Abstract
Posttranscriptional processes are important for regulation of gene expression in plant mitochondria. DEAD-box proteins, which form a huge protein family with members from all kingdoms, are fundamental components in virtually all types of processes in RNA metabolism. Two members of this protein family, designated PMH1 and PMH2 (for PUTATIVE MITOCHONDRIAL RNA HELICASE), were analyzed and characterized in mitochondria of Arabidopsis (Arabidopsis thaliana). Green fluorescent protein tagging with N-terminal PMH1 and PMH2 sequences supports the mitochondrial localization of these proteins. Northern experiments, as well as histochemical β-glucuronidase staining of transgenic plants carrying respective promoter:β-glucuronidase fusion constructs, revealed differing transcription patterns for the two genes. In response to cold, however, transcript levels of both genes increased. Immunodetection analyses of mitochondrial protein complexes after two-dimensional blue native/urea SDS-PAGE and after fractionation on sucrose gradients strongly suggest that one or both proteins are part of RNA-dependent complexes. Cold treatment of cell cultures or solubilization of mitochondria in the presence of MgCl2 favored the detection of high-molecular-mass complexes. This study paves the way for detailed analysis of high-molecular-mass complexes in mitochondria of higher plants.
Various posttranscriptional processes so far described in plant mitochondria have been suggested to be important for regulation of gene expression in this subcellular compartment (Hoffmann and Binder, 2002; Binder and Brennicke, 2003; Marchfelder and Binder, 2004; Gagliardi and Binder, 2007). This includes intron splicing, RNA editing, trimming of 3′ ends, and processing at the 5′ termini of rRNA, tRNA, and mRNA, and may also apply for regulatory processes at the level of translation. Although considerable progress has been made in elucidating the proteins involved in some of these processes, the vast majority of these components are still unknown. Purification of such proteins has been found to be difficult or, in many cases, impossible, mostly due to the low abundances of these proteins, but also due to limitations in respective in vitro systems, which are required to investigate the actual participation of a purified protein in a certain process. Alternatively, genetic approaches can be used to identify genes whose products are involved in RNA processing in plant mitochondria. For instance, forward genetics searching for RESTORER OF FERTILITY (RF) genes restoring cytoplasmic male sterility (CMS) in different plant species identified several pentatricopeptide repeat (PPR) proteins, directly or indirectly involved in degradation, stability determination, or cleavage of CMS-associated transcripts (Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Kazama and Toriyama, 2003; Koizuka et al., 2003; Wang et al., 2006). Reverse-genetic approaches have been successfully applied to assign functions to in silico-identified proteins as, for instance, to a mitochondrial PNPase and an RNase II-like protein (called RNR1), respectively (Perrin et al., 2004a, 2004b). These proteins are involved in mRNA 3′ processing and RNA degradation, as well as in processing ribosomal RNA.
DEAD-box proteins form a large gene family and are present in almost all organisms from all kingdoms (Linder, 2006). In Arabidopsis (Arabidopsis thaliana), 58 genes for DEAD-box proteins have been identified (Boudet et al., 2001; Mingam et al., 2004). Most of these proteins are considered to be RNA helicases, although this particular function was actually analyzed for only a few of these proteins (Rocak and Linder, 2004). The primary function of DEAD-box RNA helicases is to rearrange inter- or intramolecular RNA structures or to dissolve RNA-protein complexes (Rocak and Linder, 2004; Linder, 2006). These actions, dependent on ATP hydrolysis, are often prerequisites for subsequent processing steps and RNA helicases are often associated with other proteins. Consequently, DEAD-box proteins have been implicated to participate in all kinds of processes dealing with RNA, including RNA synthesis, RNA modification, RNA cleavage, RNA degradation, as well as ribosome biogenesis and translation initiation (Linder, 2006). DEAD-box proteins can thus be ideal starting points for the detection and purification of multiprotein complexes involved in gene expression in plant mitochondria. In addition, DEAD-box proteins and their genes can be readily identified by their typical conserved motifs (Rocak and Linder, 2004; Linder, 2006).
To identify proteins involved in the RNA metabolism in higher plant mitochondria, we have now analyzed two DEAD-box proteins in Arabidopsis. We determined their subcellular localization and investigated transcript levels as well as the promoter activities of these two genes. We find that these proteins are integrated in RNA-dependent high-molecular-mass complexes.
RESULTS
Posttranscriptional processes play a key role in gene expression in plant mitochondria (Gagliardi and Binder, 2007). To date, still little is known about the components involved in the individual RNA maturation steps. To get hold of such proteins in plant mitochondria, we started to examine DEAD-box proteins in Arabidopsis. The genes and gene products for these studies, At3g22310 and At3g22330, were described in previous global in silico studies of this class of proteins in this plant model species (Aubourg et al., 1999) and were detected in proteomic investigations of mitochondria (Millar et al., 2001; Heazlewood et al., 2004). The latter and our own in silico analyses (data not shown) indicated these proteins to be targeted to mitochondria, suggesting them to be good candidates for studying components of mitochondrial RNA metabolism. These proteins, designated PUTATIVE MITOCHONDRIAL RNA HELICASE1 (PMH1 [At3g22310]) and PMH2 (At3g22330), contain almost all motifs typical for DEAD-box proteins (Supplemental Fig. S1; Rocak and Linder, 2004), with the exception of motif IV. PMH1 and PMH2 are very similar to each other, with 77% identical amino acids, both carrying a Ser/Gly-rich C terminus. As seen from the AGI numbers, the genes are encoded on chromosome III, only separated by the gene (At3g22320) encoding the 24.3-kD subunit of RNA polymerases I to III (Arabidopsis Genome Initiative, 2000).
