Abstract
The fundamental principles underlying voltage sensing, a hallmark feature of electrically excitable cells, are still enigmatic and the subject of intense scrutiny and controversy. Here we show that a novel prokaryotic voltage-gated K+ (Kv) channel from Listeria monocytogenes (KvLm) embodies a rudimentary, yet robust, sensor sufficient to endow it with voltage-dependent features comparable to those of eukaryotic Kv channels. The most conspicuous feature of the KvLm sequence is the nature of the sensor components: the motif is recognizable; it appears, however, to contain only three out of eight charged residues known to be conserved in eukaryotic Kv channels and accepted to be deterministic for folding and sensing. Despite the atypical sensor sequence, flux assays of KvLm reconstituted in liposomes disclosed a channel pore that is highly selective for K+ and is blocked by conventional Kv channel blockers. Single-channel currents recorded in symmetric K+ solutions from patches of enlarged Escherichia coli (spheroplasts) expressing KvLm showed that channel open probability sharply increases with depolarization, a hallmark feature of Kv channels. The identification of a voltage sensor module in KvLm with a voltage dependence comparable to that of other eukaryotic Kv channels yet encoded by a sequence that departs significantly from the consensus sequence of a eukaryotic voltage sensor establishes a molecular blueprint of a minimal sequence for a voltage sensor.
INTRODUCTION
Voltage-gated channels are assemblies of modular membrane protein subunits consisting of two distinct, tandemly arranged, functional modules: a voltage sensor and a pore (Montal, 1990; Bezanilla, 2000). The occurrence of a pore module is now widely accepted based on the crystal structures of KcsA (Doyle et al., 1998), MthK (Jiang et al., 2002a), BacIR (Kuo et al., 2003), KvAP (Jiang et al., 2003a), and, most recently, Kv1.2 (Long et al., 2005a). Also, the recent elucidated structure of the NaK channel from Bacillus cereus (Shi et al., 2006), a nonselective tetrameric channel with an overall similar architecture to KcsA, argues in favor of a conserved pore module design. Two Kv channel structures have been solved at high resolution: the bacterial KvAP, and its isolated voltage sensor (Jiang et al., 2003a), and the mammalian Kv1.2 (Long et al., 2005a). These structures have lent credence to the occurrence of a voltage sensor module and have provided a structural framework to rationalize a wealth of sequence, mutational, and biophysical information. However, they have also raised major concerns about the mechanism of voltage sensing. It has been generally surmised that voltage sensing involved the interaction of the transmembrane potential with charged amino acids on the channel protein confined within the lipid bilayer. Multiple sequence alignments of the postulated components of the voltage sensor reveal the striking conservation of charged residues present within transmembrane helices. Particularly noteworthy are the highly conserved acidic residues on S2 and S3, and the basic residues that occur at three residue intervals on S4. The conservation of charged residues suggests that they form a network of ion pairs within the hydrophobic core of the membrane that work to stabilize the sensor fold (Papazian et al., 1995; Planells-Cases et al., 1995; Seoh et al., 1996; Tiwari-Woodruff et al., 1997; Tiwari-Woodruff et al., 2000; Myers et al., 2004).
The observed conservation of these charged residues in the sequences of both depolarization-activated and hyperpolarization-activated Kv channels has made difficult the identification of distinct sequence motifs for each of the sensors suggesting that they fold to homologous structures, and that Kv channel diversity arose by different combinatorial arrangements of the “same” modules. This notion is supported by evidence that activating voltage polarities are transposable using a limited number of mutations on either or both of the modules (Miller, and Aldrich, 1996; Zhao et al., 2004), and that sensor movement is conserved with respect to the polarity of the applied pulse: depolarization always results in an outward S4 movement relative to its resting position (Larsson et al., 1996; Miller, and Aldrich, 1996; Mannikko et al., 2002; Sesti et al., 2003; Starace, and Bezanilla, 2004; Zhao et al., 2004; Posson et al., 2005).
Understanding the minimal sequence underlying a voltage sensor fold is an unsolved question (Jiang et al., 2003a, 2004; Cuello et al., 2004; Long et al., 2005a). A mapping of voltage sensor phenotypes against naturally occurring sequence variations at conserved positions, a task now amenable given the wealth of genomic data currently available, can provide answers. Here we show that KvLm, a novel prokaryotic Kv channel, embodies an incipient voltage sensor design. In KvLm, only three out of eight positions known to be deterministic for folding (Papazian et al., 1995; Tiwari-Woodruff et al., 1997; Tiwari-Woodruff et al., 2000; Sato et al., 2003a,b; Myers et al., 2004) or sensing (Stuhmer et al., 1989; Papazian et al., 1991; Perozo et al., 1992; Shao, and Papazian, 1993; Perozo et al., 1994; Stefani et al., 1994; Planells-Cases et al., 1995; Aggarwal, and MacKinnon, 1996; Seoh et al., 1996; Bezanilla, 2000; Ahern, and Horn, 2004) in eukaryotic voltage sensors are conserved. Yet this sensor is sufficient to confer to the pore module voltage-dependent features comparable to those measured for eukaryotic Kv channels.
In the accompanying paper, we examine the interactions between highly conserved residues in the sensor components of KvLm by selective replacement of conserved voltage-sensing S4 residues and compensatory paired residue substitutions on the S2 and S3 components of the sensor. Together, our findings imply that the design of the voltage sensor is conserved among the Kv channel superfamily.
MATERIALS AND METHODS
Sequence Identification and Alignment
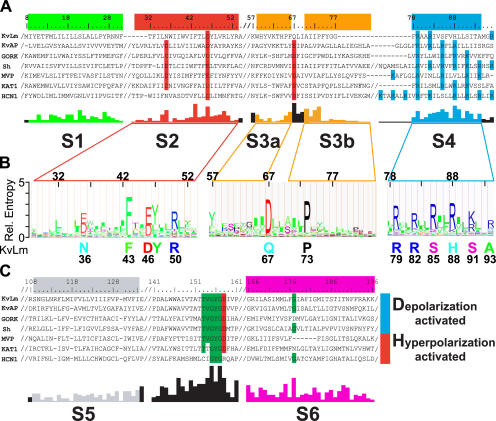
Candidate bacterial Kv channel sequences were identified from hits of a BLAST (tblastn) search of all bacterial genomes at the NCBI database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) using the Shaker Kv channel (Sh, Drosophila melanogaster, gi 13432103) sequence as query. The selection criteria were the presence of the minimal sequence identifiers for both a voltage sensor (R/KXXR/K in S4) and K+ selectivity filter (XXGY/FGD where X is mostly S/T). 54 sequences from bacterial genomes sharing <90% sequence identity by pairwise alignment were identified. To this group, 19 sequences of known hyperpolarization (11) and depolarization (8)-activated Kv channels were added to bring the total number of sequences to be aligned to 73. Of these 19 Kv channel sequences, two are from bacterial genomes (MVP from Methanococcus jannaschii and KvAP from Aeropyrum pernix, also identified in BLAST search), six from a plant genome (Arabidopsis thaliana), and 11 from other eukaryotic genomes. The master alignment was generated with Clustal X (v.1.83) (Thompson et al., 1997) using the Gonnet PAM series of protein weight matrices (Gonnet et al., 1992), refined by minimal manual adjustments, and exclusion of loops guided by the crystal structure of KvAP (Jiang et al., 2003a; see Fig. S1, available at http://www.jgp.org/cgi/content/full/jgp.200609572/DC1). From this master alignment, a profile hidden Markov model (HMM) (Eddy, 1998) was constructed using a web server application (http://bioweb.pasteur.fr/seqanal/interfaces/hmmbuild.html). The consensus sequence generated by the HMM was converted to an HMM logo using a web server application (http://logos.molgen.mpg.de/cgi-bin/logomat-m.cgi) (Fig. 1 B).
