | 5′-GCATAACGTCGCAAGACCAAAG-3′ | This study |
| RT16S Rb | 5′-TTCTTCATACACGCGGCATGG-3′ | This study |
| gyrA | ||
| gyrA Fa | 5′-AAATCTGCCCGTGTCGTTGGT-3′ | 20 |
| gyrA Ra | 5′-GCCATACCTACGGCGATACC-3′ | 20 |
| parC | ||
| parC Fa | 5′-CTGAATGCCAGCGCCAAATT-3′ | 20 |
| parC Ra | 5′-GCGAACGATTTCGGATCGTC-3′ | 20 |
| aac(6′)-Ib | ||
| aac Fa | 5′-ATATGCGGATCCAATGAGCAACGCAAAAACAAAGTTAG-3′ | 19 |
| aac Ra | 5′-ATATGCGAATTCTTAGGCATCACTGCGTGTTCGCTC-3′ | 19 |
| blaCTX-M | ||
| CTX-M-G1-F | 5′-AGT GCA AAC GGA TGA TGT-3′ | This study |
| CTX-M-G1-R | 5′-GGC TGG GTA AAA ATA GGT C-3′ | This study |
| qnrA promoter | ||
| Promoter F | 5′-CCGGATCCCGACCCCAAATCCAACA-3′ | This study |
| Promoter R | 5′-GGGAAGCTTACGGCTTCCTTTAATCAG-3′ | This study |
| pKK232-8 | ||
| pKK-SQ | 5′-TCCGGATGAGCATTCATCAG-3′ | This study |
Primers were used for amplification and sequencing.
Primers were used to carry out real-time PCR.
In order to determine if quinolone resistance was transferable in the strains with qnrA-bearing plasmids, conjugation experiments were carried out in LB broth with E. coli J53AzR as the recipient as previously described (21). Transconjugants were selected on Trypticase soy agar plates containing sodium azide (100 μg/ml) for counterselection and sulfamethoxazole (300 μg/ml), cefotaxime (10 μg/ml), gentamicin (10 μg/ml), or chloramphenicol (50 μg/ml) to select for plasmid-encoded resistance.
Plasmid DNAs were isolated with the QIAGEN (Hilden, Germany) Plasmid Midi kit, and plasmid size was estimated as previously described (22).
MIC determination.
MICs for the donor, recipient, transconjugant, and transformant strains were measured by agar dilution in accordance with the guidelines of the CLSI (2) for ciprofloxacin, ampicillin, cefotaxime, chloramphenicol, gentamicin, kanamycin, levofloxacin, streptomycin, sulfamethoxazole, tetracycline, tobramycin, and trimethoprim. The Etest (Biodisk AB, Solna, Sweden) was used to detect minimal changes in ciprofloxacin, levofloxacin, and chloramphenicol susceptibility.
QRDR sequencing.
The PCR amplifications were carried out with the TaKaRa LA PCR kit, and incubation conditions were those suggested by the manufacturer. Primers (Table 2) were used to amplify gyrA and parC. Purified PCR products were sequenced on both strands, and QRDR DNA sequences of gyrA and parC for each of the qnrA-positive isolates were compared with the QRDR DNA sequences of E. cloacae, E. coli, and C. freundii (GenBank accession numbers were AF052256, NC000913, and AF052253 for gyrA and D88981, NC000913, and AB003914 for parC, respectively).
Detection of aminoglycoside acetyltransferase aac(6′)-Ib.
The aac(6′)-Ib gene was PCR amplified with primers aac F and aac R (Table 2). PCR products were purified prior to sequencing on both strands.
ESBL genes (CTX-M, SHV, TEM, VEB, PER, and SFO) and plasmid-mediated AmpC β-lactamase genes were sought by PCR with specific primers (primers for the amplification of blaCTX-M are shown in Table 2, and primers for AmpC genes were previously described by Perez-Perez and Hanson [14]). PCR products were sequenced on both strands.
RNA preparation and real-time RT-PCR.
