Abstract
Plants contain more genes encoding core cell cycle regulators than other organisms but it is unclear whether these represent distinct functions. D-type cyclins (CYCD) play key roles in the G1-to-S-phase transition, and Arabidopsis (Arabidopsis thaliana) contains 10 CYCD genes in seven defined subgroups, six of which are conserved in rice (Oryza sativa). Here, we identify 22 CYCD genes in the poplar (Populus trichocarpa) genome and confirm that these six CYCD subgroups are conserved across higher plants, suggesting subgroup-specific functions. Different subgroups show gene number increases, with CYCD3 having three members in Arabidopsis, six in poplar, and a single representative in rice. All three species contain a single CYCD7 gene. Despite low overall sequence homology, we find remarkable conservation of intron/exon boundaries, because in most CYCD genes of plants and mammals, the first exon ends in the conserved cyclin signature. Only CYCD3 genes contain the complete cyclin box in a single exon, and this structure is conserved across angiosperms, again suggesting an early origin for the subgroup. The single CYCD gene of moss has a gene structure closely related to those of higher plants, sharing an identical exon/intron structure with several higher plant subgroups. However, green algae have CYCD genes structurally unrelated to higher plants. Conservation is also observed in the location of potential cyclin-dependent kinase phosphorylation sites within CYCD proteins. Subgroup structure is supported by conserved regulatory elements, particularly in the eudicot species, including conserved E2F regulatory sites within CYCD3 promoters. Global expression correlation analysis further supports distinct expression patterns for CYCD subgroups.
Cell cycle progression in eukaryotes is controlled by the Ser-Thr directed protein kinase activity of cyclin-dependent kinase (CDK) complexes composed of catalytic CDK and regulatory cyclin subunits (Morgan, 1997). The binding of different cyclins confers substrate specificity and regulation at different cell cycle transitions and, in particular, at the two main control points at the G1-to-S-phase and G2-to-mitosis transitions. Homologs of many key mammalian regulatory genes involved in cell cycle progression are also present in higher and lower plants (Umen and Goodenough, 2001; Rensing et al., 2002; Inze and De Veylder, 2006). These include A-, B-, and D-type cyclins (CYCA, CYCB, and CYCD), which are generally involved in controlling S-phase, G2-to-M-phase, and G1-phase, respectively. Cyclin E, a key regulator of the G1-to-S-phase transition in animals, is not present in plants.
Ancestral cell cycle regulators are often represented by single genes in invertebrates and lower plants, but there is a general trend in more complex eukaryotes for an increase both in the number of homologous subgroups and the number of genes within these discrete subgroups. Such events, reflecting gene duplication and diversification, appear to be kingdom specific. Plant cyclins in particular are encoded by larger numbers of genes than in animals, with even the small genome of Arabidopsis (Arabidopsis thaliana) encoding 10 CYCA, 11 CYCB, and 10 CYCD genes (Nieuwland et al., 2007).
Plant CYCD sequences show low protein sequence similarity to animal CYCD and form a separate clade (Wang et al., 2004) but nevertheless share key features reflecting the function of CYCD in a pathway that is conserved between animals and plants, involving the retinoblastoma (RB; animals) or RB-related gene (RBR; plants) and E2F/DP transcription factors. The latter are involved in regulating expression of many genes required for cell cycle progression, S-phase entry, and DNA replication. In nondividing cells, E2F/DP is bound by the RBR protein, which itself recruits histone deacetylases to prevent expression of such genes. Phosphorylation of RBR by CYCD-CDK complexes results in its dissociation from promoter-bound E2F/DP complexes, allowing expression of their target genes and progression of the cell into S phase (Uemukai et al., 2005). This targeted phosphorylation is dependent on a specific RBR-binding motif present near the N terminus of both animal and plant CYCD proteins and consisting of the amino acid sequence LxCxE (where x represents any amino acid; Soni et al., 1995; Ach et al., 1997; Huntley et al., 1998). Plant and animal CYCD also share the conserved cyclin_N domain and conserved cyclin signature involved in CDK binding (Nugent et al., 1991). In animals, CYCD have the specific partner CDK4, but in plants, they associate in vivo with the archetypal CDK called CDKA, a direct homolog of animal and yeast (Schizosaccharomyces pombe) CDK1/cdc2 (Healy et al., 2001).
CYCD in both animals and plants frequently respond to mitogenic and other signals that promote division and are therefore involved in the early stages of commitment of the cell to mitotic division (Sherr, 1993, 1995; Dewitte and Murray, 2003). CYCD are frequently unstable proteins degraded by specific proteolysis mechanisms as part of the cells' dynamic response to their environment (Sherr, 1993; Planchais et al., 2004). Indeed, in plants, CYCD expression is rate limiting for cell division (Dewitte et al., 2003, 2007; Menges et al., 2006; Qi and John, 2007) and, in certain cases, growth (Cockcroft et al., 2000; Mizukami and Fischer, 2000).
In Arabidopsis, the 10 CYCD genes have been classified into six or seven subgroups (Oakenfull et al., 2002; Vandepoele et al., 2002). Using this classification, the CYCD3 subgroup has three members, the CYCD4 family two, and the other groups all have a single member. The CYCD4 subgroup has relatively high homology with CYCD2;1 and it has previously been proposed that the CYCD4 cyclins should be regarded as members of the CYCD2 subgroup (Huntley and Murray, 1999; Oakenfull et al., 2002; Wang et al., 2004). In rice (Oryza sativa), which is separated from Arabidopsis by at least 150 million years (Myr) of evolution, the 14 CYCD genes fall into the same conserved subgroups (Wang et al., 2004; La et al., 2006; Guo et al., 2007). The rice CYCD2/CYCD4 subgroup has five members and the CYCD1 and CYCD5 subgroups three members each, whereas CYCD3, CYCD6, and CYCD7 are represented by a single gene (see Table I). It should be noted that here we use the original nomenclature of Ma and coworkers (Wang et al., 2004), which differs in some cases from that proposed later by La et al. (2006). For clarity, we provide a summary of the corresponding gene names and IDs in Table I.
Table I.