Figure 1.
A prokaryotic Kv channel with essential sequence conservation of the voltage sensor. (A and C) Representative Clustal X alignments of the voltage sensor module (A) and pore module (C) sequence of KvLm with sequences of six known Kv channels from bacteria (KvAP and MVP), plant (GORK and KAT1), and other eukaryotes (Sh and HCN1) (A and C). Conserved residues, functional in voltage sensing or K+ selectivity, are shaded red (acidic), blue (basic), and green (uncharged). Residue numbers are for KvLm; “/” indicates sequence breaks. Abbreviated names, accession nos., and genome source are as follows: KvLm, gi 16411529, Listeria monocytogenes; MVP, gi 15668309, Methanococcus jannaschii; KAT1, gi 10177705, Arabidopsis thaliana; HCN1, gi 29840778, Mus musculus; KvAP, gi 14601099, Aeropyrum pernix; GORK, gi 30693099, Arabidopsis thaliana; Sh, gi 13432103, Drosophila melanogaster. The conservation score below the alignment was calculated for the master alignment encompassing 19 known Kv channels and 54 Kv channel candidates from prokaryotic genomes (Fig. S1). Color bars (green for S1, red for S2, yellow for S3a and S3b, blue for S4, gray for S5, and magenta for S6) above sequence alignment and colored conservation score plot under alignment delimit transmembrane α-helices as inferred from the structure of KvAP (Jiang et al., 2003a; Cuello et al., 2004). (B) Hidden Markov model (HMM) profile for S2, S3, and S4 calculated from master alignment. The height of the residues is proportional to conservation. Below profile, at sequence positions found to be highly conserved (relative entropy > 2), the corresponding residue in KvLm is shown.
Gene Cloning, Overexpression, Purification, and Reconstitution
The gene encoding KvLm was PCR cloned from Listeria monocytogenes eGDE genomic DNA (P. Cossart, Institute Pasteur, Paris, France) and inserted into the E. coli protein expression vector pQE-70 (QIAGEN) between the SphI and BglII restriction endonuclease sites. Expression of the KvLm gene from this construct generates KvLm protein with a six-histidine (6xHis) C-terminal tag. Channel protein was expressed in E. coli XL-1 Blue cells (Stratagene), grown in LB medium to an OD550 of 0.5, and induced with 1 mM isopropyl-β-d-thyogalactopiranoside (IPTG) for 2–3 h at 37°C (OD550 1.1–1.3). Cells were harvested, resuspended in 100 mM NaCl, 5 mM KCl, 50 mM MOPS, pH 7.0, containing 1 mg/ml lysozyme and protease inhibitors (1 mM PMSF, 2 μM leupeptin A, 2 μM pepstatin), and lysed by sonication on ice for 5 min, using 30 s on/30 s off cycles. Membranes were collected by centrifugation at 110,000 g for 45 min and resuspended in 100 mM NaH2PO4 (NaPi), 5 mM KCl, pH 7.0. Membranes were solubilized in the aforementioned buffer in the presence of 10 mM dodecyl maltoside (DM) (ACROS organics) for 1 h at 4°C and the protein extract applied to a BD Talon Co2+-affinity column (BD Biosciences; CLONTECH Laboratories, Inc.). Unbound protein was washed using 10 mM imidazole, in 50 mM NaPi, 300 mM NaCl, 1 mM DM, pH 7.0; KvLm eluted at 150 mM imidazole. Purified KvLm was reconstituted into asolectin (Avanti Polar Lipids) liposomes (10 mg/ml) suspended in 450 mM KCl, 10 mM HEPES, pH 7.4, by incubating the mixture (lipid/protein ratios up to 5,000:1 [mol/mol]) at 4°C for 30 min followed by sonication on ice for 2 min. Detergent was removed by passing control (protein free) or KvLm proteoliposomes through BIO-Gel P-6 DG columns (Bio-Rad Laboratories) equilibrated in 450 mM KCl, 10 mM HEPES, pH 7.4.
Protein Analysis
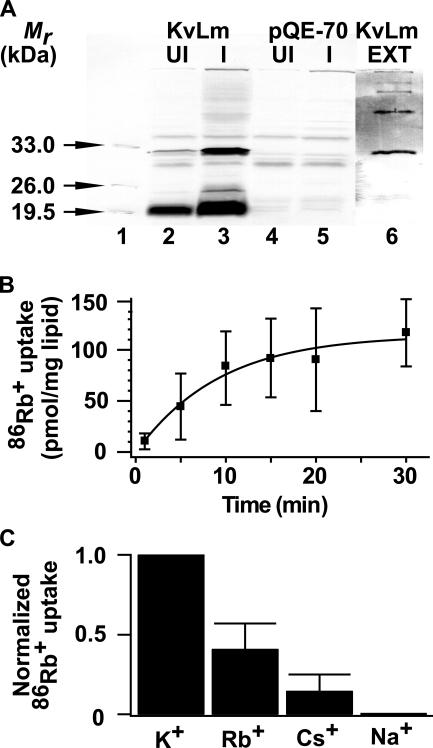
Cells overexpressing KvLm and purified KvLm were separated on 12% SDS-PAGE and transferred into PVDF membranes (Fig. 2 A). Blots were blocked in 5% de-fatted milk in Tris-buffered saline, pH 7.0 (TBS), supplemented with 0.1% Tween-20. A primary overnight incubation with anti-His tag probe (Santa Cruz Biotechnology, Inc.) in TBS (1:1,000) was followed by incubation with alkaline phosphatase-conjugated goat anti-rabbit probe (Jackson ImmunoResearch Laboratories); blots were developed using NBT/BCIP reagents (MP Biomedicals). Purified KvLm was lyophilized, resuspended in trifluoroethanol with 5% water, 0.1% trifluoroacetic acid, and subjected to electrospray ionization mass spectometry. The reconstruct produced a mass estimate of 26,025 D (C. Becker, personal communication).