Total RNA was prepared using the TaKaRa RNAiso reagent, treated with DNase I, and purified according to the manufacturer's protocol. TaqMan reverse transcription reagents (Applied Biosystems) were used to synthesize cDNA from RNA samples. A total of 1.0 μg total RNA was used in a 50-μl reverse transcription (RT) reaction, and incubation conditions were those suggested by the manufacturer: 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min. Following RT, 2 μl cDNA reaction products was used per 25 μl of real-time PCR employing SYBR green detection. All reactions were performed in tetraplicate. Amplification of an endogenous control, the 16S rRNA gene, was performed to standardize the amount of sample RNA or DNA added to a reaction. Primer sequences used for each gene target are presented in Table 2.
Real-time PCR was performed using SYBR green PCR master mix (Applied Biosystems) and carried out using an ABI Prism 7300 sequence detection system. PCR cycling conditions were 1 cycle at 50°C for 2 min, which was followed by 1 cycle at 95°C for 10 min, which was then followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Results were analyzed with the ABI Prism 7300 sequence detection system software. Relative quantification was determined by the 2−ΔΔCT or ΔΔCT method. Expression of the endogenous control gene, 16S rRNA, was used to normalize data.
Cloning of the qnrA promoters in pKK232-8.
The promoter-like sequences of qnrA (9) and the common-region right-hand boundary were amplified with PCR by using primers promoter F, containing the BamHI site, and promoter R, containing the HindIII site, from qnrA-bearing plasmids pHS3, pHS4, pHS5, and pHS6. The PCR products with sizes of 274 bp or 281 bp were cut with BamHI plus HindIII, resulting in a 262-bp or 269-bp fragment, and cloned into pKK232-8, creating pKK232-8-HS3, pKK232-8-HS4, pKK232-8-HS5, and pKK232-8-HS6 (Table 1). All promoter fragments were sequenced using primer pKK-SQ after their final insertion into plasmids to verify that no mutations occurred during PCR or cloning. After cloning of the qnrA promoters into pKK232-8, these plasmids and pKK232-8 were transformed into HB101.
RESULTS
Screening for the qnrA gene and conjugation.
The qnrA gene was detected in 7 of the 541 ciprofloxacin-resistant or -intermediate clinical strains consecutively isolated from a teaching hospital in Shanghai, China, including 4 of 13 (30.8%) E. cloacae, 2 of 98 (2.0%) E. coli, and 1 of 9 (11%) Citrobacter sp. strains. qnrA was not found in 169 strains of K. pneumoniae and 20 strains of other Enterobacteriaceae. All seven qnrA-positive strains, five strains isolated from sputum and one each from wound exudate and urine, were isolated from inpatients, including six patients with nosocomial infections and one patient with asymptomatic bacteriuria. These seven strains were isolated from five different wards (Table 3). Antimicrobial agents were used in six patients, and fluoroquinolones (ciprofloxacin or levofloxacin) were used in five patients before the isolation of qnrA-positive strains. The patient with bacteriuria had not been exposed to an antimicrobial agent prior to isolation of the qnrA-positive C. freundii isolate. qnrA was not found in any of the 128 strains of P. aeruginosa, 77 strains of A. baumannii, and 27 strains of other nonfermenting bacillus strains.
TABLE 3.
Characteristics of qnrA-positive clinical strains
| Organism and strain | Ward of isolation | MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|---|
| CIP | CTX | CAZ | GEN | SMZ | ||
| E. cloacae | ||||||
| 63 | Geriatricsb | 32 | 64 | 32 | 4 | ≥512 |
| 91 | Neurosurgeryc | 4 | 32 | 128 | 8 | ≥512 |
| 113 | Geriatricsb | 128 | 64 | 128 | 8 | ≥512 |
| 641 | Worldwide medical centerd | 4 | 32 | 64 | 4 | ≥512 |
| E. coli | ||||||
| 633 | Neurosurgeryc | ≥128 | ≥128 | 4 | ≥128 | ≥512 |
| 650 | Geriatricsb | 32 | 1 | 4 | ≥128 | ≥512 |
| C. freundii | ||||||
| 64 | Nephrology | 32 | 2 | 4 | 32 | ≥512 |
CIP, ciprofloxacin; CTX, cefotaxime; CAZ, ceftazidime; GEN, gentamicin; SMZ, sulfamethoxazole.