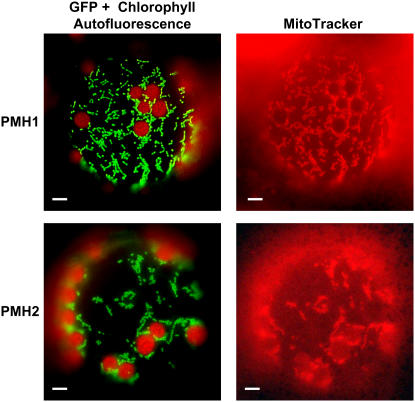
Subcellular Localization of PMH1 and PMH2
PMH2 has previously been found in proteome analyses of plant mitochondria indicating that this protein is located in this organelle (Millar et al., 2001; Heazlewood et al., 2004). In addition, in silico prediction of subcellular targeting by prediction programs TargetP (http://www.cbs.dtu.dk/services/TargetP) and Predotar (http://urgi.infobiogen.fr/predotar/predotar.html) strongly suggest mitochondrial localization of both PMH1 and PMH2. To address this question experimentally, two cDNA fragments of each gene (corresponding to the N-terminal 69 and 100 amino acids of PMH1 and 62 and 104 amino acids of PMH2, respectively) were fused upstream in frame to the reading frame encoding the GFP (Davis and Vierstra, 1998). The constructs were transformed into tobacco (Nicotiana tabacum) protoplasts stained with MitoTracker Red, a dye that specifically accumulates in mitochondria. Subcellular localization of the fusion proteins was monitored by fluorescent microscopy. Colocalization of the green fluorescence of all four fusion proteins with the fluorescence of MitoTracker Red revealed that the PMH:GFP fusion proteins are imported into these organelles (Fig. 1; data not shown). This strongly suggests that also in vivo PMH1 and PMH2 are targeted to mitochondria, which is further substantiated by the detection of these proteins in mitochondrial lysates (see below; Millar et al., 2001; Heazlewood et al., 2004).
Figure 1.
PMH1 and PMH2 are located in mitochondria. Images of tobacco protoplasts expressing the PMH1:GFP (69-amino acid construct) and the PMH2:GFP (104-amino acid construct) fusion proteins, respectively, were taken with filters allowing the detection of GFP fluorescence and chlorophyll autofluorescence (left) and a filter optimized for visualization of MitoTracker fluorescence (right). Bars correspond to 10 μm.
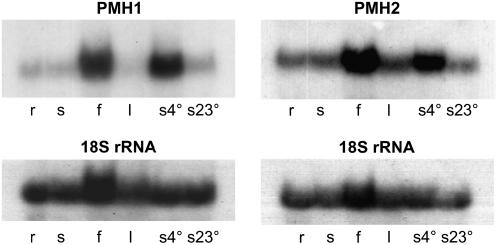
PMH1 and PMH2 Show Differing Expression Patterns, But Both Are Induced by Cold Treatment
The high similarity of these proteins suggests that they might have similar functions, which can complement each other. As a first step, we investigated whether they are expressed in different tissues and developmental stages. In northern analyses, performed with gene-specific probes covering the 5′-untranslated regions as well as the unique regions of the mitochondrial targeting sequences, PMH1 steady-state transcripts are mainly found in flowers, at very low levels in roots and seedlings, but are hardly detectable in leaves (Fig. 2, left). Because many RNA helicases from bacteria and various eukaryotic organisms show enhanced expression at low temperatures (Owttrim, 2006), we also analyzed PMH1 mRNA levels in seedlings grown at 4°C. A strong signal in the northern analysis demonstrates high expression of PMH1 under these conditions. The analogous northern experiment performed with the PMH2 probe also detected predominant transcription of this gene in flowers (about 3-fold higher than in seedlings), but revealed also PMH2 transcripts in roots, seedlings, and leaves (Fig. 2, right). Similar to PMH1, an elevated amount of PMH2 mRNAs was found after cultivation of seedlings for 24 h in the cold (approximately 2.7-fold higher), although induction seems to be less strong (Fig. 2, right).
Figure 2.
Transcription of PMH1 and PMH2. Northern-blot analysis shows that PMH1 transcripts are predominantly present in flowers (f), whereas PMH2 mRNA is almost equally present in roots (r), seedlings (s), and leaves (l) and is approximately 3-fold more abundant in flowers (compared to seedlings; top). Steady-state transcript levels of both genes are elevated in cold-treated seedlings (4°C for 24 h, s4°) in comparison to control plants cultivated at 23°C (s23°). The quality of the RNA and the loading of the gel were checked by hybridization with an 18S rRNA-specific oligonucleotide probe (bottom).
Enhanced PMH1 transcription after cold treatment was further analyzed and quantitatively resolved over time. Approximately a 7-fold induction is seen 12 h after transfer of the seedlings to 4°C. A 3-fold increased mRNA level is still detectable after 48 h. Maximal induction is observed after 36 h with approximately a 10-fold higher amount of PMH1 mRNAs in comparison to the control plants grown under identical conditions at 23°C (data not shown).
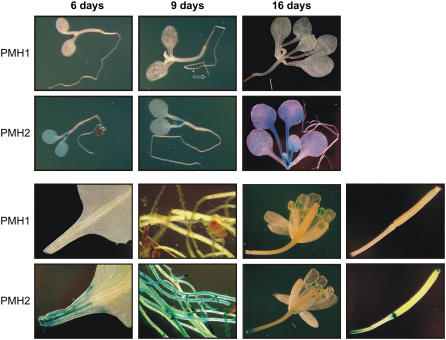
Transcriptional activity was studied in more detail using the GUS reporter gene. To this end, 2.0 kb upstream of the ATG containing potential promoter sequences of both genes were fused to the GUS reading frame. This region upstream of PMH1 includes the preceding gene At3g22300 and 331 bp of its promoter. The PMH2 upstream region comprises the gene At3g22320, which is encoded on the opposite strand.