Figure 2.
Expression, purification, and functional reconstitution of KvLm in proteoliposomes. (A) Immunoblots of E. coli cell lysates: lane 1, Mr standards; lanes 2 and 3, uninduced (UI) and IPTG-induced (I) cultures transformed with pQE-70 containing the gene encoding KvLm; lanes 4 and 5, UI and I control cultures transformed with empty vector. A prominent band on lane 3 is at an anticipated Mr ∼29 kD for KvLm. Lane 6, immunoblot of purified KvLm; the 29 kD protein retains propensity to oligomerize in SDS. (B) Time course of 86Rb+ uptake by KvLm reconstituted in proteoliposomes at 10 μg protein/mg phospholipid. (C) Ionic selectivity of KvLm-evoked 86Rb+ uptake. Bars represent uptake at 30 min into proteoliposomes prepared in 0.45 M Cl− salts of the indicated cations.
Flux Assay
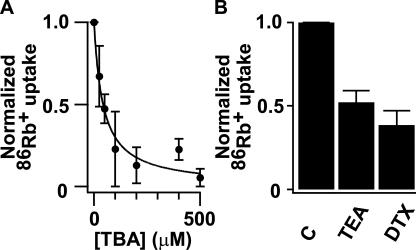
86Rb+ (DuPont New England Nuclear) flux assay was performed as previously described in detail (Heginbotham et al., 1998). 86Rb+ uptake was initiated by mixing KvLm proteoliposomes or control liposomes equilibrated in detergent-free reconstitution buffer (450 mM KCl, 10 mM HEPES, pH 7.4) with two volumes of medium containing 400 mM sorbitol, 10 mM HEPES pH 7.4 (SHEPES), 30 μM KCl, 5 μM RbCl, supplemented with 0.5 μCi/ml 86Rb+, and incubated at the indicated conditions. Extra-liposomal 86Rb+ was removed by passing aliquots (100 μl) of the mixture through Dowex cation exchange columns equilibrated in SHEPES. Liposomes containing trapped 86Rb+ were eluted with 2 ml SHEPES directly into scintillation vials and radioactivity was measured. To assay equilibrium uptake into control liposomes and KvLm proteoliposomes, the K+-selective ionophore valinomycin (1 μM) was added to each one of the remaining samples. After 1 min, 150-μl aliquots were filtered through Dowex columns and liposomes eluted with 2 ml SHEPES. To compare results under different conditions, KvLm-evoked fluxes were normalized with respect to the maximum uptake, which was elicited by valinomycin. Uptake in control liposomes was subtracted from each data point (Fig. 2 B). To establish a relative selectivity sequence (Fig. 2 C), proteoliposomes were reconstituted in 0.45 M Cl− salts of the test cations. In experiments to assay block (Fig. 3), each of the candidate channel blockers was added at the indicated concentrations either to the SHEPES uptake buffer or to the liposome reconstitution buffer.
Figure 3.
Block of KvLm-evoked 86Rb+ uptake by TBA, TEA, and DTX. (A) Dose–response curve for TBA-block measured at 20 min. Solid line depicts the fit to normalized 86Rb+ uptake = (1 + [TBA]/K d)−1 with a K d = 43 ± 6 μM (K d ± SD). (B) Bars represent uptake at 20 min in presence of 10 mM TEA, and 500 nM DTX relative to control (C).
Single Channel Recordings from Spheroplasts
Spheroplasts were prepared as previously described (Martinac et al., 1990). Excised patches in the inside-out configuration were used (Gambale, and Montal, 1990). Pipette solution contained 150 mM KCl, 210 mM sorbitol, 10 mM HEPES, and 40 mM MgCl2 titrated to pH 7.2 with KOH; bath solution was the same as the pipette solution but contained 410 mM sorbitol. For selectivity measurements, both chloride and methanesulfonate salts of K+, Na+, and Rb+ (Alfa Aesar) at 150 mM replaced KCl in both bath and pipette solutions. Pipette glass was Drummond Scientific 100-μl calibrated pipettes pulled to yield resistances of 7–17 MΩ when immersed in recording solution. Records were acquired at a sampling frequency >5 kHz and filtered online to 3 kHz with a 3-pole Bessel filter. Data were analyzed using Clampfit v.9.2 software (Axon Instruments) and IGOR Pro (Wavemetrics). For all records, the holding voltage was 0 mV and the interpulse interval was >3 s. Single channel conductance was calculated from Gaussian fits to current histograms. Probability density analysis provides the number of open and closed states and the channel open (τo) and closed (τc) lifetimes. The voltage dependence of channel opening was calculated from measurements of the fraction of time that the channel is open (P o) as a function of voltage, either by integration of conductance histograms, or from dwell time histograms according to P o = Nτo /T, where N is the number of openings, τo is the mean open time, and T is the total recording time at a given voltage (Labarca et al., 1985; Keller et al., 1986). All statistical values represent means ± SEM, unless otherwise indicated. n and N denote number of experiments and number of events, respectively. Control experiments on TK2420 transformed with empty pQE-70 (n > 20) failed to show consistent channel activity of reproducible unitary conductance.
Online Supplemental Material
The online supplemental material (Fig. S1) is available at http://www.jgp.org/cgi/content/full/jgp.200609572/DC1. The figure shows a prokaryotic Kv with essential sequence conservation of the voltage sensor. Clustal X alignment of the sequence of a prokaryotic Kv candidate from the genome of L. monocytogenes (KvLm) with 19 sequences of known Kv channels and 53 sequences from prokaryotic genomes postulated to encode for Kv channels.