These three strains were isolated from the same ward from March 7 to July 5.
These two strains were isolated from two different wards of the neurosurgery department.
This inpatient had peritonitis because of appendicitis with perforation.
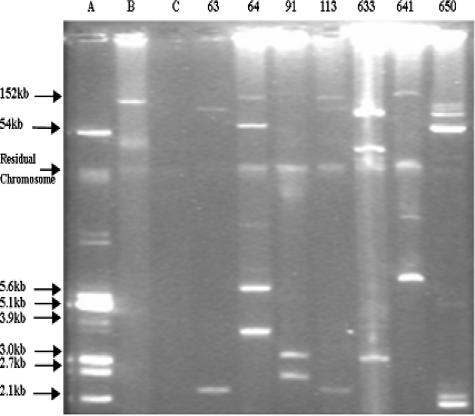
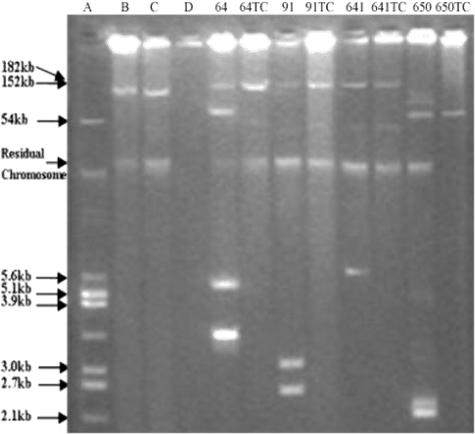
Out of seven qnrA-positive strains, four strains transferred quinolone resistance onto plasmids of different sizes by conjugation. Among the transferable plasmids, pHS3, pHS4, and pHS5 were greater than 150 kb in size, while pHS6 was approximately 65 kb. Each strain had a unique plasmid profile (Fig. 1 and 2). The MICs of ciprofloxacin against transconjugants demonstrated an 11-fold difference, from 0.094 μg/ml to 1 μg/ml, or were 12- to 125-fold higher than the recipient E. coli strain J53 (Table 4). Three other qnrA-bearing strains failed to produce transconjugants, although high-molecular-weight plasmids were visualized in donor strains (Fig. 1), and multiple agents were used for selection.
FIG. 1.
Plasmid DNAs from clinical and reference strains. (A) E. coli V517. (B) E. coli J53 plac. (C) E. coli J53. 63, 64, 91, 113, 633, 641, and 650 are clinical strains.
FIG. 2.
Plasmid DNA of donors and transconjugants. (A) E. coli V517. (B) E. coli J53 plac. (C) E. coli J53 R27. (D) E. coli J53. 64, 64TC, 91, 91TC, 641, 641TC, 650, and 650TC are clinical strains and transconjugants.
TABLE 4.
Characteristics of four qnrA-bearing plasmids in E. coli J53a
| Transconjugant | Original clinical strain | Size of plasmid (kb) | MIC (μg/ml)
|
Presence of aac(6′)-Ib-cr | Relative expression level of qnrA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LEV | AMP | CTX | CAZ | GEN | STR | KAN | AMK | TOB | SMZ | TMP | |||||
| E. coli J53 pHS3 | C. freundii 64 | >150 | 0.25 | 0.19 | 512 | ≤0.06 | 0.125 | 8 | 2 | 8 | 2 | 8 | ≥512 | ≤0.06 | + | 1.2 |
| E. coli J53 pHS4 | E. cloacae 91 | >150 | 0.094 | 0.19 | ≥512 | 8 | 8 | 2 | 8 | 64 | 4 | 8 | ≥512 | ≥512 | − | 1.0 |
| E. coli J53 pHS5 | E. cloacae 641 | >150 | 0.125 | 0.19 | ≥512 | 8 | 32 | 2 | 8 | 64 | 4 | 8 | ≥512 | ≥512 | − | 2.5 |
| E. coli J53 pHS6 | E. coli 650 | ∼65 | 1.0 | 0.38 | 512 | 0.125 | 0.125 | 0.25 | 1 | 16 | 2 | 8 | ≥512 | 0.25 | + | 32.5 |
| Recipient J53AzR | NA | NA | 0.008 | 0.012 | 8 | ≤0.06 | 0.125 | 0.25 | 1 | 0.5 | 0.5 | 0.25 | 8 | ≤0.06 | NA | NA |
CIP, ciprofloxacin; AMP, ampicillin; CAZ, ceftazidime; CTX, cefotaxime; GEN, gentamicin; STR, streptomycin; KAN, kanamycin; AMK, amikacin; TOB, tobramycin; SMZ, sulfamethoxazole; TMP, trimethoprim; NA, not applicable.
