Abstract
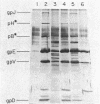
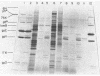
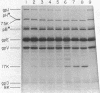
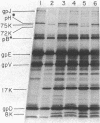
Previous studies have shown that bacteriophage lambda initially binds to liposomes bearing its receptor protein by the tip of the tail fiber (type 1 complex). It then associates more directly so that the hollow tail tube is in direct contact with the membrane (type 2 complex). DNA can be injected across the lipid bilayer into the liposome from type 2 complexes. We show here that gpJ, the tail fiber protein, becomes more sensitive to proteolytic degradation in type 2 complexes, indicating that the tail fiber does not pass into the liposome and that the tail fiber may undergo a conformational change in type 2 complexes. Another bacteriophage protein, pH, is sensitive to proteolytic degradation in free bacteriophage, type 1 complexes, or type 2 complexes formed with free receptor, but is resistant to proteinases in type 2 complexes formed with liposomes. This finding suggests that pH associates with the membrane. We suggest that this association is part of the mechanism by which a transmembrane hole for DNA entry is formed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bleviss M., Easterbrook K. B. Self-assembly of bacteriophage lambda tails. Can J Microbiol. 1971 Jul;17(7):947–954. doi: 10.1139/m71-151. [DOI] [PubMed] [Google Scholar]
- Buchwald M., Siminovitch L. Production of serum-blocking material by mutants of the left arm of the lambda chromosome. Virology. 1969 May;38(1):1–7. doi: 10.1016/0042-6822(69)90121-4. [DOI] [PubMed] [Google Scholar]
- Casjens S. R., Hendrix R. W. Locations and amounts of major structural proteins in bacteriophage lambda. J Mol Biol. 1974 Sep 15;88(2):535–545. doi: 10.1016/0022-2836(74)90500-2. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Hendrix R. W., Casjens S. R. Protein cleavage in bacteriophage lambda tail assembly. Virology. 1974 Sep;61(1):156–159. doi: 10.1016/0042-6822(74)90250-5. [DOI] [PubMed] [Google Scholar]
- Katsura I. Morphogenesis of bacteriophage lambda tail. Polymorphism in the assembly of the major tail protein. J Mol Biol. 1976 Nov 5;107(3):307–326. doi: 10.1016/s0022-2836(76)80007-1. [DOI] [PubMed] [Google Scholar]
- Katsura I. Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion. J Mol Biol. 1981 Mar 15;146(4):493–512. doi: 10.1016/0022-2836(81)90044-9. [DOI] [PubMed] [Google Scholar]
- Konopa G., Taylor K. Coliphage lambda ghosts obtained by osmotic shock or LiCl treatment are devoid of J- and H-gene products. J Gen Virol. 1979 Jun;43(3):729–733. doi: 10.1099/0022-1317-43-3-729. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mount D. W., Harris A. W., Fuerst C. R., Siminovitch L. Mutations in bacteriophage lambda affecting particle morphogenesis. Virology. 1968 May;35(1):134–149. doi: 10.1016/0042-6822(68)90313-9. [DOI] [PubMed] [Google Scholar]
- Randall-Hazelbauer L., Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973 Dec;116(3):1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa M., Scandella D. Multiple steps during the interaction between coliphage lambda and its receptor protein in vitro. Virology. 1976 Jul 1;72(1):182–194. doi: 10.1016/0042-6822(76)90322-6. [DOI] [PubMed] [Google Scholar]
- Roessner C. A., Struck D. K., Ihler G. M. Morphology of complexes formed between bacteriophage lambda and structures containing the lambda receptor. J Bacteriol. 1983 Mar;153(3):1528–1534. doi: 10.1128/jb.153.3.1528-1534.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Scandella D., Arber W. An Escherichia coli mutant which inhibits the injection of phage lambda DNA. Virology. 1974 Apr;58(2):504–513. doi: 10.1016/0042-6822(74)90084-1. [DOI] [PubMed] [Google Scholar]
- Scandella D., Arber W. Phage lambda DNA injection into Escherichia coli pel- mutants is restored by mutations in phage genes V or H. Virology. 1976 Jan;69(1):206–215. doi: 10.1016/0042-6822(76)90207-5. [DOI] [PubMed] [Google Scholar]
- Schwartz M. Reversible interaction between coliphage lambda and its receptor protein. J Mol Biol. 1975 Nov 25;99(1):185–201. doi: 10.1016/s0022-2836(75)80167-7. [DOI] [PubMed] [Google Scholar]
- Tsui L. C., Hendrix R. W. Proteolytic processing of phage lambda tail protein gpH: timing of the cleavage. Virology. 1983 Mar;125(2):257–264. doi: 10.1016/0042-6822(83)90199-x. [DOI] [PubMed] [Google Scholar]