After transformation of the respective constructs into Arabidopsis, five different plant lines originating from five individual integration events per construct were examined by histochemical GUS staining. Substantial staining is consistently observed in transgenic plants expressing GUS under the control of the PMH2 promoter. Strong promoter activities are indicated in cotyledons, emerging leaves, expanded leaves, as well as root tips of 6-, 9-, and 16-d-old seedlings grown on Murashige and Skoog medium (Fig. 3) or soil (data not shown). Transcriptional activity is also observed in roots, anthers, and the basal parts of siliques of adult plants. In addition, pronounced staining is found in tissue parts close to the cut sites of detached leaves and siliques, as well as of parts of flowering stalks, suggesting that transcription of PMH2 is triggered by wounding (Fig. 3). This is confirmed by experiments in which the leaf blades were mechanically wounded by a cut or a pin prick. All of these mechanical injuries consistently trigger promoter activity in tissues adjacent to the site of treatment (Supplemental Fig. S2).
Figure 3.
Histochemical analysis of transgenic Arabidopsis plants expressing the GUS reporter gene under the PMH1 and PMH2 promoters. Six-, 9-, and 16-d-old seedlings grown on Murashige and Skoog medium (top), as well as different organs of adult plants cultivated on soil (bottom, from left to right: basis of leaves, roots, flowers, and siliques), were analyzed. Further details are given in the text.
A different pattern of activity was found in plants expressing GUS governed by the PMH1 promoter. Whereas no staining at all is detectable in seedlings grown on soil (data not shown), very weak promoter activity is indicated in the root tips of seedlings cultivated on Murashige and Skoog medium under otherwise identical conditions (Fig. 3, top, indicated by an arrow). GUS activity is not seen in any part of the adult plants, except for substantial promoter activity restricted to anthers. In contrast to PMH2, no induction by mechanical treatment is observed for PMH1, indicating that both genes respond differently to this external stimulus. All in all, the results of northern analysis are consistent with the observations in the histochemical analyses, both indicating different expression patterns for each gene, suggesting that the two proteins have partially different roles in the plant.
Both PMH Proteins Are Expressed in Arabidopsis Cell Suspension Culture
For further analysis of the PMH proteins, two PMH1-derived peptides were synthesized and used for antibody production in rabbits (Supplemental Fig. S1). The antiserum should detect both proteins and indeed also binds to recombinant PMH2 (data not shown).
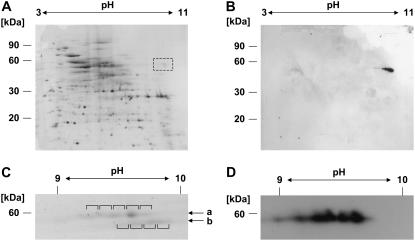
To investigate whether both proteins are expressed in an Arabidopsis cell suspension culture, mitochondria were isolated and purified from this tissue. Proteins from these organelles were separated by isoelectric focusing (IEF) on immobilized nonlinear pH gradients ranging from pH 3 to 11 followed by standard SDS-PAGE. Staining with Coomassie Blue revealed a protein pattern similar to those observed in previous proteome analyses of mitochondria (Kruft et al., 2001). Several weak spots corresponding to proteins of 60 kD are present between pH 9 and 10, where PMH2 has been found previously (Fig. 4A, highlighted by a dashed box; Millar et al., 2001). Proteins in this range were then inspected by immunodetection with the PMH1/PMH2 antiserum. A single series of spots is observed exactly at the expected position corresponding to proteins of about 60 kD with a pI between pH 9 and 10 (Fig. 4B). To obtain higher resolution in this range, mitochondrial proteins were separated on a gradient spanning pH 7 to 11. This revealed two rows of protein spots in the expected area in the stained gel (Fig. 4C, arrows a and b). Immunodetection analysis of such a gel detected at least five different spots, which correspond to the top row (Fig. 4D). To unambiguously identify the proteins detected by the PMH1/PMH2 antiserum, spots of both rows were analyzed by mass spectrometry (MS; Fig. 4C, white rectangles). In spots 1 to 4 of the top row, predominantly peptide masses matching fragments specific for PMH1 or PMH2 were found, whereas spot 5 contains only peptides corresponding to PMH2. In the spots of the bottom row, peptides predominantly matching translation elongation factors (At1g07920, At1g07930, At1g07940) were identified. These proteins have also been found previously in the mitochondrial proteome of Arabidopsis (Heazlewood et al., 2004).
Figure 4.
The two DEAD-box proteins are detectable in mitochondria from cell suspension cultures. Total mitochondrial protein is analyzed by IEF (first dimension) and Tricine-SDS-PAGE (second dimension). A, Coomassie staining revealed spots in the area where PMH2 is expected from previous studies (pH 10/60 kD; dashed box). B, Immunodetection analysis with the PMH1/PMH2 antiserum detects specific spots in this area. C and D, Enhanced resolution of the proteins was achieved on pH 7 to 11 gradients. A section of the stained gel (C) shows spots in expected size and pH range (indicated by arrows a and b). Relevant spots (indicated by brackets) were excised from the gel and analyzed by MS. In all spots of the top row, PMH1 and/or PMH2 is the predominant protein. D, Immunodetection analysis indicates the presence of the PMH proteins in the top row of the proteins seen between pH 9 to 10.
Taken together, this analysis shows that PMH1 as well as PMH2 are expressed in this Arabidopsis cell suspension culture. In addition, these experiments confirm that the antiserum recognizes the PMH proteins and can be used for selective detection of these proteins in mitochondrial lysates.