qnrA contained in all seven strains was identical to the qnrA sequence in the reported Shanghai E. coli strains (22) (GenBank accession numbers AY259085 and AY259086), which has been designated qnrA1 (11).
Of seven qnrA1-positive clinical strains, four strains (strains 91, 113, 633, and 641) with cefotaxime MICs of 32 to ≥128 μg/ml carried ESBL genes (three strains with CTX-M-9 and one strain with CTX-M-14); strains 63 and 633 also carried the ampC β-lactamase genes ACT-1 and DHA-1, respectively. The other two strains with cefotaxime MICs of 1 to 2 μg/ml were negative for all β-lactamase genes tested. Transconjugants J53 pHS4 and J53 pHS5, with cefotaxime MICs of 8 μg/ml, were positive for CTX-M-9, and the other two transconjugants with cefotaxime MICs of ≤0.06 to 0.125 μg/ml were negative for the β-lactamase genes tested.
QRDR sequencing.
The MICs of ciprofloxacin in qnrA1-bearing clinical strains were much higher than those of the corresponding transconjugants, with MICs of 4 to ≥128 μg/ml and 0.094 to 1 μg/ml, respectively. QRDRs in clinical strains and transconjugants were sequenced in order to determine if target modification occurs in qnrA-bearing clinical strains as it can in laboratory strains (10). Point mutations in gyrA were found with the resulting amino acid substitutions Ser83Ile, Ser83Leu, and Thr83Ile in E. cloacae, E. coli, and C. freundii, respectively, and Asp87Asn in E. coli. In parC, the substitution Ser80Ile was found in five strains. Five clinical strains with ciprofloxacin MICs of 32 to ≥128 μg/ml had two to three mutations in the gyrA and parC genes, and two strains of E. coli (strains 91 and 641) with ciprofloxacin MICs of 4 μg/ml exhibited no mutation in either gene.
Detection of aac(6′)-Ib.
aac(6′)-Ib and aac(6′)-Ib-cr were evaluated to see if aac(6′)-Ib-cr caused the 11-fold difference in ciprofloxacin MICs among the transconjugants. The aac(6′)-Ib-cr gene, a variant of aac(6′)-Ib that confers reduced susceptibility to ciprofloxacin by N acetylation of its piperazinyl amine, was present in E. coli J53AzR pHS3 and E. coli J53AzR pHS6. E. coli J53AzR pHS4 and E. coli J53AzR pHS5 also contained an aac(6′)-Ib gene but not the aac(6′)-Ib-cr variant. The two J53 transconjugants with aac(6′)-Ib-cr had higher ciprofloxacin MICs (0.25 or 1.0 μg/ml) than the transconjugants with aac(6′)-Ib (MICs of 0.094 to 0.125 μg/ml), suggesting that aac(6′)-Ib-cr contributed to the decrease in ciprofloxacin susceptibility. The MICs of levofloxacin conferred by pHS3 were the same as those conferred by pHS4 and pHS5, indicating that the resistance against levofloxacin, which lacks an unsubstituted piperazinyl nitrogen, was unaffected by aac(6′)-Ib-cr (19) (Table 4).
qnrA RNA expression.