CYCD genes of Arabidopsis (http://www.tigr.org/tdb/e2k1/ath1/, TIGR annotation version 5.0, June 2007), poplar (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html, Populus genome release 1.1, June 2007), and rice (http://www.tigr.org/tdb/e2k1/osa1/, TIGR rice annotation release 5, June 2007), showing the corresponding accession number
| CYCD | Species | Accession No. |
|---|---|---|
| CycD1 group | ||
| Arath;CYCD1;1 | Arabidopsis | At1g70210 |
| Orysa;CYCD1;1 | Rice | Os09g21450 |
| Orysa;CYCD1;2 | Rice | Os06g12980 |
| Orysa;CYCD1;3 | Rice | Os08g32540 |
| Poptr;CYCD1;1 | Poplar | AM746109 |
| Poptr;CYCD1;2 | Poplar | AM746110 |
| Poptr;CYCD1;3 | Poplar | AM746111 |
| Poptr;CYCD1;4 | Poplar | AM746112 |
| Poptr;CYCD1;5 | Poplar | AM746113 |
| CycD2/4 group | ||
| Arath;CYCD2;1 | Arabidopsis | At2g22490 |
| Orysa;CYCD2;1 | Rice | Os07g42860 |
| Orysa;CYCD2;2 | Rice | Os06g11410 |
| Orysa;CYCD2;3 | Rice | Os03g27420 |
| Poptr;CYCD2;1 | Poplar | AM746114 |
| Poptr;CYCD2;2 | Poplar | AM746115 |
| Arath;CYCD4;1 | Arabidopsis | At5g65420 |
| Arath;CYCD4;2 | Arabidopsis | At5g10440 |
| Orysa;CYCD4;1 | Rice | Os09g29100 |
| Orysa;CYCD4;2 | Rice | Os08g37390 |
| CycD3 group | ||
| Arath;CYCD3;1 | Arabidopsis | At4g34160 |
| Arath;CYCD3;2 | Arabidopsis | At5g67260 |
| Arath;CYCD3;3 | Arabidopsis | At3g50070 |
| Orysa;CYCD3;1 | Rice | Os09g02360 |
| Poptr;CYCD3;1 | Poplar | AM746116 |
| Poptr;CYCD3;2 | Poplar | AM746117 |
| Poptr;CYCD3;3 | Poplar | AM746118 |
| Poptr;CYCD3;4 | Poplar | AM746119 |
| Poptr;CYCD3;5 | Poplar | AM746120 |
| Poptr;CYCD3;6 | Poplar | AM746121 |
| CycD5 group | ||
| Arath;CYCD5;1 | Arabidopsis | At4g37630 |
| Orysa;CYCD5;1 | Rice | Os12g39830 |
| Orysa;CYCD5;2 | Rice | Os03g42070 |
| Orysa;CYCD5;3 | Rice | Os03g10650 |
| Poptr;CYCD5;1 | Poplar | AM746122 |
| Poptr;CYCD5;2 | Poplar | AM746123 |
| Poptr;CYCD5;3 | Poplar | AM746124 |
| CycD6 group | ||
| Arath;CYCD6;1 | Arabidopsis | At4g03270 |
| Orysa;CYCD6;1 | Rice | Os07g37010 |
| Poptr;CYCD6;1 | Poplar | AM746125 |
| Poptr;CYCD6;2 | Poplar | AM746126 |
| Poptr;CYCD6;3 | Poplar | AM746127 |
| Poptr;CYCD6;4 | Poplar | AM746128 |
| Poptr;CYCD6;5 | Poplar | AM746129 |
| CycD7 group | ||
| Arath;CYCD7;1 | Arabidopsis | At5g02110 |
| Orysa;CYCD7;1 | Rice | Os11g47950 |
| Poptr;CYCD7;1 | Poplar | AM746130 |
| Moss CycD | ||
| Phypa;CYCD | P. patens | AJ488282 |
The genome sequence of poplar (Populus trichocarpa) has recently become available, providing a model system for tree genomics and a further eudicot species that diverged from Arabidopsis approximately 120 Myr ago. To date, most of the poplar genome sequence (approximately 480 Mb) has been assigned to the 19 chromosomes, with only a few small fragments still unmapped. Here, we identify the poplar CYCD genes and consider in more detail the structure and conservation of CYCD subgroups in the two eudicot species (poplar and Arabidopsis) and the monocot rice. We present evidence of striking conservation suggestive of distinct functions for different CYCD subgroups between monocots and eudicots and between annual herbs and perennial woody plants. Global expression correlation analysis of Arabidopsis and rice CYCD genes supports the proposal of distinct functions for CYCD subgroups.
RESULTS AND DISCUSSION
Identification of Poplar CYCD Genes
The genome sequence of the poplar species black cottonwood (Tuskan et al., 2006; http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html; Populus genome release 1.1, June 2007) was searched both for annotated cyclin genes and by BLAST searching with Arabidopsis CYCD protein sequences. This resulted in identification of 22 candidate CYCD genes, which were further analyzed to identify subgroup relationships to Arabidopsis and rice CYCD.
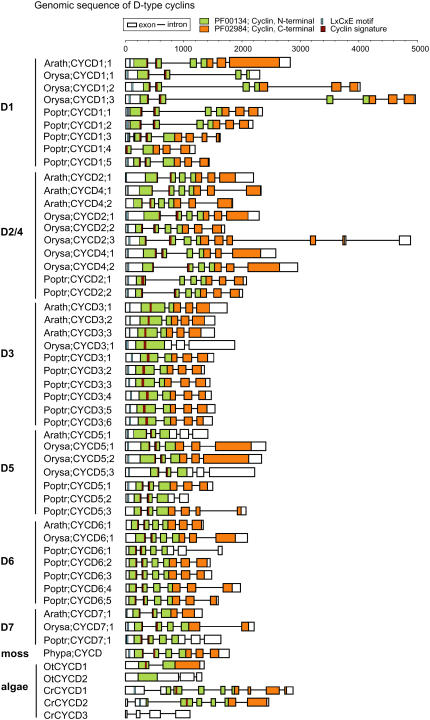
Predicted protein sequence alignment and phylogenetic tree analysis using the full CYCD sequences from Arabidopsis, rice, and poplar clearly shows that the poplar CYCD cluster with putative orthologs from Arabidopsis and rice within each of the clades corresponding to the CYCD1, CYCD2/CYCD4, CYCD3, CYCD5, CYCD6, and CYCD7 subgroups (Fig. 1). Six poplar CYCD3 genes are identified consisting of three pairs of closely related genes arising from genome duplication (see below), which all show closer homology to CYCD3 genes in Arabidopsis than to the single CYCD3 gene of rice. The poplar genome encodes five members of both the CYCD1 and the CYCD6 subgroups, the latter of which has only a single member in both Arabidopsis and rice. The CYCD5 family has three members, and, based on sequence similarity and highest similarity to other plant species, we identify two CYCD2 genes in poplar. As in Arabidopsis and rice, the CYCD7 group has only a single member.