RESULTS
Identification and Functional Reconstitution of KvLm
We took a reductionist approach aimed at identifying primitive Kv channels using a tblastn search of the database of 356 bacterial and 35 archaeal genomes. Using the archetypal Shaker Kv channel sequence as query, targets were screened for the signature sequences of a minimal voltage sensor, namely S4 (R/KXXR/K) and the K+ selectivity filter (XXGY/FGD). The Kv channel identified from the genome of L. monocytogenes, here denoted as KvLm, was selected for functional characterization because of its interesting sequence in the sensor (Fig. 1, A and B). Inspection of an alignment of 73 prokaryotic and eukaryotic Kv channel sequences (Fig. S1) reveals similarities and striking differences, highlighted in Fig. 1 by three prototypes of depolarization-activated channels (KvAP [Jiang et al., 2003a; Ruta et al., 2003], GORK [Ache et al., 2000], and Shaker [Tempel et al., 1987]) and hyperpolarization-activated channels (MVP [Sesti et al., 2003)], KAT [Schachtman et al., 1992], and HCN1 [Bell et al., 2004]) from bacteria, plants, and animals, respectively. Analysis of the KvLm sequence predicts a canonical fold with six transmembrane segments, presumably α-helical. Notable are the deviations at conserved positions on the voltage sensor (S2, S3, and S4) (Papazian et al., 1995; Planells-Cases et al., 1995; Seoh et al., 1996; Nelson et al., 1999) magnified in Fig. 1 B. Cases in point are the occurrence of polar residues on S2 (N36) and S3 (Q67) in place of the highly conserved acidic residues and, remarkably, the presence of only the first two N-terminal arginines on S4 (R79 and R82), the rest occupied by polar serine (S85 and S91) and histidine (H88) residues. That other highly conserved residues in the sensor sequence of Kv channels such as F43, D46, Y47, and R50 in S2 and P73 in S3 are also conserved in the sensor of KvLm suggests that KvLm harbors a deviant sensor design within a common sensor architecture. The pore sequence of KvLm also exhibits homology to the consensus Kv pore sequence including the characteristic signature sequence for K+ selectivity (TVGYGD) in the loop between S5 and S6, and the conservation of a glycine residue in the middle of S6 (G177) proposed to participate in Kv gating (Jiang et al., 2002a,b; Shealy et al., 2003; Magidovich, and Yifrach, 2004; Ding et al., 2005). These structural inferences derived from sequence analysis are consistent with the modularity of Kv channels (Nelson et al., 1999; Patten et al., 1999; Caprini et al., 2001, 2005; Lu et al., 2001; Long et al., 2005b; Yu et al., 2005).
Is KvLm a K+ channel? To demonstrate that KvLm is a K+ channel, we expressed it in a heterologous system and examined its channel activity. KvLm was expressed in bacteria and purified by Co2+-chelate chromatography in the presence of dodecylmaltoside; it migrates in SDS gels with an Mr ∼29 kD, consistent with a 247-residue protein (Fig. 2 A). Channel activity of purified KvLm was evaluated using an established flux assay based on the concentrative uptake of 86Rb+ into reconstituted proteoliposomes (Heginbotham et al., 1998). The time course of 86Rb+ uptake by KvLm-containing proteoliposomes exhibits a half-time of ∼7 min (Fig. 2 B); by contrast, control liposomes display low permeability to 86Rb+. KvLm sharply discriminates between K+ and Na+, displaying an apparent selectivity sequence of K+ > Rb+ > Cs+ ≫ Na+ (Fig. 2 C). KvLm is sensitive to dendrotoxin (DTX), tetrabutylammonium (TBA), and TEA (Fig. 3), blockers of noninactivating Kv channels, exhibiting a similar potency profile (Ferroni et al., 1992; Imredy, and MacKinnon, 2000; Harvey, 2001; Hille, 2001). A dose–response curve is illustrated for TBA (Fig. 3 A); a K d = 43 ± 6 μM is in fair agreement with that of Shaker (Ding, and Horn, 2002). Together, these findings are consistent with the predicted K+ channel activity of the KvLm protein.
KvLm Is a Depolarization-activated K+ Channel
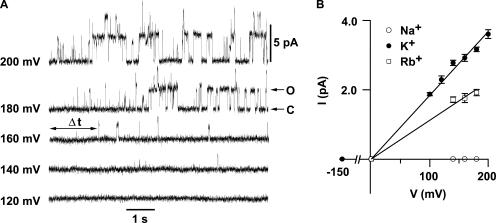
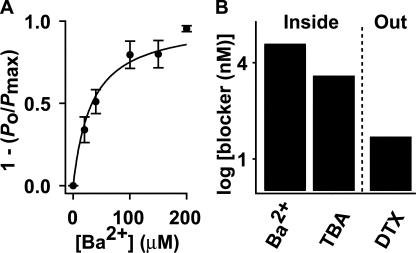
Electrical measurements of channel activity using patch clamp recordings require expression of KvLm in a suitable heterologous system. A favorite strategy involves expression of KvLm in amphibian oocytes or mammalian cells. Expression of KvLm, however, was undetectable in oocytes and in a number of popular cells, including HEK-293 and CHO-K1. Next, we selected the K+ uptake-deficient TK2420 E. coli strain, which does not grow in low K+ media (Rhoads et al., 1976). We reliably obtained single channel recordings on membrane patches excised from transformed bacterial spheroplasts (Martinac et al., 1990); Fig. 4 A shows a family of K+ currents in response to a series of 8-s depolarizing steps, from a holding potential (Vh) of 0 mV to 200 mV. Brief channel openings are discernible at voltages >100 mV and become progressively longer and more frequent with depolarization, reaching a plateau at ∼200 mV. The KvLm channel rectifies (Fig. 4 B); it exhibits ohmic behavior at depolarizing voltages, whereas it is nonconductive at hyperpolarizing voltages. The mean single channel conductance (γ), determined from the positive branch of the I-V curve, is 18.4 ± 0.3 pS in symmetric 0.15 M KCl or 0.15 M potassium methanesulfonate solutions, indicating that KvLm is a cation-selective channel. Equivalent measurements for Rb+ and Na+ show for Rb+ a γ = 11.2 ± 0.5 pS, whereas no channel currents were detected in Na+, all indicative of the high K+ selectivity of KvLm. KvLm is blocked by Ba2+ and TBA from the intracellular side, and by DTX from the extracellular solution (Fig. 5). The dose–response curve for Ba2+ determined at V = 140 mV yields a K d = 33 ± 4 μM; corresponding values for TBA and DTX are 3.9 ± 0.7 μM and ≤50 nM (Fig. 5 B). The blocker potency profile and the ionic selectivity sequence are in agreement with those obtained for purified KvLm reconstituted in proteoliposomes (Fig. 3), thereby establishing the fidelity of the assays and validating KvLm as a member of the Kv channel superfamily (Fig. 1).
Figure 4.
KvLm is a depolarization-activated K+-selective channel. (A) Currents from excised TK2420 E. coli patches were evoked from a Vh of 0 mV to the indicated voltages. Upward deflections indicate transitions from closed (C) to open (O) state; two channels were present in this particular patch. (B) Ionic selectivity of KvLm. Single channel current–voltage (I-V) relationships in 0.15 M symmetric Cl− or MES− solutions of K+ (n = 17, N = 10,560), Rb+ (n = 4, N = 203), and Na+ (n = 9, N =0).
Figure 5.
Block of KvLm by Ba2+, TBA, and DTX at 140 mV. (A) Dose–response curve for Ba2+ block from the bath, as the fraction of blocked channels (1 – (P o/P max)) (n = 6, N = 3,774) at an imposed [Ba2+]. Solid line is a fit of (1 – (P o/P max)) = (1 + K d/ [Ba2+])−1 to the data with K d = 33 ± 4 μM. (B) Bars represent K d ± SD values for Ba2+ and TBA (K d = 3.9 ± 0.7 μM, n = 7, N = 1,148) applied to the bath (Inside); for DTX, lowest [DTX] (K d = 50 nM, n = 8, N = 1710) applied from the pipette (Out) at which >50% block occurred.