Detection of PMH1 and/or PMH2 in RNA-Containing Polypeptide Complexes
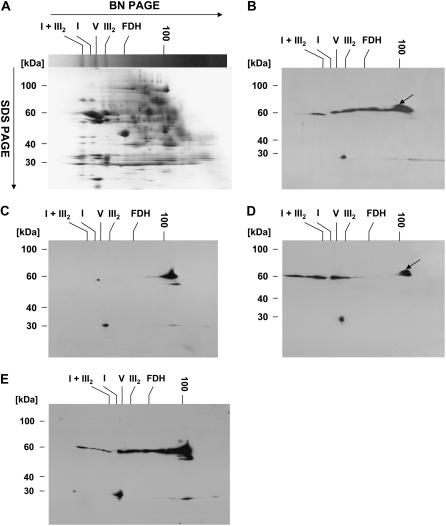
PMH1/PMH2 antiserum was then used to examine whether these DEAD-box proteins are associated with other proteins. To follow this issue, 1 mg of total mitochondrial protein isolated from the cell suspension culture was solubilized with dodecylmaltoside (DDM) and separated by two-dimensional blue native (BN)/urea SDS-PAGE. Coomassie staining of proteins after the separation revealed a pattern in which distinct complexes can be identified by comparison with the results of analogous separations (Fig. 5A; Eubel et al., 2003). Immunodetection analysis of analogously separated proteins with the PMH1/PMH2 antibody revealed protein complexes with apparent sizes ranging between <100 kD up to 1,500 kD (Fig. 5B). The detected proteins or complexes do not correspond to any of the highly abundant protein complexes visible on the Coomassie-stained gel (Fig. 5, A and B). The signal corresponding to a protein size <100 kD most likely represents monomeric PMH1 or PMH2 or a mixture of both proteins. In addition, a slightly larger protein can be seen, which is also detected in the first and second fractions of the Suc gradient (Fig. 5, B and D, arrow; see below). Furthermore, a single protein with an apparent molecular mass of 30 kD also cross-reacts with the PMH1/PMH2 antiserum.
Figure 5.
PMH1/PMH2 are bound in RNA-dependent high-molecular-mass complexes. DDM-solubilized total mitochondrial protein from cell suspension culture was separated in the first dimension by BN-PAGE followed by urea SDS-PAGE in the second dimension. A to E, Coomassie-stained gel (A); immunodetection analysis of DDM-solubilized protein with PMH1/PMH2 antibodies (B); as B after separation of RNase A-treated mitochondrial lysates (C); immunodetection analysis with PMH1/PMH2 antibody after cold treatment of the suspension culture for 18 h (D); and solubilization in the presence of 10 mm MgCl2 (E). Sizes in the first dimension are estimated from complexes I + III2 (1,500 kD), I (1,000 kD), V (580 kD), III2 (480 kD), formate dehydrogenase (FDH; 200 kD), and aconitase (100 kD; Eubel et al., 2005).
DEAD-box proteins are involved in multiple processes that require interaction of these proteins with RNA. We thus tested whether the integrity of the PMH1/PMH2-containing complexes depends on the presence of RNA. To this end, solubilized total mitochondrial protein was treated with RNase A prior to separation by two-dimensional BN/urea SDS-PAGE. Digestion of the RNA almost completely disassembled the high-molecular-mass complexes (Fig. 5C). Beside a weak signal corresponding to a size of about 580 kD (ATP synthase), only the signal most likely representing the monomeric protein can now be detected (Fig. 5C). This strongly suggests that the stability or maintenance of the detected complexes or at least the association of PMH1 or PMH2 with the complexes depends on the presence of RNA.
Because cold treatment enhanced the steady-state transcript levels of both PMH proteins, we also examined the influence of cold on complex composition and/or size. The cell suspension culture was grown under normal conditions and then incubated for 18 h at 4°C under otherwise identical conditions. Mitochondria were isolated and proteins were investigated as in the previous experiments. This treatment decreased the abundance of the complexes with sizes below 480 kD (corresponding to the size of complex III; Fig. 5D). In contrast, substantially more complexes are detectable that are even larger than those observed in mitochondria from normally grown cultures (Fig. 5D). A similar complex pattern is apparent when the mitochondrial protein is solubilized in the presence of 10 mm MgCl2, which can stabilize ribonucleoprotein particles (Fig. 5E). Again, a shift toward higher molecular masses is seen under these conditions.
In summary, our analysis suggests that PMH1 and/or PMH2 is part of RNA-dependent high-molecular-mass complexes. The size of the protein complexes or at least the interaction of the PMH proteins with complexes of higher molecular masses increases upon cultivation of the cells at 4°C or by the presence of MgCl2 during solubilization.
PMH1 and/or PMH2 Is Associated with a Very Large Complex
To investigate the complex association of the PMH proteins by an independent experimental procedure, DDM-solubilized mitochondrial protein was fractionated in discontinuous Suc gradients. In addition, this experiment will allow conclusions about potential interaction of the PMH proteins with ribosomes. Thus, 100 mg of mitochondria were solubilized with DDM under conditions optimized for the enrichment of intact mitochondrial ribosomes and polysomes (Raczynska et al., 2006). The lysate was size fractionated on a Suc step gradient with steps of 5% ranging from 15% to 55% (w/v) Suc; each step was collected separately and proteins and RNA were analyzed.
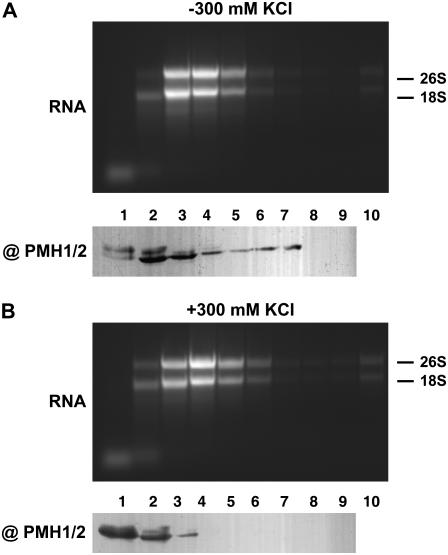
Proteins were investigated by immunodetection analysis with the PMH1/PMH2 antiserum after SDS-PAGE of 50 μL of each fraction. PMH1 and/or PMH2 is detected in fractions 1 to 7 and a slightly larger protein is found mainly in fractions 1 and 2 (Fig. 6A, top). This protein has also been detected in BN/SDS-PAGE, but had not been seen in the IEF/SDS-PAGE analysis, which might be attributed to conditions during IEF.
Figure 6.