Figure 1.
Phylogenetic tree analysis of the 52 CYCD protein sequences from Arabidopsis (AtCYCD), poplar (PtCYCD), rice (OsCYCD), moss (P. patens, Phypa;CYCD), and algae (C. reinhardtii, CrCYCD; O. tauri, OtCYCD). Full protein sequences were analyzed with ClustalW (Thompson et al., 1994) and bootstrap values calculated in PHYML (http://atgc.lirmm.fr/phyml/; Guindon and Gascuel, 2003). Bootstrap values are shown at each node of the constructed unrooted tree. Corresponding gene IDs of all Arabidopsis, poplar, rice, and moss cyclins are listed in Table I (Wang et al., 2004; La et al., 2006). [See online article for color version of this figure.]
Renaudin et al. (1996) defined the accepted nomenclature for plant cyclins to avoid confusion in these large gene families. According to the guidelines of the then Committee for Plant Gene Nomenclature, this consists of the species abbreviation as used in SWISSPROT (a two-letter abbreviation being insufficient for the range of plants for which gene sequences exist), the gene name according to cyclin class (CYCD) and subgroup CYCD1 to CYCD7, and a member (allele) number used to distinguish multiple genes in the same subgroup in a species. While subgroup numbers indicate homology, allele numbers are arbitrary according to the order of their cloning or identification (Renaudin et al., 1996). A summary of the proposed CYCD gene nomenclature in poplar according to these guidelines is provided in Table I, together with accession numbers for the predicted cDNA sequences.
Evolutionary analysis suggests that the monocot-eudicot divergence happened approximately 140 to 150 Myr ago (Chaw et al., 2004), and the lineage of the two dicot species Arabidopsis and poplar split some 30 to 50 Myr later (approximately 100–120 Myr ago). Consistent with the identification of CYCD subgroups, poplar CYCD3, CYCD5, CYCD6, and CYCD7 groups all show higher similarity to orthologous genes in Arabidopsis compared to rice (Fig. 1).
As discussed above, previous analysis of both Arabidopsis and rice CYCD genes has led to the conclusion that the CYCD4 cyclins should be regarded as members of the CYCD2 subgroup (Huntley and Murray, 1999; Oakenfull et al., 2002; Wang et al., 2004). This similarity between the CYCD2 and CYCD4 subgroups was further confirmed by our phylogenetic analysis incorporating the poplar genes, which revealed that no clear specific CYCD4 homolog could be identified but that the six groups corresponding to CYCD1, CYCD2/4, CYCD3, CYCD5, CYCD6, and CYCD7 are recognizable. This supports the identification of six common ancestors of these groups predating the separation of monocots and eudicots. Indeed, Arabidopsis CYCD2;1 is as related to Arabidopsis CYCD4;1 and CYCD4;2 as to orthologous rice and poplar genes, and the lack of evidence for distinctive orthologs within the CYCD2/CYCD4 subgroup between eudicots and monocots supports the origin of a combined CYCD2/CYCD4 group before the split (Wang et al., 2004). However, the present nomenclature is now well established for Arabidopsis and its revision would cause considerable confusion.
We extended our analysis with further CYCD sequences from moss and green algae (Fig. 1). The oldest fossil record of green algae dates to 700 to 750 Myr ago, whereas mosses appeared approximately 450 to 500 Myr ago. The CYCD sequence from the moss Physcomitrella patens (Phypa;CYCD, AJ488282), as well as a total of five CYCD genes from green algae, three from Chlamydomonas reinhardtii (CrCYCD1 = C_140186; CrCYCD2 = C_290120; CrCYCD3 = C_1460039; Bisova et al., 2005), and two from Ostreococcus tauri (OtCYCD1 = AY675099; OtCYCD2 = e_gw1.18.00.99.1; Robbens et al., 2005; http://genome.jgi-psf.org/cgi-bin/searchGM?db=Ostta4) were included. Phylogenetic tree analysis using full protein sequences shows that Phypa;CYCD clearly clusters closer to the CYCD groups of other angiosperm species, whereas CYCD from green algae are outliers with a more distant relationship to the CYCD of vascular plants and cannot be assigned as members of specific higher plant CYCD subgroups.
Chromosomal Distribution of Cyclin D Genes in Poplar
Analysis of the poplar genome suggests that the most recent duplication event in the Salicaceae occurred 60 to 65 Myr ago and affected approximately 92% of the Populus genome, although the slow rate of accumulated nucleotide divergence in Populus means that this equates to only 8 to 13 Myr in Arabidopsis (Tuskan et al., 2006). Further analysis of the 22 CYCD genes of poplar shows that they are distributed over 14 of the 19 poplar chromosomes (Fig. 2). On chromosome II, we find three CYCD genes of different subgroups, all of which are represented on duplicated regions in three distinct locations in the genome, although in the case of Poptr;CYCD3;1 this is on a as-of-yet-unattributed scaffold fragment (Scaf40). Chromosomes I, V, VII, IX, and XI each have two CYCD genes, and chromosomes III, IV, VI, VIII, X, XIII, XIV, and XIX contain a single CYCD gene. Homologous CYCD genes located on different chromosomes in poplar that are found in regions that resulted from the salicoid segmental duplication are highlighted (Fig. 2; Tuskan et al., 2006). As expected, all duplicated pairs are more closely related than other genes of the same subgroup (Fig. 1). Poptr;CYCD5;1 and Poptr;CYCD7;1 are located in regions unaffected by the genome duplication (Tuskan et al., 2006). Poptr;CYCD1;5 and Poptr;CYCD6;1 are also unpaired genes in single copy although lying within regions that are otherwise apparently duplicated.
Figure 2.