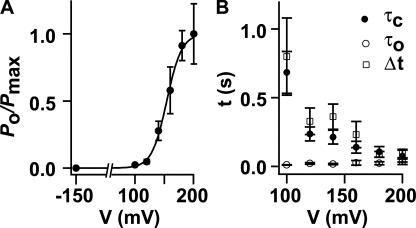
Next we focused on the voltage dependence of KvLm. Inspection of Fig. 4 A shows that the open channel probability (P o) sharply increases with depolarization (Fig. 6 A), a hallmark of depolarization-activated Kv channels. Two parameters were extracted from the fit of a two-state Boltzmann distribution to the data (Bezanilla, 2000): V 1/2, the voltage at which half of the channels are open (P o/P max = 0.5), and the apparent gating charge z, the estimated number of charges displaced within the membrane in response to the field. For KvLm, V 1/2 = 154 ± 1 mV and z = 2.0 ± 0.2 e. This z value is in accord with reports for a number of Kv channels (Bezanilla, 2000); specifically, it is between that reported for the depolarization-activated channels KvAP (z = 3.2 e) (Ruta et al., 2003) and Shaker (z = 3.2 e) (Yifrach, and MacKinnon, 2002), and the hyperpolarization-activated channels MVP (z = 1.1 e) (Sesti et al., 2003), KAT1 (z = 1.1 e) (Latorre et al., 2003), and HCN1 (z = 2.2 e) (Bell et al., 2004). The V 1/2 value is shifted positively compared with Shaker (V 1/2 = −27 mV) (Yifrach, and MacKinnon, 2002), yet it is similar to that of the voltage- and Ca2+-activated K+ (BK) channel (V 1/2 = 178 mV) (Niu et al., 2004). This analysis shows that despite its minimal sequence, KvLm contains a robust voltage sensor. This is also manifested in the time course of the current (Fig. 6 B); Δt, defined as the time interval between the onset of a pulse and the appearance of the first opening (Fig. 4 A), is shortened with depolarization. And, it correlates with the drastic reduction of the channel residence time in the closed state (τc) with depolarization (Fig. 6 B). Taken together, the findings provide compelling evidence that KvLm is a depolarization-activated K+ channel.
Figure 6.
Voltage dependence of KvLm opening. (A) A fit of the Boltzmann distribution (P o/P max) = (1 + exp[−zF(V − V 1/2)/RT])−1 with z = 2.0 ± 0.2 e and V 1/2 = 154 ± 1 mV. (B) Voltage dependence of Δt (n = 17, N = 10,560), τc, and τo (n = 6, N = 895). Records in which only one channel was present were included in this analysis.
DISCUSSION
Voltage Sensor Design
Having identified a novel bacterial voltage-gated channel, the next question is what this tells us about voltage sensor design. The following inferences can be derived from our analysis. (a) That five of the eight charges conserved in Kv channels are replaced in KvLm by polar residues implies that the architecture of the KvLm sensor core is stabilized predominantly by hydrophobic and H-bonding rather than electrostatic interactions. (b) An apparent gating charge of two for KvLm (Fig. 6 A) is consistent with the presence of two moving charges in the sensor, presumably the conserved R79 and R82 on S4. This is in accord with evidence derived from accessibility and gating current measurements that the conserved four N-terminal arginines of S4 in Shaker move with the field with a z value of 3.2 (Aggarwal, and MacKinnon, 1996; Seoh et al., 1996; Bezanilla, 2000; Ahern, and Horn, 2004). (c) D46 on S2 is the only acidic residue required for a minimal voltage sensor, in agreement with results from gating current measurements of Shaker that show a large contribution to charge movement from this position and little from the two other conserved acidic positions, namely 36 on S2 and 67 on S3 (Seoh et al., 1996). (d) Voltage sensing is critically dependent on the coupling between sensor and pore modules to the extent of dictating the polarity of sensing; this argues for the presence of a single primordial sensor and pore whose interaction surfaces were minimally perturbed by multiple evolution rounds to generate depolarization- and hyperpolarization-activated channels. A case in point is the L226P mutant of the Bacillus halodurans voltage-gated Na+ channel (NaChBac) (Ren et al., 2001; Zhao et al., 2004), in which replacement of a leucine for a proline on S6 of the pore module transforms NaChBac from a depolarization-activated channel into a hyperpolarization-activated channel (Zhao et al., 2004). This concurs with evidence that sensors move in the same direction in both depolarization- and hyperpolarization-activated channels, implying that the sensor is conserved, but coupling with the pore differs in a crucial and deterministic way (Aggarwal, and MacKinnon, 1996; Bezanilla, 2000; Mannikko et al., 2002; Latorre et al., 2003; Sesti et al., 2003; Elliott et al., 2004).
Paddle Conservation in a Voltage Sensor
The intriguing “voltage sensor paddle” model of sensing, based on the structure of KvAP, is proposed to be a helix-loop-helix motif formed by the C-terminal half of S3, (denoted S3b), a short linker and S4 (Jiang et al., 2003a,b; Ruta et al., 2005). This amphipathic cationic structure is conjectured to traverse the lipid bilayer core in response to the field in a manner akin to the translocation of hydrophobic cations and, accordingly, acts as the voltage sensor (Jiang et al., 2003b; Ruta et al., 2005). Analysis shows that the sequence corresponding to the S3b helix on KvAP is mostly absent in KvLm: it is at most eight residues long (not 16 as in KvAP) (Fig. 1). S3b in KvLm appears truncated at a highly conserved proline (P73); the sequence that follows (FYGG) and immediately precedes the beginning of S4 likely folds into a short linker loop, thereby predictably shortening S3b in KvLm to only four residues (A70 to P73). It is clear, therefore, that neither length nor character as present in the S3b of KvAp need be conserved as an essential component of a minimal sensor, now defined by KvLm. This finding is in accord with the observations that the length and sequence of S3b are not conserved across the Kv channel family (Cohen et al., 2003; Gonzalez et al., 2005) (see also Fig. 1, A and B). Further, S4 in KvLm is barely recognizable and is predicted to be a discontinuous helix with a kink C terminus of the voltage-sensing S91, in agreement with site-directed spin labeling and EPR data for KvAP (Cuello et al., 2004). The disparity in length and sequence of the S3b and S4 components of the paddle across the Kv channel superfamily suggests that they contribute significantly to the functional diversity of depolarization-activated Kv channels. In this framework, S3b may act as a physical linker whose length and not sequence is most important for voltage sensing, presumably by placing the paddle in its correct position. This inference is supported by comparison of the KvAP and Kv1.2 sensor structures (Long et al., 2005b), which shows that despite the significant difference in length of the S3b and S4 (two helical turns longer in Kv1.2), the short distance between conserved acidic residues on S2 and S3 and interacting basic residues in S4 is conserved (Papazian et al., 1995; Planells-Cases et al., 1995; Tiwari-Woodruff et al., 1997; Tiwari-Woodruff et al., 2000).