Fractionation of mitochondrial ribonucleoprotein complexes. Total mitochondrial protein solubilized in the absence (A) and presence (B) of 300 mm KCl was fractionated on discontinuous Suc gradients. Fraction 1 corresponds to the sample buffer loaded onto the gradient. Fraction 2 corresponds to 15% Suc, 3 to 20%, 4 to 25%, 5 to 30%, 6 to 35%, 7 to 40%, 8 to 45%, 9 to 50%, and 10 to 55% Suc, respectively. Proteins of fractions 1 to 9 were analyzed by immunodetection with the @PMH1/PMH2 antibody. RNA of fractions 1 to 10 was inspected by agarose gel electrophoresis and ethidium bromide staining (RNA). Fractionation was done in buffer optimized for the separation of intact ribosomes and polysomes. Positions of 26S rRNA (26S) and 18S rRNA (18S) are indicated.
To determine the localization of ribosomes or polysomes in the gradient, total RNA was extracted from 800 μL of each Suc gradient fraction and inspected by agarose gel electrophoresis. The rRNAs were found predominantly in fractions 3 to 5, but only found in minor amounts in fraction 2, where the PMH proteins are highly abundant (Fig. 6A). This experiment demonstrates that a large portion of the PMH proteins does not cofractionate with ribosomes. When mitochondria are lysed in the presence of 300 mm KCl (Fig. 6B), the ribosomal RNA is again found in the same fractions, indicating that they remained intact as expected from a previous analysis (Fig. 6B, top; Raczynska et al., 2006). In contrast, most of the PMH proteins are now found in fractions 1 and 2, indicating that they are released from their complexes (Fig. 6B, bottom). These experiments strongly suggest that PMH proteins are not integrated into or stably associated with ribosomes, but we can presently not exclude weak or transient interaction of the PMH proteins with the translation machinery.
DISCUSSION
PMH1 and PMH2 Are Mitochondrial Proteins
In this study, we analyze two DEAD-box proteins encoded in the nuclear genome of Arabidopsis. We provide evidence for localization of the two proteins in mitochondria (Fig. 1). Fusion proteins consisting of different N-terminal parts of these polypeptides and GFP were consistently transported into these organelles. In addition, the putative RNA helicases were repeatedly detected in mitochondrial protein fractions (Figs. 4–6). This cofractionation has been observed previously in proteomic analyses applying gel-based and liquid chromatography-based separations all supporting localization of the two proteins in mitochondria (Millar et al., 2001; Heazlewood et al., 2004). However, it can presently not be excluded that one or both of these proteins can additionally be present in another subcellular compartment because PMH2 (At3g22330) has recently been identified in a proteome analysis of nucleoli, but this localization has not been further analyzed (Pendle et al., 2005).
In the mitochondrial lysate, several spots are detected after IEF and SDS-PAGE. Presently, we can only speculate about the appearance of different protein spots, which were observed in all experiments performed. These might be experimental artifacts, such as protein carbamylation or oxidation, even though IEF conditions were chosen to avoid such effects. In addition, the differential presence of PMH proteins in these spots suggests that they represent PMH proteins with differing posttranslational modifications. However, further experiments are required to clarify this issue.
Distinct Expression Patterns Suggest Different Roles of PMH1 and PMH2
PMH1 and PMH2 share 77% identical amino acids and thus are very similar to each other. In addition, each protein contains a characteristic C terminus, which is in both proteins predominantly composed of Ser and Gly. These similarities suggest similar functions of both proteins; however, there are clear differences in terms of the spatiotemporal transcription patterns of each gene. Also, the response of PMH1 expression to cold is stronger than the induction of PMH2 and only PMH2 promoter activity is induced by wounding. From these experiments, it can be concluded that, despite the high similarity of the two proteins, the polypeptides might fulfill different roles in plant mitochondria. PMH1 is expressed only in certain situations and might support PMH2 in its function. For instance, during flowering, which is an energy-consuming process, elevated mitochondrial activity is required. Likewise, cold stress might be a situation in which PMH2 activity is needed and in which it is supported by PMH1, which is more strongly induced under this environmental condition than PMH2. Although the Genevestigator expression analysis tool (https://www.genevestigator.ethz.ch) cannot discriminate between PMH1 and PMH2 genes (both genes are represented by a single oligonucleotide on the ATH1 gene chip), cold induction is confirmed by a meta-profile analysis. This investigation also reveals enhanced expression after heat stress in seeds, particularly 3 h after imbibition, and in stems, conditions, and tissues not inspected in our expression studies. In contrast to our northern and histochemical GUS-staining analyses, strong expression in flowers or parts of them is not indicated by the meta-profile analysis. Likewise, no response to wounding is found by this gene expression analysis, although enhanced PMH mRNA levels are found upon biotic stress caused by different pathogens. However, nothing is known about a function of a DEAD-box protein in response to wounding or pathogen attack.
PMH1 and PMH2 Contain Ser- and Gly-Rich C Termini
Both PMH proteins contain approximately 100-amino-acid-long C termini rich in Arg, Gly, and Ser. Such termini, although in different lengths, are widespread among RNA-binding proteins (Lorkovic and Barta, 2002; Vermel et al., 2002), and, in Arabidopsis, there are a number of putative RNA helicases, which also contain such particular C termini (Aubourg et al., 1999). One of these proteins, At5g26743, shares 62% identical amino acids with the core regions of PMH1 and PMH2 and has a 30-amino-acid-long Ser/Gly-rich stretch within the C terminus. This protein is predicted to be targeted to chloroplasts, where its homolog in spinach (Spinacia oleracea) has indeed very recently been detected in a proteomic approach investigating plastid proteins (Baginsky et al., 2007). This protein might thus be the plastid ortholog of the two mitochondrial PMH proteins. Approximately a 50-amino acid Gly/Arg-rich C terminus is also present in the nuclear DEAD-box protein PRH75 (At5g62190). The recombinant extension of this protein was found to bind to RNA and is therefore considered to be an independent RNA-binding module (Lorkovic et al., 1997). The same function can be assumed for the C termini of PMH1 and PMH2, suggesting direct interaction of these proteins with RNA, which is consistent with the loss of the association of the PMH proteins from the complexes upon RNase treatment. In Arabidopsis, at least the mitochondrial RNA-binding protein RBP1a containing a 30-amino-acid-long Gly-rich C terminus is also inducible by cold (Vermel et al., 2002), and it would be interesting to see whether this protein is present in one of the PMH1/PMH2-containing complexes.