Genomic localization of poplar CYCD genes. The schematic view of chromosome reorganization in poplar after the most recent genome-wide salicoid-specific duplication event 65 Myr ago is adapted from Tuskan et al. (2006). Regions that are assumed to correspond to homologous genome blocks are shaded gray and connected with lines. To illustrate the genomic distribution of the poplar CYCD genes (Poptr;CYCDs), 17 of the 19 poplar chromosomes are presented as vertical or horizontal bars and indicated by their linkage group number (I–XIX). In addition, one unattributed scaffold (Scaf40) containing Poptr;CYCD3;1 gene is shown. Poptr;CYCD genes are represented by colored boxes. Recent duplication events between paralogous CYCD genes are indicated using colored dashed lines within the gray-filled trapezoids. [See online article for color version of this figure.]
We conclude that the six major subgroups of plant CYCD genes are conserved across angiosperm evolution in both ephemeral herbs and woody perennials and that all three model species contain at least one representative member of each of the subgroups. This conservation suggests distinct and important functions of each subgroup.
Exon and Intron Organization of CYCD Are Conserved in Angiosperms and Moss But Not Algae
We examined the exon-intron organization for a total of 52 CYCD genes from angiosperms, moss, and algae. In most cases, striking conservation of gene structure is found. Moss Phypa;CYCD consists of six exons (exon length: 1, 276 bp; 2, 87 bp; 3, 99 bp; 4, 202 bp; 5, 128 bp, 6, 291 bp), and this distribution and length of exons and introns in moss is remarkably conserved with most angiosperm CYCD subgroups (Fig. 3; Supplemental Fig. S1; Supplemental Table S1). The ancestral CYCD gene structure is characterized by the first exon ending at a constant position within the highly conserved cyclin signature, essential for cyclin binding to its partner CDK. Without an intact cyclin signature, cyclins are nonfunctional. All CYCD genes except those of the algae and the CYCD3 subgroup (see below) and two others (Poptr;CYCD1;4, potentially a pseudogene and Poptr;CYCD2;1) have conserved this feature. Furthermore, in almost all cases, exon 4 contains the junction between the cyclin N-terminal and C-terminal homology. We therefore propose that the distribution of CYCD genes over six exons is representative of the structure of the first plant ancestral CYCD gene to appear on land 450 Myr ago. This ancestral CYCD gene has diverged into the various CYCD subgroups, with subsequent duplication of genes within each subgroup. In contrast, available algae CYCD gene sequences do not show conservation of structure with moss and higher plants.
Figure 3.
Genomic domain structure of 52 CYCD genes of plants, moss, and algae. The PFAM motifs (http://pfam.janelia.org/hmmsearch.shtml) for the Cyclin, N terminal (green boxes) and Cyclin, C terminal (orange boxes), LxCxE motif (blue box), and cyclin signature (burgundy boxes) are shown as indicated. All plant CYCDs contain a cyclin_N domain with a distinct cyclin signature close to the N terminus at the end of exon 1 and start of exon 2, with the exception of the CYCD3s, in which the cyclin signature is found wholly in exon 1. All but Arath;CYCD5;1, Orysa;CYCD1;1, Poptr;CYCD6;1, Orysa;CYCD3;1, Orysa;CYCD5;3, and Poptr;CYCD7;1 contain a cyclin_C domain. Nearly all CYCD have a LxCxE responsible for RBR binding close to the N terminus, with Arath;CYCD5;1 showing a variant motif (for detail see Supplemental Table S2). [See online article for color version of this figure.]
This ancestral structure is conserved overall for most members of the CYCD1, CYCD2/4, and CYCD6 groups. Nine of the 10 CYCD3 genes, however, have only four exons (Orysa;CYCD3;1 has three exons), with a conserved length of both central exons (exon 2, 202 bp; exon 3, 131 bp; Fig. 3; Supplemental Table S1). Strikingly, all CYCD3 genes in all three species, including the single rice Orysa;CYCD3;1, do not have their cyclin signature split by an intron, and the first exon represents exons 1 to 3 from the ancestral gene structure, suggesting these fused into one exon when the original CYCD3 gene arose in angiosperms. A similar phenomenon is seen in all CYCD5 genes, where the ancestral exons 3 and 4 have similarly fused into one exon.
Whereas exon length, particularly of the central exons, is conserved or very similar between members of CYCD subgroups, intron length can show variation. This is notable in the CYCD1 and CYCD2/4 subgroups, which also both tend to have a higher degree of genomic structural differences from the common pattern. However, the overall evolutionary conservation observed for the exon-intron arrangement of CYCD genes in various subgroups between Arabidopsis, poplar, and rice is consistent with the appearance of the various subgroups before the monocot-eudicot divergence approximately 140 to 150 Myr ago (Chaw et al., 2004) and the proposal that these evolved from an ancestral gene represented by the single CYCD gene present in moss.
To understand the wider implications of CYCD gene evolution, we also compared the structure of the three human CYCD genes, which are representative of the genes from other vertebrates, with Arabidopsis. Human cyclin D1 shares only 24% to 28% identity (45%–49% similarity; data not shown) with Arabidopsis CYCD1;1, CYCD2;1, and CYCD3;1, but Figure 4 shows that two key exon/intron boundaries are also conserved in vertebrates, corresponding to the end of exon 1 in the cyclin signature and the highly conserved splice junction within the cyclin_C domain (boundary of exons 4 and 5 of the ancestral plant gene; Fig. 4). These highly conserved sites that interrupt key functional domains of the protein might reduce the chance of partial genes with damaging ectopic function, and the broad pattern we observe in plants is consistent with occasional intron loss in the evolution of certain CYCD subgroups. In contrast, the algae appear to show entirely distinct intronic distribution with no relic of an ancestral gene structure.
Figure 4.
Protein sequence relationships and exon structures of Arabidopsis, moss, and human CYCDs. A, Aligned protein sequences are presented with the regions corresponding to successive exons colored according to their homology to the six moss exons. The cyclin signature is boxed, and the five most highly conserved residues required for cyclin function are indicated with stars. Note the conservation of exon boundaries in the cyclin signature and between exons IV and V. B, Cluster and bootstrap analysis for aligned sequences as shown in A. [See online article for color version of this figure.]