Determinants of Gating Polarity
One approach to identify residues that determine gating polarity in depolarization-activated and hyperpolarization-activated Kv channels is by functional characterization of Kv channels with sequences, such as that of KvLm, that markedly deviate at conserved positions from the consensus Kv sensor sequence (Fig. 1 B). Functional annotation of these sequences may allow for a grouping of sensor sequences into two distinct pools. And absolutely conserved residues likely to stabilize the sensor fold would be readily identified. This least common denominator approach to identify the minimal sequence determinants for a sensor fold has led us to establish a new set of minimal constraints embodied in KvLm. What about observed differences between the consensus sequences of these two groups? Inspection of the sequence alignment of the bacterial hyperpolarization-activated Kv channel MVP and depolarization-activated Kv channels KvLm and KvAP points to key divergences in the S3 and S4 region. Specifically, in MVP, a thoroughly conserved proline in S3b (P73 in KvLm) is replaced by an alanine, and two conserved basic residues in S4 (corresponding to R81 and H88 in KvLm) are occupied by asparagine and valine. It is clear now that an incompletely charged S4 as present in KvLm, MVP, and many other sequences in the master alignment does not constitute a signature of hyperpolarization-activated channels. In contrast, only four other sequences (denoted Cauran, Smelil, Ssp, and Vchole) in the master alignment other than MVP lack the conservation of the aforementioned proline in S3b. These observations highlight a new focus toward the identification of gating polarity determinants in the sensor fold.
Modularity of the Voltage Sensor
Our findings raise significant questions about the sequence-structure determinism. The incipient nature of the sensor fold in KvLm suggests that it embodies a minimal design sufficient to endow it with voltage-dependent features comparable to those of eukaryotic Kv channels. That such sensor motif occurs as an independently folded module and operates as a voltage-gated, proton-selective, and Zn2+-sensitive channel was recently demonstrated (Ramsey et al., 2006; Sasaki et al., 2006). Furthermore, that such sensor module is present in other membrane protein families, excluding ion channels, was elegantly shown in the Ciona intestinalis voltage sensor–containing phosphoinositide phoshatase (Murata et al., 2005). The provocative prospect of establishing the occurrence of minimum units of structure with specific functional attributes opens a testable path to assess the origin of channel diversity emerging from module shuffling and optimization by mutation events, and to an understanding of the surface features underlying the compatibility between the two modules responsible for the functional coupling of the sensor to the pore and, ultimately, of channel gating.
Conservation of Structure in KvLm
Coupled to a sensor with an unconventional sequence, KvLm discloses a pore with conduction properties similar to those of Shaker (Tempel et al., 1987), the most intensively studied eukaryotic Kv channel. The conservation of a presumably important gating residue (G177) in Kv channels (Jiang et al., 2002b; Shealy et al., 2003; Magidovich, and Yifrach, 2004; Ding et al., 2005), comparable single channel conductance (Heginbotham, and MacKinnon, 1993), and TBA (Ding, and Horn, 2002), Ba2+ (Hurst et al., 1996), and DTX (Imredy, and MacKinnon, 2000) blocking affinities is interpreted as indicative of major structural similarity between the Shaker (Long et al., 2005a) and KvLm pores, which, given the observed diversity at the sensor, suggests that pore and sensor modules evolved separately, in accord with the modularity of Kv channels (Nelson et al., 1999).
Given the intriguing features identified in the KvLm sensor sequence, we pursue an analysis of the interacting residues that may underlie the differences between eukaryotic and prokaryotic Kv sensors. This analysis is presented in the accompanying paper (see Lundby et al. on p. 293 of this issue).
Supplemental Material
Acknowledgments
We thank the members of M. Montal's group for valuable help and discussions throughout this work; the members of Ching Kung's laboratory for advice on spheroplast patch clamping; Pascale Cossart for L. monocytogenes genomic DNA; Wolfgang Epstein (The University of Chicago, Chicago, IL) for the TK2420 E. coli strain; Christian Becker for mass spectral analysis, and members of Ricardo Olcese's laboratory for oocyte expression studies.
This work was supported by a grant from the National Institutes of Health (R01-GM49711 to M. Montal and T32-NS07220 to N. Spitzer).
David C. Gadsby served as editor.
A. Lundby's present address is Department of Medical Physiology, The Panum Institute, University of Copenhagen, Blegdamsvej 3, DK-2200 Copenhagen N, Denmark.
C. Zazueta's present address is Instituto Nacional de Cardiologia Dr. Ignacio Chavez, Department of Biochemistry, Juan Badiano No. 1, Colonia Seccion XVI, Mexico, D.F., 14080, Mexico
Abbreviations used in this paper: DTX, dendrotoxin; HMM, hidden Markov model; TBA, tetrabutylammonium.