PMH1 and/or PMH2 Is Associated with Large RNA-Containing Complexes
As indicated by two-dimensional BN/SDS-PAGE and discontinuous Suc gradient centrifugation, both, or at least one, of the PMH proteins are part of large protein complexes. The exact composition of these complexes is presently unclear, but extrapolating from other DEAD-box protein-containing complexes, it can be assumed that several different proteins are present in these complexes. Certainly, RNA is a component of the complexes as clearly indicated by the observation that RNase digestion disrupts the complexes themselves or at least prevents the interaction of the PMH proteins with the complexes. In the latter case, the DEAD-box proteins would directly interact with the RNA, which would be possible through the Gly-rich C termini as mentioned above, whereas the former would indicate a central role of RNA in the architecture of the complex. Recently, another RNA-dependent protein complex has been observed in plant mitochondria, which contains a FLAG-tagged version of the PPR protein RF592, the restorer of the pcf-associated CMS in petunia (Petunia hybrida; Gillman et al., 2007). The other components of this complex are still unknown.
The dependence of complex formation on RNA might also explain the size variability of the complex, which ranges from the monomeric form up to 1,500 kD as estimated from BN/SDS-PAGE. Within this size range, distinct intermediate complexes are reproducibly observed. In addition, size variability is also indicated by the presence of the PMH proteins in different fractions of the Suc gradients, suggesting that size variation is not the result of a particular preparative procedure. This distribution is highly reproducible and independent from the presence of aminocaproate, which can destabilize protein complexes (data not shown; Novakova et al., 2006).
Several explanations are possible for this size variation. First, the different complexes could represent different degradation intermediates, but the high reproducibility and the influences of MgCl2 and cold do not support this explanation. Second, the pattern might be attributed to assembly intermediates, but such intermediates are typically rather low in abundance and should not be detectable at such high levels. Third, this pattern might indicate the presence of mature complexes with varying associated components, different RNA molecules, different proteins, or both. This has, for instance, been observed for p53. This multifunctional protein is present in different complexes as indicated by a western-blot analysis of whole-cell extracts also separated by two-dimensional BN/SDS-PAGE (Camacho-Carvajal et al., 2004). Here, protein spots and a smear are also observed in a horizontal line indicative of protein being present in several distinct complexes.
Searching for a Function of the PMH1 and PMH2 DEAD-Box Proteins
The functions of PMH1 and PMH2 remain unclear. Distribution of these proteins and of the rRNAs in the discontinuous Suc gradients is too different to reflect stable association with mature ribosomes, although weak interaction of PMH1 and PMH2 is possible (Fig. 6). Thus, a function of these proteins in translation initiation cannot be excluded. To obtain more information about the functional role of these proteins, we established the respective knockout mutants. However, preliminary characterization of these plants grown under normal conditions as well as under different temperatures did not reveal any obvious phenotype (data not shown). Thus, further detailed studies are required to uncover the physiological function of these PMH proteins.
Maybe PMH proteins are involved in RNA secondary structure rearrangements, which are required for maintenance of cellular functions at reduced and elevated temperatures, as has been suggested for prokaryotic DEAD-box RNA helicases (Owttrim, 2006). Such a scenario could also be relevant for plant mitochondria, which are exposed to highly differing environmental temperatures. The studies reported here will form a convenient platform for further detailed investigations to elucidate the exact function of the mitochondrial DEAD-box proteins PMH1 and PMH2.
MATERIALS AND METHODS
Subcellular Localization Studies
DNA fragments representing the N-terminal parts of PMH1 and PMH2, respectively, were amplified with the following primer pairs: pmh1GFP1.5 (5′-CACTCTCTGGGATCCGAAAATG)/pmh1GFP1.3 (5′-GCGGATCCTAACATGAAAGTCTCTCAC), pmh1GFP1.5/pmh1GFP2.3 (5′-CGAATCTCGGATCCCTCCATCG), pmh2GFP1.5 (5′-CACCTGGATCCGAAAATGATCAC)/pmh2GFP1.3 (5′-ATGGATCCCTTTCGCTTCAACACC), and pmh2GFP1.5/pmh2GFP2.3 (5′-GCGGATCCCAAGCCCATCACCACCAAC). PCR was performed using oligo(dT)-primed cDNA from Arabidopsis (Arabidopsis thaliana) total RNA as template and BD Advantage 2 polymerase following the instructions given by the manual (TaKaRa). The cDNA products, which code for 69 and 100 amino acids of the PMH1 protein and 62 and 104 amino acids of the PMH2 polypeptide, respectively, were digested with BamHI and ligated into the corresponding site in the psmGFP4 vector (Davis and Vierstra, 1998). After sequence analysis, correct clones were transformed into tobacco (Nicotiana tabacum) protoplasts. Protoplast isolation, transformation, staining, and inspection by fluorescence microscopy were done as described previously (Koop et al., 1996; Däschner et al., 2001).
Northern Hybridization
Arabidopsis ecotype Col-0 was grown on standard soil supplemented with Osmocote Exact Mini (Scotts) or on Murashige and Skoog medium in a growth chamber under a 16-h-light (160–200 μmol m−2 s−1, 23°C)/8-h-dark (21°C) regime. Tissues from adult plants were harvested from 3- to 4-week-old plants. Seedlings were grown for 7 d and then transferred to cold or kept at 23°C as control. RNA was isolated using an RNeasy plant mini kit following the manual given by the manufacturer (Qiagen). For northern analysis, about 10 μg of total RNA were denatured with glyoxale, separated on agarose gels, transferred to Hybond N nylon membranes, and hybridized according to standard protocols or to a protocol provided by the manufacturer (GE Healthcare). Probes used for hybridizations correspond to the long cDNA fragments used for studying subcellular targeting. Loading of the gels was monitored by hybridization with oligonucleotide P18SrRNA (5′-AAGCATATGACTACTGGCAGG) complementary to nuclear/cytoplasmic 18S rRNA sequences.