Cyclin Domain and Cyclin Box Protein Sequences
Plant cyclins display the same typical structural organization as cyclins from other eukaryotes, with a conserved region of 250 amino acids called the cyclin core consisting of the cyclin_N and cyclin_C domains (Nugent et al., 1991). The cyclin_N domain is about 120 amino acids long and spans the CDK-binding region with a conserved cyclin signature of eight amino acids (Supplemental Table S2; Wang et al., 2004; Nieuwland et al., 2007). The cyclin_N domain spans five helices (H1–H5) of the cyclin A protein structure (Jeffrey et al., 1995), and is also called the cyclin box. It is the defining domain for cyclins, and contains five noncontiguous highly conserved residues shown to be essential for activity in mitotic cyclins, which are also highly conserved in CYCD (Fig. 5). Almost all of the 46 Arabidopsis, rice, and poplar CYCD have a conserved Arg (R) residue before the conserved Trp (W) residue as part of the distinctive box corresponding to the helix H1, a conserved Asp (D) residue as part of a modified DRF-motif corresponding to helix H2, a conserved Leu (L) and Lys (K) residue as part of a modified QL- and K(V/M)EE-motif corresponding to helix H3, and a Glu (E) residue corresponding to helix H5 in mitotic cyclins (for CYCD3 group as example, see Fig. 5; Renaudin et al., 1996). The Trp residue at the start of the cyclin core motif within helix H1 is present in all subgroups with the exception of CYCD6. However, the position of all other five core residues in the cyclin_N domain is highly conserved in CYCD6, indicating that the overall helical structure is maintained. The cyclin_C domain is less conserved and is present in most, but not all, cyclins, suggesting a specific but perhaps not critical function of this domain (Wang et al., 2004). It is present in all but seven of the 47 CYCD sequences examined here (Arabidopsis CYCD5;1; rice Orysa;CYCD1;1, Orysa;CYCD3;1, and Orysa;CYCD5;3; and poplar Poptr;CYCD5;2, Poptr;CYCD6;1, and Poptr;CYCD7;1).
Figure 5.
Aligned sequences of CYCD3 cyclins from Arabidopsis, rice, and poplar in the cyclin_N domain as indicated in Supplemental Table S2. The bold stars highlight highly conserved residues proven to be essential for cyclin activity and boxes indicate further regions of high conservation within plant CYCD (Renaudin et al., 1996). The lines drawn above the sequences indicate the helix domains of the structure of bovine cyclin A (Jeffrey et al., 1995), as adapted from Renaudin et al. (1996).
LxCxE RBR-Binding Motif
Animal cyclin D and most plant CYCD have the amino acid motif LxCxE near their amino terminus (Ewen et al., 1993; Soni et al., 1995), required for binding to the RB or RBR protein. This is part of a longer sequence xLxCxExxx in which the positively charged residues Lys or Arg significantly reduce the interaction with RBR (Singh et al., 2005). The interaction of CYCD1, CYCD2, and CYCD3 with RBR has been demonstrated in vitro and early studies have shown that it depends on an intact LxCxE motif (Ach et al., 1997; Huntley et al., 1998; Nakagami et al., 1999, 2002; Boniotti and Gutierrez, 2001; Koroleva et al., 2004; Kawamura et al., 2006).
In Arabidopsis, CYCD4;2 and CYCD6;1 have no canonical LxCxE motif within their coding sequence, and CYCD5;1 has a slightly divergent motif (LxxCxE; Supplemental Table S2; Vandepoele et al., 2002). Despite this, CYCD4;2 has similar effects when overexpressed to other CYCD (Kono et al., 2006). Whether the presence of the related motifs LxSxE (LESEE) and LxxSxD (LVNSFDD) within CYCD4;2 may be involved in RBR binding is unknown but conceivable. In rice, 13 cyclins have the conserved LxCxE motif in their sequence with only OsCYCD6;1 missing this structural feature (Supplemental Table S2; Figs. 3 and 6). Indeed, no CYCD6 protein from any of the species has the LxCxE motif, including all five of the poplar CYCD6 genes, indicating that this is a specific and identifying feature of this subgroup (Figs. 3 and 6). All other plant CYCDs contain the LxCxE motif except Poptr;CYCD1;4 and Poptr;CYCD5;3, although these may represent truncated genes.
Figure 6.
Protein structure and relationships of vascular plant CYCD. Left, Phylogenetic tree analysis of CYCD genes in each of the six defined subgroups was performed as described in Figure 1 using the PHYML algorithm after multiple sequence alignment (ClustalW v 1.83, Guindon and Gascuel, 2003). Right, Graphic representation of protein domain structure and analyzed motifs in plant CYCD. Potential PEST regions were predicted by the program PESTfind (https://emb1.bcc.univie.ac.at/toolbox/pestfind/pestfind-analysis-webtool.htm; Rechsteiner and Rogers, 1996), and strong PEST sequences are shaded gray. *, Putative phosphorylation sites for CDKs and other Pro-directed kinases, based on the presence of S/TP dipeptides. The equivalent to the functional T191 of tobacco CYCD3;3 is indicated by a lavender-colored asterisk (Nakagami et al., 2002). For detailed analysis results, see Table II. Blue box, LxCxE motif; green box, cyclin N_domain; orange box, cyclin_C domain; burgundy box, cyclin signature. [See online article for color version of this figure.]
Distinct variants of the LxCxE motif are also present in different subgroups. The consensus CYCD1 sequence is LLCGE, for CYCD2 LLCAE, CYCD3 LYCEE, and CYCD7 LLC(D/E)E. CYCD5 has a more variable sequence with Q, R, H, and Y at position 5, which are not present in these positions in other CYCD (apart from a single example: Orysa;CYCD2;2 has Y at position 5). These distinct variants may suggest subgroup-specific differences in RBR binding.
PEST Sequences and Potential CDK Phosphorylation Motifs
Human CYCD are degraded rapidly by ubiquitin-mediated proteolysis, a mechanism in which phosphorylation of the Thr-286 (T286) residue by glycogen synthase kinase-3β is implicated (Diehl and Sherr, 1997; Diehl et al., 1997, 1998). T286 or its equivalent is located within a hydrophilic PEST domain (rich in Pro [P], Gln [E], Ser [S], and Thr [T]), a conserved structural feature in CYCD of various organisms, including plants, and therefore a similar mechanism for the regulation of CYCD levels might be operational in plants (Rechsteiner and Rogers, 1996; Nakagami et al., 2002; Oakenfull et al., 2002). With the exception of Arath;CYCD4;2, all Arabidopsis CYCD contain a potential or poor PEST region (Table II), which is variously located N terminal or C terminal to the cyclin box or both (Fig. 6; Wang et al., 2004; Menges et al., 2006). Overall, 35 of the 47 CYCD analyzed have at least one detected poor PEST region (PESTfind score ≥ 0) within their sequence (Table II), although for only 15 of these are strong PEST sequences present. Nevertheless, PEST regions are general features of G1 cyclins and suggest that plant CYCD are likely to be short-lived proteins like their animal homologs. This has been experimentally confirmed for Arabidopsis CYCD3;1, which is degraded by a proteasome-dependent pathway (Planchais et al., 2004).