There remained, however, a fourfold difference in the MICs of ciprofloxacin (0.25 and 1.0 μg/ml, respectively) between transconjugants J53 pHS6 and J53 pHS3, which both harbored qnrA1 and aac(6′)-Ib-cr. To determine whether the difference in ciprofloxacin MICs was caused by different levels of qnrA1 transcripts, the level of qnrA1-specific transcripts was determined by quantitative RT-PCR. The relative expression levels of qnrA1 in transconjugants were similar, from 1.0 to 2.5, but the relative expression level in J53 pHS6 was 32.5, much higher than the above-mentioned three transconjugants (Table 4). This result indicated that the higher ciprofloxacin MIC in transconjugant J53 pHS6 might be related to the higher qnrA1 expression level than that in J53 pHS3.
Effect of an upstream DNA sequence on promoter strength.
A fragment containing the promoter sequence of qnrA1-bearing plasmids was cloned upstream of the promoterless cat gene in reporter plasmid pKK232-8. The resulting plasmids, pKK232-8-HS3, pKK232-8-HS4, pKK232-8-HS5, pKK232-8-HS6, and pKK232-8, were transformed into E. coli HB101 competent cells. Chloramphenicol acetyltransferase activity is proportional to the strength of the promoter sequence. Chloramphenicol acetyltransferase determination was then estimated by measuring the chloramphenicol MIC. The chloramphenicol MICs for HB101 cells and transformant with pKK232-8 were both 3 μg/ml. In contrast, the MICs of chloramphenicol were 8 μg/ml for transformants with pKK232-8-HS3, pKK232-8-HS4, and pKK232-8-HS5 and 96 μg/ml for the transformant with pKK232-8-HS6, an MIC that was 12-fold higher than that of the above-mentioned other three transformants. This result indicated that the promoter in plasmid pHS6 (ciprofloxacin MIC of 1.0 μg/ml) was 12-fold stronger than that in plasmids pHS3 (ciprofloxacin MIC of 0.25 μg/ml) and pHS4 and pHS5 (ciprofloxacin MICs of 0.094 to 0.125 μg/ml).
Sequences of qnrA1 promoters.
The sequences of the cloned fragments upstream of qnrA1 between it and ORF513 from different plasmids were sequenced (Fig. 3). The sequences upstream of qnrA1 fragments from pHS3, pHS4, and pHS5 were identical to In36 (GenBank accession number AY259085), and the sequence from pHS6 was identical to In37 (accession number AY259086). Just like the difference of ciprofloxacin MICs between plasmids pHS6 and pHS3, the ciprofloxacin MIC for a strain carrying a plasmid carrying In37 is fourfold higher than that of a strain carrying a plasmid with In36 (MIC of 1 μg/ml versus 0.25 μg/ml) (22). The sequences differed by 7 bp (GTTAGCA) between the +1 transcription initiation site and the start of qnrA1, with these 7 bp absent in pHS6 and In37 compared to pHS3 and In36 (Fig. 3).
FIG. 3.
The qnrA promoter-like sequence of integrons (In36 and In37) and plasmids pHS3, pHS4, pHS5, and pHS6. The deduced amino acid sequences are designed in single-letter code below the nucleotide. The sequences are similar to the promoter structure for qnrA expression in pQR1. The transcription orientation of the qnrA gene is indicated by the horizontal arrow. The −35 and −10 promoter sequences are boxed, as is the +1 transcription initiation site.
DISCUSSION
In this study, all consecutive gram-negative clinical strains that were resistant or intermediate to ciprofloxacin and that were isolated in a teaching hospital of Fudan University in Shanghai in 2005 were screened for qnrA, regardless of their susceptibilities to other antimicrobials. The results thus reflect the overall prevalence of qnrA in clinical isolates with reduced susceptibility to ciprofloxacin. qnrA continues to be present in consecutive clinical isolates of gram-negative bacteria in Shanghai. The prevalence now appears to be higher in Enterobacter spp. (31% ) than in other species of clinical strains. qnrA was detected in 11% to 17% of strains of Enterobacter spp. isolated in the United States recently (17, 18). qnrA was positive in 15 of 47 (32%) blood culture isolates of Enterobacteriaceae resistant to both ciprofloxacin and cefotaxime collected in the United Kingdom. Of 15 qnrA-positive strains, 9 strains were E. cloacae, with a positive rate 56% (9/16) (3). An outbreak of E. cloacae infections occurred in a medical center in The Netherlands in 2002. All 83 outbreak strains were resistant to tobramycin and ceftriaxone, and 43% were both resistant and intermediate resistant to ciprofloxacin. qnrA1 was present in 78 (94%) of these 83 isolates (12). In a recent report (23), qnr genes were positive in 16.3% (86/526) of clinical strains of E. cloacae isolated in Taiwan. qnrA1, qnrB2, and qnrS1 genes were detected alone or in combination in 0.6%, 10.1%, and 6.5% of isolates, respectively. In this study, qnrA1 was detected in 2% of 98 strains of E. coli and in 8% of those that were highly resistant to ciprofloxacin (MICs of 8 to ≥256 μg/ml) collected in the same hospital from 2000 to 2001 (22). qnrA was not found in a relatively large number of strains (232 strains) of nonfermenting bacilli including 128 strains of P. aeruginosa and 77 strains of A. baumannii in this population.