Histochemical GUS Staining
To study in vivo promoter activities, potential promoter regions corresponding to sequences from −1,981 to +14 (PMH1) and −2,067 to +3 (PMH2) with respect to the ATG (+1) were amplified with primer pairs pmh1prom5′ (5′-ATAGTCGACTCAGAAACTCTAGAATCC)/pmh1prom3′ (5′-ATAGTCGACTGTGCTAATCATTTTCAG) and pmh2prom5′Xba2 (5′-ACTCTAGAATGGTAGCCATCTCAACACC)/pmh1prom3′Xba (5′-TATCTAGACATTTTCAGATTCAGGTGTTC) on total DNA extracted with a DNeasy plant mini kit as outlined by the manufacturer (Qiagen). After amplification, PCR fragments were cloned upstream of the GUS genes into vector pBecks19/101, transformed into Arabidopsis plants by floral dip, and selected on kanamycin-containing Murashige and Skoog medium (Clough and Bent, 1998). Histochemical staining was done with plants derived from five independent transformation events per gene as given in a previously described protocol (Hull and Devic, 1995).
BN/Urea SDS-PAGE
Mitochondria were isolated and purified from an Arabidopsis Col-0 cell suspension culture as described before (Klein et al., 1998). To investigate the response to cold treatment, cell cultures were first grown at normal growth conditions and then kept at 4°C for 18 h prior to isolation of mitochondria. Two-dimensional BN/SDS-PAGE was done according to a protocol established for plant mitochondria with the following specific parameters (Eubel et al., 2005). About 1 mg of mitochondrial protein (=10 mg mitochondria) was resuspended in 80 μL of solubilization buffer (750 mm aminocaproate, 50 mm BisTris, pH 7.0, 0.5 mm EDTA). Proteins were solubilized by adding 15 μL DDM (10% [w/v]) to a final concentration of 1.3% (w/v). Optionally, 10 mm MgCl2 or 1 μg RNaseA/100 μL protein lysate was added. The solution was kept on ice for 10 min and samples were then centrifuged at 18,000g for 1 h to remove insoluble constituents. After adding 20 μL of Coomassie loading buffer (10% [w/v] Coomassie in 750 mm aminocaproate) to the supernatant, the sample was loaded onto 3% to 16% (w/v) acrylamide gradient gels for separation in the first dimension. Separation in the second dimension was carried out with urea gels (12.5%). Gels were then stained with either colloidal Coomassie Blue or proteins were blotted onto polyvinylidene difluoride membranes. Immunodetection analysis was carried out with a polyclonal serum against peptides NH2-AQRERTLAGFRDGNF and NH2-ELPSIAVERGSASMFE corresponding to amino acids 397 to 411 and 485 to 500, respectively, relative to the ATG of PMH1. Peptide synthesis, immunization of rabbits, as well as recovery of the sera were done by EUROGENTEC. All procedures of immunodetection followed standard protocols (Sambrook and Russel, 2001).
IEF
IEF was performed with the IPGphor system following the manufacturer's instructions (GE Healthcare). About 20 mg of mitochondria were resolved in 200 μL lysis buffer (2 m thiourea, 5 m urea, 2% CHAPS, 2% SB 3–10, 40 mm Tris base, 2 mm Tris-bicarbonate-phosphate, 0.2 mm phenylmethylsulfonyl fluoride) for 1 h and insoluble constituents were removed by centrifugation at 18,400g for 20 min. Prior to separation by IEF, proteins were precipitated by adding 3 volumes of precipitation solution (90% [v/v] acetone, 10% [v/v] methanol, 10 mm dithiothreitol [DTT]), incubated overnight, and centrifuged for 15 min at 20,000g. Proteins were resuspended in rehydration solution (2 m thiourea, 5 m urea, 1% [w/v] CHAPS, 1% [w/v] SB 3–10, a trace of bromphenol blue, 2 mm Tris-bicarbonate-phosphate, 0.5% immobilized pH gradient buffer). Separation was done on immobiline nonlinear DryStrip gels, pH 3 to 11 and 7 to 11, respectively, under the following conditions: 50 μA per strip at 21°C. For pH 3 to 11 gradients, rehydration 12 h, step 1: step and hold, 200 V, 3 h; step 2: step and hold, 500 V, 3 h; step 3: gradient 1,000 V, 3 h; step 4: gradient, 8,000 V, 6 h; and step 5: step and hold, 8,000 V, 7 h. pH gradient 7 to 11: rehydration 24 h, step 1: step and hold, 200 V, 2 h; step 2: step and hold, 500 V, 2 h; step 3: gradient, 4,000 V, 4 h; step 4: gradient, 8,000 V, 5 h; and step 5: step and hold, 8,000 V 7 h. After focusing, gel strips were equilibrated in buffer A (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% [v/v] glycerol, 2% [w/v] SDS, a trace of bromphenol blue, 1% [w/v] DTT) or buffer B, which is identical to buffer A except that DTT is replaced by 2.5% indole acetic acid. Subsequently, Tris-Tricine PAGE was carried out according to a previously described protocol (Schagger and von Jagow, 1987), and proteins were visualized with colloidal Coomassie, investigated by immunodetection analysis, or examined by MS.
For MS, visible protein spots were excised from the gel and digested with trypsin as described previously (Shevchenko et al., 1996). Peptides were extracted, separated, and analyzed (i.e. sequenced) by nano-liquid chromatography-coupled electrospray ionization-tandem MS using a hybrid triple quadrupole/linear ion trap mass spectrometer (4000 Qtrap; Applied Biosystems) under standard conditions. For protein identification, fragment spectra were searched against the National Center for Biotechnology Information nonredundant database using Mascot as the search engine.