Table II.
Locations of PEST sequences and putative CDK phosphorylation sites
Positions of S and T in S/TP dipeptides are shown, with the equivalent to the demonstrated phosphorylation site T191 of tobacco CYCD3;3 shown in bold (Nakagami et al., 2002). Full CDK consensus phosphorylation sites (S/TPxR/K) are underlined, although it should be noted that not all such sites appear to be active substrates (Ubersax et al., 2003), and minimal S/TP dipeptides are also known to be CDK phosphorylated in certain proteins (Nash et al., 2001; Moses et al., 2007).
| CYCD | Potential PEST Region Amino Acids (Score) | Poor PEST Region Amino Acids (Score) | Putative CDK Phosphorylation Sites Amino Acid and Position |
|---|---|---|---|
| CycD1 group | |||
| Arath;CYCD1;1 | 307–321 (+12.11) | 11–60 (+2.45); 254–266 (+1.59) | S256; S318; T185; T288 |
| Orysa;CYCD1;1 | – | 1–65 (+1.27) | S63; T185 |
| Orysa;CYCD1;2 | 42–76 (+14.17) | – | S51; S114; S286; T86; T227 |
| Orysa;CYCD1;3 | 319–350 (+14.09) | 1–17 (+1.73); 17–93 (+3.46) | S51; S96; S346; T218 |
| Poptr;CYCD1;1 | 1–34 (+7.72) | – | S24; T175 |
| Poptr;CYCD1;2 | – | 1–34 (+3.83) | S24; T175 |
| Poptr;CYCD1;3 | – | – | S6; T160; T207 |
| Poptr;CYCD1;4 | – | 231–250 (+4.72) | S214; T95 |
| Poptr;CYCD1;5 | – | – | S198; S305; T9; T182; T226; T271 |
| CycD2/4 group | |||
| Arath;CYCD2;1 | – | 42–66 (+1.31); 241–266 (+0.27); 332–356 (+4.56) | S315; S344; T200 |
| Orysa;CYCD2;1 | – | – | S306; T198; T354 |
| Orysa;CYCD2;2 | 204–218 (+5.47) | – | S302; S324; T210 |
| Orysa;CYCD2;3 | 8–48 (+8.14) | – | S310; T209 |
| Poptr;CYCD2;1 | – | – | S322; T211; T228; T351 |
| Poptr;CYCD2;2 | – | 1–40 (+0.39) | S308; T196; T213; T334 |
| Arath;CYCD4;1 | – | 1–53 (+2.90) | S252; T27; T181; T281 |
| Arath;CYCD4;2 | – | – | S50; T172; T268 |
| Orysa;CYCD4;1 | – | 161–179 (+2.94) | S296; S309; S345; T206 |
| Orysa;CYCD4;2 | – | – | S334; S371; T213 |
| CycD3 group | |||
| Arath;CYCD3;1 | 28–69 (+5.08); 302–351 (+10.89) | – | S47; S310; S343; T190; T375 |
| Arath;CYCD3;2 | 297–341 (+5.12) | – | S306; S335; T199 |
| Arath;CYCD3;3 | – | 1–29 (+2.62) | S232; S282; S297; S326; S359; T189 |
| Orysa;CYCD3;1 | – | 338–356 (+3.54) | S302; T189 |
| Poptr;CYCD3;1 | – | 315–351 (+3.06) | S322; S349; T209 |
| Poptr;CYCD3;2 | – | 298–338 (+2.33) | S305; S332; T193 |
| Poptr;CYCD3;3 | – | 23–65 (+2.67); 299–340 (+2.07) | S306; S334; T192 |
| Poptr;CYCD3;4 | – | 35–81 (+4.91) | S48; S138; S351; T208; T319 |
| Poptr;CYCD3;5 | – | – | S13; S55; S311; S369; T308 |
| Poptr;CYCD3;6 | – | 304–345 (+2.61) | S13; S55; S311; S369; T308 |
| CycD5 group | |||
| Arath;CYCD5;1 | 304–318 (+5.35) | – | S314; T176; T283 |
| Orysa;CYCD5;1 | – | 318–355 (+0.38) | S165; S288; T203 |
| Orysa;CYCD5;2 | – | 323–355 (+4.75) | S297; T215 |
| Orysa;CYCD5;3 | 21–54 (+8.71) | – | S341; T13; T199; T331 |
| Poptr;CYCD5;1 | – | – | T186; T203; T285 |
| Poptr;CYCD5;2 | – | 1–35 (+0.00) | S7; S194; T177 |
| Poptr;CYCD5;3 | – | 35–458 (+2.58) | S7; S144; S195; T178 |
| CycD6 group | |||
| Arath;CYCD6;1 | – | 15–29 (+0.36) | S291; T157 |
| Orysa;CYCD6;1 | – | – | S28; S45; S207; T161; T272 |
| Poptr;CYCD6;1 | 2–15 (+5.01) | – | T153 |
| Poptr;CYCD6;2 | 274–295 (+11.20) | – | T152; T263 |
| Poptr;CYCD6;3 | 274–299 (+8.20) | 13–30 (+0.58) | T152; T263 |
| Poptr;CYCD6;4 | – | – | T20; T149; T259 |
| Poptr;CYCD6;5 | – | – | T152; T262 |
| CycD7 group | |||
| Arath;CYCD7;1 | – | 1–24 (+1.09) | S13; T16; T142 |
| Orysa;CYCD7;1 | 1–56 (+15.05) | – | S286; T25; T43; T189; T235 |
| Poptr;CYCD7;1 | – | – | S13; S301; T16; T141 |
| Moss CycD | |||
| Phypa;CYCD | 323–340 (+18.18) | – | S2; S100; S300; S312; S326; S332; T186 |
In mammalian cells, T286 phosphorylation is important for the full ability of cyclin D1 to activate CDKs and for intracellular localization, stability, and degradation (Diehl and Sherr, 1997; Diehl et al., 1997, 1998; Germain et al., 2000). In plants, Nakagami et al. (2002) showed that Thr-191 (T191) of tobacco (Nicotiana tabacum) CYCD3;3 is a putative phosphorylation site important for full kinase activity and nuclear import in tobacco BY-2 cells. In Arabidopsis CYCD3;1, there are five putative phosphorylation sites for Pro-directed kinases based on the conserved motif S/TP, of which three are found in putative PEST regions (Fig. 6; Table II; Menges et al., 2006). Indeed, Ser-343 (S343) of Arabidopsis CYCD3;1 is phosphorylated in suspension-cultured cells under the various growth states examined (Menges et al., 2006), although no evidence was found for S343 phosphorylation being involved in protein turnover.