Most qnrA-bearing plasmids in E. coli J53 conferred an MIC of ciprofloxacin of 0.25 μg/ml. However, the qnrA plasmids from clinical E. coli isolates collected in Shanghai provided different levels of ciprofloxacin MICs, from 0.125 μg/ml to 2 μg/ml (22). In a study (19) of these plasmids, a variant of aminoglycoside acetyltransferase, aac(6′)-Ib-cr, that was able to N-acetylate ciprofloxacin and norfloxacin at the secondary amino nitrogen on their piperazinyl substituent was found. aac(6′)-Ib-cr causes low-level resistance to certain fluoroquinolones and acts in concert with qnrA. aac(6′)-Ib-cr (GenBank accession number AY259086) was first reported in 2003 in a qnrA-bearing plasmid, pHSH2, but it was found that aac(6′)-Ib-cr was widespread geographically and stable over time in the United States. Nine (11%) of the 78 E. coli strains from Shanghai carried non-cr variants of aac(6′)-Ib, and 40 (51%) harbored the cr variant allele of aac(6′)-Ib (16). Among 313 strains of Enterobacteriaceae collected from North America, 50% carried aac(6′)-Ib, and of these strains, 28% carried the cr variant (13). In the present study, aac(6′)-Ib-cr was detected in two (pHS3 and pHS6) of the four qnrA-bearing plasmids. The ciprofloxacin resistance level conferred by pHS3 and pHS6 was higher than the resistance conferred by the other two cr variant-negative plasmids, pHS4 and pHS5. This result supports previous reports that aac(6′)-Ib-cr and qnrA act additively to generate ciprofloxacin resistance (19). The levofloxacin resistance level was not affected by aac(6′)-Ib-cr.
qnrA RNA expression results indicated that the different ciprofloxacin MICs conferred by two qnrA1- and aac(6′)-Ib-cr-carrying plasmids, pHS3 and pHS6, were associated with the differential transcription of the qnrA gene. Differences in qnrA transcription have also previously been claimed among qnrA plasmids (20).
Further study on the strength of the promoter indicated that different promoter strengths may cause the difference in ciprofloxacin MICs conferred by pHS6 and pHS3. Between the promoter and the start of qnrA, seven nucleotides, GTTAGCA, were missing in pHS6, a difference also found between In37 and In36, which are similarly associated with differences in MICs of ciprofloxacin. This DNA difference may contribute to the difference in promoter strength between pHS6 and pHS3. Fournier et al. (4) previously reported that mutations or other DNA changes within or around the consensus sequences of promoters caused the difference in promoter strengths. Three different mutations in the consensus sequences of promoters resulted in a 4- to 31-fold increase in promoter strength compared to that of the wild-type promoter. The same study also reported that a change from 17 to 16 bp between the −35 and −10 consensus sequences resulted in a ninefold decrease in the promoter strength. A G→A mutation 3 bp downstream of the −10 consensus sequence seemed to increase the promoter strength by about twofold (4).