References
- Ache, P., D. Becker, N. Ivashikina, P. Dietrich, M.R. Roelfsema, and R. Hedrich. 2000. GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K(+)-selective, K(+)-sensing ion channel. FEBS Lett. 486:93–98. [DOI] [PubMed] [Google Scholar]
- Aggarwal, S.K., and R. MacKinnon. 1996. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 16:1169–1177. [DOI] [PubMed] [Google Scholar]
- Ahern, C.A., and R. Horn. 2004. Specificity of charge-carrying residues in the voltage sensor of potassium channels. J. Gen. Physiol. 123:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, D.C., H. Yao, R.C. Saenger, J.H. Riley, and S.A. Siegelbaum. 2004. Changes in local S4 environment provide a voltage-sensing mechanism for mammalian hyperpolarization-activated HCN channels. J. Gen. Physiol. 123:5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla, F. 2000. The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 80:555–592. [DOI] [PubMed] [Google Scholar]
- Caprini, M., M. Fava, P. Valente, G. Fernandez-Ballester, C. Rapisarda, S. Ferroni, and A. Ferrer-Montiel. 2005. Molecular compatibility of the channel gate and the N terminus of S5 segment for voltage-gated channel activity. J. Biol. Chem. 280:18253–18264. [DOI] [PubMed] [Google Scholar]
- Caprini, M., S. Ferroni, R. Planells-Cases, J. Rueda, C. Rapisarda, A. Ferrer-Montiel, and M. Montal. 2001. Structural compatibility between the putative voltage sensor of voltage-gated K+ channels and the prokaryotic KcsA channel. J. Biol. Chem. 276:21070–21076. [DOI] [PubMed] [Google Scholar]
- Cohen, B.E., M. Grabe, and L.Y. Jan. 2003. Answers and questions from the KvAP structures. Neuron. 39:395–400. [DOI] [PubMed] [Google Scholar]
- Cuello, L.G., D.M. Cortes, and E. Perozo. 2004. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 306:491–495. [DOI] [PubMed] [Google Scholar]
- Ding, S., and R. Horn. 2002. Tail end of the s6 segment: role in permeation in shaker potassium channels. J. Gen. Physiol. 120:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S., L. Ingleby, C.A. Ahern, and R. Horn. 2005. Investigating the putative glycine hinge in Shaker potassium channel. J. Gen. Physiol. 126:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, D.A., J. Morais Cabral, R.A. Pfuetzner, A. Kuo, J.M. Gulbis, S.L. Cohen, B.T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 1998. Profile hidden Markov models. Bioinformatics. 14:755–763. [DOI] [PubMed] [Google Scholar]
- Elliott, D.J., E.J. Neale, Q. Aziz, J.P. Dunham, T.S. Munsey, M. Hunter, and A. Sivaprasadarao. 2004. Molecular mechanism of voltage sensor movements in a potassium channel. EMBO J. 23:4717–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferroni, S., R. Planells-Cases, C.M. Ahmed, and M. Montal. 1992. Expression of a genomic clone encoding a brain potassium channel in mammalian cells using lipofection. Eur. Biophys. J. 21:185–191. [DOI] [PubMed] [Google Scholar]
- Gambale, F., and M. Montal. 1990. Voltage-gated sodium channels expressed in the human cerebellar medulloblastoma cell line TE671. Brain Res. Mol. Brain Res. 7:123–129. [DOI] [PubMed] [Google Scholar]
- Gonnet, G.H., M.A. Cohen, and S.A. Benner. 1992. Exhaustive matching of the entire protein sequence database. Science. 256:1443–1445. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C., F.J. Morera, E. Rosenmann, O. Alvarez, and R. Latorre. 2005. S3b amino acid residues do not shuttle across the bilayer in voltage-dependent Shaker K+ channels. Proc. Natl. Acad. Sci. USA. 102:5020–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, A.L. 2001. Twenty years of dendrotoxins. Toxicon. 39:15–26. [DOI] [PubMed] [Google Scholar]
- Heginbotham, L., L. Kolmakova-Partensky, and C. Miller. 1998. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heginbotham, L., and R. MacKinnon. 1993. Conduction properties of the cloned Shaker K+ channel. Biophys. J. 65:2089–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, B. 2001. Ion Channels of Excitable Cells. Third edition. Sinauer, Sunderland, MA. 814 pp.
- Hurst, R.S., L. Toro, and E. Stefani. 1996. Molecular determinants of external barium block in Shaker potassium channels. FEBS Lett. 388:59–65. [DOI] [PubMed] [Google Scholar]
- Imredy, J.P., and R. MacKinnon. 2000. Energetic and structural interactions between delta-dendrotoxin and a voltage-gated potassium channel. J. Mol. Biol. 296:1283–1294. [DOI] [PubMed] [Google Scholar]
- Jiang, Q.X., D.N. Wang, and R. MacKinnon. 2004. Electron microscopic analysis of KvAP voltage-dependent K+ channels in an open conformation. Nature. 430:806–810. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B.T. Chait, and R. MacKinnon. 2002. a. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, M. Cadene, B.T. Chait, and R. MacKinnon. 2002. b. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B.T. Chait, and R. MacKinnon. 2003. a. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., V. Ruta, J. Chen, A. Lee, and R. MacKinnon. 2003. b. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 423:42–48. [DOI] [PubMed] [Google Scholar]
- Keller, B.U., R.P. Hartshorne, J.A. Talvenheimo, W.A. Catterall, and M. Montal. 1986. Sodium channels in planar lipid bilayers. Channel gating kinetics of purified sodium channels modified by batrachotoxin. J. Gen. Physiol. 88:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, A., J.M. Gulbis, J.F. Antcliff, T. Rahman, E.D. Lowe, J. Zimmer, J. Cuthbertson, F.M. Ashcroft, T. Ezaki, and D.A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- Labarca, P., J.A. Rice, D.R. Fredkin, and M. Montal. 1985. Kinetic analysis of channel gating. Application to the cholinergic receptor channel and the chloride channel from Torpedo californica. Biophys. J. 47:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, H.P., O.S. Baker, D.S. Dhillon, and E.Y. Isacoff. 1996. Transmembrane movement of the shaker K+ channel S4. Neuron. 16:387–397. [DOI] [PubMed] [Google Scholar]
- Latorre, R., R. Olcese, C. Basso, C. Gonzalez, F. Munoz, D. Cosmelli, and O. Alvarez. 2003. Molecular coupling between voltage sensor and pore opening in the Arabidopsis inward rectifier K+ channel KAT1. J. Gen. Physiol. 122:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S.B., E.B. Campbell, and R. MacKinnon. 2005. a. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. [DOI] [PubMed] [Google Scholar]
- Long, S.B., E.B. Campbell, and R. MacKinnon. 2005. b. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 309:903–908. [DOI] [PubMed] [Google Scholar]
- Lu, Z., A.M. Klem, and Y. Ramu. 2001. Ion conduction pore is conserved among potassium channels. Nature. 413:809–813. [DOI] [PubMed] [Google Scholar]
- Lundby, A., J.S. Santos, C. Zazueta, and M. Montal. 2006. Molecular template for a voltage sensor in a novel K+ channel. II. Conservation of a eukaryotic sensor fold in a prokaryotic K+ channel. J. Gen. Physiol. 128:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidovich, E., and O. Yifrach. 2004. Conserved gating hinge in ligand- and voltage-dependent K+ channels. Biochemistry. 43:13242–13247. [DOI] [PubMed] [Google Scholar]
- Mannikko, R., F. Elinder, and H.P. Larsson. 2002. Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature. 419:837–841. [DOI] [PubMed] [Google Scholar]