We provide an overview of putative CDK phosphorylation sites in the CYCD of Arabidopsis, rice, and poplar in Figure 6 and Table II. The majority of the potential phosphorylation sites identified are located C terminal in the protein sequence. Strikingly, we find that independent of overall protein length, almost all (42/47) of the CYCD sequences have a conserved phosphorylation site located immediately at the beginning of the cyclin_C domain (Fig. 6; Table II). This corresponds to T191 of tobacco CYCD3;3, suggesting this highly conserved site is important for CYCD, probably for full kinase activity and nuclear import.
We also note that the phosphorylated residue identified in Arabidopsis CYCD3;1 at S343 is close to the carboxy terminus in a region rich in hydrophilic amino acids, particularly Ser and Glu, and several other CYCD contain sites at similar positions in regions rich in these amino acids or Asn. Several CYCD3 also contain a possible CDK phosphorylation site (TP) at their extreme carboxy terminus, but the functionality and potential role of this is unknown.
Global Correlation Analysis of CYCD Expression
Genome-wide analysis of cell cycle genes in Arabidopsis (Vandepoele et al., 2002; Menges et al., 2005) and rice (Guo et al., 2007) suggests that plants contain a much larger number of genes encoding potential cell cycle regulators (up to 100 genes in Arabidopsis; Vandepoele et al., 2002; Torres Acosta et al., 2004; Wang et al., 2004; Menges et al., 2005), but for most of these genes, it is still unclear whether they represent diverse functions, duplicated functions, or display tissue-specific expression. We previously analyzed 327 publicly available microarray data sets and found that most core cell cycle genes are expressed across almost all plant tissues, although some show strong tissue specificity (Menges et al., 2005). Further analysis using the expression map of Arabidopsis development (Schmid et al., 2005) and data from specific cell types derived from cell sorting (Birnbaum et al., 2003) shows that all three Arabidopsis CYCD3 genes and CYCD6;1 are highly correlated in their expression with mitotic cyclins, indicating that this may reflect the concentration of dividing cells in the samples (Menges and Murray, 2007). CYCD1;1 shows the highest abundance in flower samples and the shoot apex and also appears to be particularly expressed in shoot samples, whereas CYCD3;3 and CYCD4;1 appear to be more highly expressed in roots. CYCD2;1, although more or less constantly expressed, shows the highest expression in mature pollen (Menges and Murray, 2007).
We sought to extend the analysis of cell cycle genes and to understand further whether the distinct CYCD subgroups correlate with expression profile differences. Using publicly available data for about 1,730 Affymetrix ATH1 GeneChip arrays (Supplemental Table S3), we correlated the expression levels of each of the 86 core cell cycle regulators represented by probe sets on the array with all other unique genes across the wide range of tissues and experimental conditions represented by the available arrays and clustered the linear correlation coefficients (Fig. 7; Supplemental Fig. S2 for detailed heatmap). The resulting cluster tree clearly confirms that all three CYCD3 and CYCD6;1 show the closest relationship to other known mitosis-related core cell cycle genes, most notably CYCA and CYCB, and the mitosis-specific CDKB2 genes (Fig. 7). CYCD1;1, CYCD4;1, CYCD5;1, and notably CYCD2;1 are more distantly related to the mitotic cluster (as is cyclin CYCD4;2, which is poorly detected), suggesting roles distinct from CYCD3 and CYCD6.
Figure 7.
The linear Pearson coefficient of 86 core cell cycle genes with all genes represented by unique probes on the Affymetrix ATH1 GeneChip array over 1,730 experiments was calculated as described in “Materials and Methods” and imported into GeneMaths (version 2.01) for hierarchical cluster analysis (right). The heatmap represents a compressed picture of all 21,692 unique genes represented by probes on the ATH1 array (left to right), with the color representing the degree of correlation from white/blue (low) to red/black (high) with each of these probe sets (for an uncompressed and more detailed picture see Supplemental Fig. S2). The cluster tree on the right represents the similarity of expression of each cell cycle regulator across all probe sets with other cell cycle genes. Two specific groups of coregulated genes are indicated (pollen specific and mitosis associated). For more information on core cell cycle gene function, see Menges et al. (2005).
Extensive microarray data is not available for rice or poplar, but datasets derived from massively parallel signature sequencing (MPSS) are available for Arabidopsis (http://mpss.udel.edu/at/; Meyers et al., 2004) and rice (http://mpss.udel.edu/rice/; Nakano et al., 2006). We analyzed the available data for expression of CYCD genes, which in the case of Arabidopsis (Supplemental Table S4) broadly confirms the data from the microarray expression atlas of development (Menges and Murray, 2007) and confirms strongly different transcript distributions in different tissues. The rice MPSS data shows that transcripts of several CYCD genes were not detected, and others are again strongly tissue specific (Supplemental Table S5). For example, CYCD2;1 is strongly expressed in developing seed and seed germinating in the light (as also previously reported for Arabidopsis CYCD2;1; Masubelele et al., 2005) but absent in the dark. The only other rice CYCD gene detected as expressed in germinating seeds was CYCD5;1, which otherwise was only detected in some mature root samples and ovary.