Mutations in the QRDR of gyrA and parC were detected in five of seven qnrA-bearing clinical strains with higher ciprofloxacin MICs. This finding is consistent with other reports. A previously reported study indicated that 25 of 28 (89%) clinical strains of E. cloacae and other enterobacterial species that harbored qnrA had mutations in the GyrA gyrase subunit and ParC topoisomerase subunit (8). A ciprofloxacin-resistant (MIC > 32 μg/ml) E. coli isolate, isolate 1B, was isolated from a urinary specimen of a Canadian patient treated with norfloxacin for infection due to a ciprofloxacin-susceptible isolate, isolate 1A. Both isolates harbored qnrA1 and blaVEB-1 genes. Isolate 1B had amino acid substitutions in gyrase and topoisomerase, already known to be responsible for resistance to quinolones (15).
In conclusion, qnrA continues to be present in clinical isolates of gram-negative bacilli in Shanghai, China. The prevalence of qnrA now appears to be higher in E. cloacae (31%) than in other species. qnrA has not yet been found in nonfermenting bacilli. The coexistence of both qnrA and aac(6′)-Ib-cr in a single plasmid and the differential transcription of the qnrA gene appear to contribute to differences in the levels of plasmid-encoded ciprofloxacin resistance.
Acknowledgments
This work was supported by grant no. 2005CB0523101 (to M.W.) from the National Basic Research Program of China from the Ministry of Science and Technology, China, and grant no. 30440061 and 30572229 (to M.W.) from National Natural Science Foundation of China.
We thank George A. Jacoby and David C. Hooper for providing reference strains UAB1, E. coli J53AzR, E. coli V517, E. coli R1, E. coli plac, and E. coli R27 and for critical reviews of the manuscript.
Footnotes
Published ahead of print on 27 August 2007.
REFERENCES
- 1.Cambau, E., C. Lascols, W. Scougakoff, C. Bebear, R. Bonnet, J. D. Cavallo, L. Gutmann, M. C. Ploy, V. Jarlier, C. J. Soussy, and J. Jobert. 2006. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002-2005. Clin. Microbiol. Infect. 12:1013-1020. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing, 15th informational supplement (M100-S15). Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Corkill, J. E., J. J. Anson, and C. A. Hart. 2005. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from blood cultures in Liverpool, UK. J. Antimicrob. Chemother. 56:1115-1117. [DOI] [PubMed] [Google Scholar]
- 4.Fournier, B., A. Gravel, D. C. Hooper, and P. H. Roy. 1999. Strength and regulation of the different promoters for chromosomal β-lactamases of Klebsiella oxytoca. Antimicrob. Agents Chemother. 43:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49:801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2):S120-S126. [DOI] [PubMed] [Google Scholar]
- 7.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, Oh, H., A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lascols, C., J. Robert, V. Cattoir, C. Bebear, J. D. Cavallo, I. Podglajen, M. C. Ploy, R. Bonnet, C. J. Soussy, and E. Cambau. 2007. Type II topoisomerase mutations in clinical isolates of Enterobacter cloacae and other enterobacterial species harbouring the qnrA gene. Int. J. Antimicrob. Agents 29:402-409. [DOI] [PubMed] [Google Scholar]
- 9.Mammeri, H., M. V. De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 11.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 12.Paauw, A., A. C. Fluit, J. Verhoef, and M. A. Leverstein-van Hall. 2006. Enterobacter cloacae outbreak and emergence of quinolone resistance gene in Dutch hospital. Emerg. Infect. Dis. 12:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Perez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbial. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., J. D. Pitout, L. Calvo, J. M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum β-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 17.Robicsek, A., D. F. Sahm, J. Strahilevitz, G. A. Jacoby, and D. C. Hooper. 2005. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 49:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robicsek, A., J. Strahilevitz, D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Martínez, J. M., C. Velasco, A. Pascual, I. Garcia, and L. Martinez-Martinez. 2006. Correlation of quinolone resistance levels and differences in basal and quinolone-induced expression from three qnrA-containing plasmids. Clin. Microbiol. Infect. 12:440-445. [DOI] [PubMed] [Google Scholar]
- 21.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48:1295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, M., J. H. Tran, G. A. Jacoby, Y. Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, J. J., W. C. Ko, S. H. Tsai, and J. J. Yan. 2007. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob. Agents Chemother. 51:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]