Fractionation on Suc Gradients
For subfractionation of mitochondrial protein complexes, 100 mg of purified organelles were dissolved in solubilization buffer (see above) in the presence of 200 mm Tris-HCl, pH 8.0, 35 mm MgCl2, 25 mm EGTA, 200 mm Suc, 40 mm EDTA, 2% (w/v) DDM, 0.5 mg/mL heparin, 10 mm β-mercaptoethanol, 500 mm chloramphenicol, and optional 300 mm KCl (Raczynska et al., 2006). The supernatant containing the mitochondrial proteins was then loaded onto discontinuous Suc gradients, with Suc concentrations ranging in 5% steps from 15% to 55% in gradient buffer containing 40 mm Tris-HCl, pH 8.0, 20 mm KCl, 10 mm MgCl2, 1 mm EDTA; 0.5 mg/mL heparin; 10 mm β-mercaptoethanol; and optional 100 μg/mL chloramphenicol. Gradients were centrifuged at 217,000g for 4 h at 4°C. Fractions of 1 mL were taken from top to bottom, each corresponding to a complete step. Proteins (50 μL) from each of the fractions were directly analyzed by immunodetection analysis, whereas RNA was precipitated from 800 μL of each fraction by the addition of 1 volume of 8 m guanidine HCl and 4 volumes of 100% ethanol and an incubation −20°C overnight. A second precipitation was done by addition of 0.1 volume 3 m sodium acetate, pH 5.2, and 2 volumes of 100% ethanol. RNA was inspected by agarose gel electrophoresis and ethidium bromide staining.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of PMH1 and PMH2 amino acid sequences.
Supplemental Figure S2. Wound induction of PMH2 promoter activity.
Supplementary Material
Acknowledgments
We thank Friedrich Ossenbühl, Jesco Heinemeyer, and Hans-Peter Braun for their technical advice in BN-PAGE. We are also very grateful to Bärbel Weber for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. Bi 590/7–1 and 7–2) and the Studienstiftung des deutschen Volkes (fellowship to J.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stefan Binder (stefan.binder@uni-ulm.de).
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A (1999) The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Grossmann J, Gruissem W (2007) Proteome analysis of chloroplast mRNA processing and degradation. J Proteome Res 6 809–820 [DOI] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Brennicke A (2003) Gene expression in plant mitochondria: transcriptional and posttranscriptional control. Philos Trans R Soc Lond B Biol Sci 358 181–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet N, Aubourg S, Toffano-Nioche C, Kreis M, Lecharny A (2001) Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res 11 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35 262–272 [DOI] [PubMed] [Google Scholar]
- Camacho-Carvajal MM, Wollscheid B, Aebersold R, Steimle V, Schamel WW (2004) Two-dimensional Blue native/SDS gel electrophoresis of multi-protein complexes from whole cellular lysates: a proteomics approach. Mol Cell Proteomics 3 176–182 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–742 [DOI] [PubMed] [Google Scholar]
- Däschner K, Couee I, Binder S (2001) The mitochondrial isovaleryl-coenzyme a dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36 521–528 [DOI] [PubMed] [Google Scholar]
- Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F, et al (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep 4 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Braun HP, Millar AH (2005) Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods 1 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Jansch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol 133 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D, Binder S (2007) Expression of the plant mitochondrial genome. In D Logan, ed, Plant Mitochondria. Blackwell Publishing, Ames, IA, pp 50–95
- Gillman JD, Bentolila S, Hanson MR (2007) The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J 49 217–227 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Binder S (2002) Functional importance of nucleotide identities within the pea atp9 mitochondrial promoter sequence. J Mol Biol 320 943–950 [DOI] [PubMed] [Google Scholar]
- Hull GA, Devic M (1995) The beta-glucuronidase (gus) reporter gene system. Gene fusions, spectrophotometric, fluorometric, and histochemical detection. In H Jones, ed, Methods in Molecular Biology, Vol 49. Humana Press, Totowa, NJ, pp 125–141 [DOI] [PubMed]
- Kazama T, Toriyama K (2003) A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett 544 99–102 [DOI] [PubMed] [Google Scholar]
- Klein M, Binder S, Brennicke A (1998) Purification of mitochondria from Arabidopsis. Methods Mol Biol 82 49–53 [DOI] [PubMed] [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34 407–415 [DOI] [PubMed] [Google Scholar]
- Koop HU, Steinmuller K, Wagner H, Rossler C, Eibl C, Sacher L (1996) Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta 199 193–201 [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol 127 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Linder P (2006) Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res 34 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Herrmann RG, Oelmuller R (1997) PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants. Mol Cell Biol 17 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchfelder A, Binder S (2004) Plastid and plant mitochondrial RNA processing and RNA stability. In H Daniell, C Chase, eds, Molecular Biology and Biotechnology of Plant Organelles. Springer, Dordrecht, The Netherlands, pp 261–294
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Mingam A, Toffano-Nioche C, Brunaud V, Boudet N, Kreis M, Lecharny A (2004) DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J 2 401–415 [DOI] [PubMed] [Google Scholar]
- Novakova Z, Man P, Novak P, Hozak P, Hodny Z (2006) Separation of nuclear protein complexes by blue native polyacrylamide gel electrophoresis. Electrophoresis 27 1277–1287 [DOI] [PubMed] [Google Scholar]
- Owttrim GW (2006) RNA helicases and abiotic stress. Nucleic Acids Res 34 3220–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Lange H, Grienenberger JM, Gagliardi D (2004. a) AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res 32 5174–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D (2004. b) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem 279 25440–25446 [DOI] [PubMed] [Google Scholar]
- Raczynska KD, Le Ret M, Rurek M, Bonnard G, Augustyniak H, Gualberto JM (2006) Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett 580 5641–5646 [DOI] [PubMed] [Google Scholar]
- Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5 232–241 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166 368–379 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68 850–858 [DOI] [PubMed] [Google Scholar]
- Vermel M, Guermann B, Delage L, Grienenberger JM, Marechal-Drouard L, Gualberto JM (2002) A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc Natl Acad Sci USA 99 5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, et al (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.