- Martinac, B., J. Adler, and C. Kung. 1990. Mechanosensitive ion channels of E. coli activated by amphipaths. Nature. 348:261–263. [DOI] [PubMed] [Google Scholar]
- Miller, A.G., and R.W. Aldrich. 1996. Conversion of a delayed rectifier K+ channel to a voltage-gated inward rectifier K+ channel by three amino acid substitutions. Neuron. 16:853–858. [DOI] [PubMed] [Google Scholar]
- Montal, M. 1990. Molecular anatomy and molecular design of channel proteins. FASEB J. 4:2623–2635. [DOI] [PubMed] [Google Scholar]
- Murata, Y., H. Iwasaki, M. Sasaki, K. Inaba, and Y. Okamura. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243. [DOI] [PubMed] [Google Scholar]
- Myers, M.P., R. Khanna, E.J. Lee, and D.M. Papazian. 2004. Voltage sensor mutations differentially target misfolded K+ channel subunits to proteasomal and non-proteasomal disposal pathways. FEBS Lett. 568:110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R.D., G. Kuan, M.H. Saier Jr., and M. Montal. 1999. Modular assembly of voltage-gated channel proteins: a sequence analysis and phylogenetic study. J. Mol. Microbiol. Biotechnol. 1:281–287. [PubMed] [Google Scholar]
- Niu, X., X. Qian, and K.L. Magleby. 2004. Linker-gating ring complex as passive spring and Ca(2+)-dependent machine for a voltage- and Ca(2+)-activated potassium channel. Neuron. 42:745–756. [DOI] [PubMed] [Google Scholar]
- Papazian, D.M., X.M. Shao, S.A. Seoh, A.F. Mock, Y. Huang, and D.H. Wainstock. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301. [DOI] [PubMed] [Google Scholar]
- Papazian, D.M., L.C. Timpe, Y.N. Jan, and L.Y. Jan. 1991. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature. 349:305–310. [DOI] [PubMed] [Google Scholar]
- Patten, C.D., M. Caprini, R. Planells-Cases, and M. Montal. 1999. Structural and functional modularity of voltage-gated potassium channels. FEBS Lett. 463:375–381. [DOI] [PubMed] [Google Scholar]
- Perozo, E., D.M. Papazian, E. Stefani, and F. Bezanilla. 1992. Gating currents in Shaker K+ channels. Implications for activation and inactivation models. Biophys. J. 62:160–168, discussion 169–171. [DOI] [PMC free article] [PubMed]
- Perozo, E., L. Santacruz-Toloza, E. Stefani, F. Bezanilla, and D.M. Papazian. 1994. S4 mutations alter gating currents of Shaker K channels. Biophys. J. 66:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planells-Cases, R., A.V. Ferrer-Montiel, C.D. Patten, and M. Montal. 1995. Mutation of conserved negatively charged residues in the S2 and S3 transmembrane segments of a mammalian K+ channel selectively modulates channel gating. Proc. Natl. Acad. Sci. USA. 92:9422–9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posson, D.J., P. Ge, C. Miller, F. Bezanilla, and P.R. Selvin. 2005. Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature. 436:848–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, I.S., M.M. Moran, J.A. Chong, and D.E. Clapham. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D., B. Navarro, H. Xu, L. Yue, Q. Shi, and D.E. Clapham. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. [DOI] [PubMed] [Google Scholar]
- Rhoads, D.B., F.B. Waters, and W. Epstein. 1976. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67:325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta, V., J. Chen, and R. MacKinnon. 2005. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 123:463–475. [DOI] [PubMed] [Google Scholar]
- Ruta, V., Y. Jiang, A. Lee, J. Chen, and R. MacKinnon. 2003. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 422:180–185. [DOI] [PubMed] [Google Scholar]
- Sasaki, M., M. Takagi, and Y. Okamura. 2006. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 312:589–592. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Y. Hosoo, M. Sakaguchi, and N. Uozumi. 2003. a. Requirement of negative residues, Asp 95 and Asp 105, in S2 on membrane integration of a voltage-dependent K+ channel, KAT1. Biosci. Biotechnol. Biochem. 67:923–926. [DOI] [PubMed] [Google Scholar]
- Sato, Y., M. Sakaguchi, S. Goshima, T. Nakamura, and N. Uozumi. 2003. b. Molecular dissection of the contribution of negatively and positively charged residues in S2, S3, and S4 to the final membrane topology of the voltage sensor in the K+ channel, KAT1. J. Biol. Chem. 278:13227–13234. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P., J.I. Schroeder, W.J. Lucas, J.A. Anderson, and R.F. Gaber. 1992. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science. 258:1654–1658. [DOI] [PubMed] [Google Scholar]
- Seoh, S.A., D. Sigg, D.M. Papazian, and F. Bezanilla. 1996. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 16:1159–1167. [DOI] [PubMed] [Google Scholar]
- Sesti, F., S. Rajan, R. Gonzalez-Colaso, N. Nikolaeva, and S.A. Goldstein. 2003. Hyperpolarization moves S4 sensors inward to open MVP, a methanococcal voltage-gated potassium channel. Nat. Neurosci. 6:353–361. [DOI] [PubMed] [Google Scholar]
- Shao, X.M., and D.M. Papazian. 1993. S4 mutations alter the single-channel gating kinetics of Shaker K+ channels. Neuron. 11:343–352. [DOI] [PubMed] [Google Scholar]
- Shealy, R.T., A.D. Murphy, R. Ramarathnam, E. Jakobsson, and S. Subramaniam. 2003. Sequence-function analysis of the K+-selective family of ion channels using a comprehensive alignment and the KcsA channel structure. Biophys. J. 84:2929–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, N., S. Ye, A. Alam, L. Chen, and Y. Jiang. 2006. Atomic structure of a Na+- and K+-conducting channel. Nature. 440:570–574. [DOI] [PubMed] [Google Scholar]
- Starace, D.M., and F. Bezanilla. 2004. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 427:548–553. [DOI] [PubMed] [Google Scholar]
- Stefani, E., L. Toro, E. Perozo, and F. Bezanilla. 1994. Gating currents of cloned Shaker potassium channels. In Handbook of Membrane Channels: Molecular and Cellular Physiology. C. Peracchia, editor. Academic Press, San Diego, CA. 29–40.
- Stuhmer, W., F. Conti, H. Suzuki, X.D. Wang, M. Noda, N. Yahagi, H. Kubo, and S. Numa. 1989. Structural parts involved in activation and inactivation of the sodium channel. Nature. 339:597–603. [DOI] [PubMed] [Google Scholar]
- Tempel, B.L., D.M. Papazian, T.L. Schwarz, Y.N. Jan, and L.Y. Jan. 1987. Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila. Science. 237:770–775. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin, and D.G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff, S.K., M.A. Lin, C.T. Schulteis, and D.M. Papazian. 2000. Voltage-dependent structural interactions in the Shaker K+ channel. J. Gen. Physiol. 115:123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff, S.K., C.T. Schulteis, A.F. Mock, and D.M. Papazian. 1997. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys. J. 72:1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifrach, O., and R. MacKinnon. 2002. Energetics of pore opening in a voltage-gated K(+) channel. Cell. 111:231–239. [DOI] [PubMed] [Google Scholar]
- Yu, F.H., V. Yarov-Yarovoy, G.A. Gutman, and W.A. Catterall. 2005. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol. Rev. 57:387–395. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., T. Scheuer, and W.A. Catterall. 2004. Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc. Natl. Acad. Sci. USA. 101:17873–17878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.