In the case of the Arabidopsis microarray data, global correlation analysis is possible. Genes most closely correlating with each CYCD subgroup were examined in more detail, using cutoff scores depending on the degree of correlation with other genes from ≥0.5 (CYCD2;1, CYCD4;2, and CYCD5;1) to ≥0.75 (CYCD3). Several known cytokinesis-related genes are found in the cluster of top correlators to all three CYCD3 genes and CYCD6;1, such as the syntaxin-encoding gene KNOLLE (Lukowitz et al., 1996; Lauber et al., 1997), the kinesin-like protein AtNACK1/HINKEL (Nishihama et al., 2002; Strompen et al., 2002; Rashotte et al., 2003), and further kinesin-like genes and microtubule-associated proteins (Table III; Supplemental Table S6 for detailed list of selected genes). In an earlier study, we identified 82 genes as showing a mitosis-specific expression during cell cycle progression (Menges et al., 2005), most of which contain a mitosis-specific activator (MSA) regulatory site in their promoter (Ito et al., 2001). Notably, we find 56% of these mitosis-specific genes (46 genes) in the set of 308 correlators to CYCD3;3, 52% (43/233 genes) in the CYCD3;1 correlation set, and 44% (36/243 genes) in the CYCD3;2 set. The close correlation of CYCD3 genes with this mitotic gene set does not indicate that CYCD3 is involved in mitosis itself, but rather that it is specifically expressed in cells engaged in mitotic cycles. This is further supported by analysis of the CYCD3 expression patterns and function (Riou-Khamlichi et al., 1999; Swaminathan et al., 2000; Dewitte et al., 2003, 2007; Menges et al., 2006). In contrast, expression of CYCD4;1 shows high correlation with ribosomal protein gene expression and CYCD2;1 has no significant correlators, suggesting that posttranslational regulation may be important in controlling its activity (Table III; Healy et al., 2001).
Table III.
Global expression correlators of CYCD
For each Arabidopsis CYCD gene, the highest correlating genes (see “Materials and Methods” for detailed analysis) were subjected to GO analysis using the BiNGO software (Maere et al., 2005). GOC, GO category; GO-ID, GO identifier. GO terms in the table were selected for their highest significance based on P values in each GO category. The cluster frequency corresponding to the correlator set of each CYCD gene represents the number of times each GO-ID occurs compared to the total number in the correlator set that belong to an assigned GOC (actual numbers and percent values). Note that not all genes have an assigned BP, MF, or CC. The total frequency for all annotations in the Arabidopsis genome is also given.
| Gene | Correlators | GOC | GO-ID | Description | P Value | Cluster Frequency | Total Frequency |
|---|---|---|---|---|---|---|---|
| CYCD1;1 | 220 (≥0.6) | BP | 7169 | Transmembrane receptor protein Tyr kinase signaling pathway | 8.25E-10 | 13/162 8.0% | 115/20,763 0.5% |
| 7010 | Cytoskeleton organization and biogenesis | 1.45E-05 | 10/162 6.1% | 145/20,763 0.6% | |||
| 7049 | Cell cycle | 2.20E-05 | 8/162 4.9% | 87/20,763 0.4% | |||
| MF | 4674 | Protein Ser/Thr kinase activity | 8.35E-08 | 15/176 8.5% | 232/22,033 1.0% | ||
| 3677 | DNA binding | 4.23E-05 | 32/176 18.1% | 1,600/22,033 7.2% | |||
| CC | 31225 | Anchored to membrane | 2.01E-06 | 12/168 7.1% | 189/21,632 0.8% | ||
| CYCD2;1 | 19 (≥0.5) | None | |||||
| CYCD3;1 | 233 (≥0.75) | BP | 7010 | Cytoskeleton organization and biogenesis | 2.41E-17 | 21/182 11.5% | 145/20,763 0.6% |
| 7018 | Microtubule-based movement | 1.74E-13 | 12/182 6.5% | 43/20,763 0.2% | |||
| 74 | Regulation of progression through cell cycle | 1.21E-09 | 10/182 5.4% | 54/20,763 0.2% | |||
| 6263 | DNA-dependent DNA replication | 1.04E-08 | 8/182 4.3% | 33/20,763 0.1% | |||
| MF | 3777 | Microtubule motor activity | 2.28E-15 | 14/179 7.8% | 56/22,033 0.2% | ||
| 3753 | Cyclin-dependent protein kinase regulator activity | 1.07E-07 | 6/179 3.3% | 16/22,033 0.0% | |||
| 4674 | Protein Ser/Thr kinase activity | 1.25E-05 | 12/179 6.7% | 232/22,033 1.0% | |||
| CC | 5875 | Microtubule associated complex | 1.39E-14 | 13/176 7.3% | 52/21,632 0.2% | ||
| 31225 | Anchored to membrane | 5.84E-06 | 11/176 6.2% | 189/21,632 0.8% | |||
| 43228 | Non-membrane-bound organelle | 7.54E-06 | 19/176 10.7% | 621/21,632 2.8% | |||
| CYCD3;2 | 243 (≥0.75) | BP | 7010 | Cytoskeleton organization and biogenesis | 5.13E-11 | 16/181 8.8% | 145/20,763 0.6% |
| 7018 | Microtubule-based movement | 8.06E-09 | 9/181 4.9% | 43/20,763 0.2% | |||
| 6263 | DNA-dependent DNA replication | 1.70E-08 | 8/181 4.4% | 33/20,763 0.1% | |||
| 74 | Regulation of progression through cell cycle | 1.20E-05 | 7/181 3.8% | 54/20,763 0.2% | |||
| MF | 3777 | Microtubule motor activity | 1.83E-10 | 11/186 5.9% | 56/22,033 0.2% | ||
| 3753 | Cyclin-dependent protein kinase regulator activity | 8.12E-06 | 5/186 2.6% | 16/22,033 0.0% | |||
| 5488 | Binding | 5.96 E-05 | 71/186 38.1% | 5,061/22,033 22.9% | |||
| 4674 | Protein Ser/Thr kinase activity | 8.75E-05 | 11/186 5.9% | 232/22,033 1.0% | |||
| CC | 5875 | Microtubule-associated complex | 8.35E-10 | 10/179 5.5% | 52/21,632 0.2% | ||
| 31225 | Anchored to membrane | 7.84E-08 | 13/179 7.2% | 189/21,632 0.8% | |||
| CYCD3;3 | 308 (≥0.75) | BP | 6996 | Organelle organization and biogenesis | 1.76E-16 | 34/238 14.2% | 427/20,763 2.0% |
| 7010 | Cytoskeleton organization and biogenesis | 6.28E-14 | 20/238 8.4% | 145/20,763 0.6% | |||
| 7018 | Microtubule-based movement | 3.23E-12 | 12/238 5.0% | 43/20,763 0.2% | |||
| 31497 | Chromatin assembly | 3.70E-09 | 11/238 4.6% | 63/20,763 0.3% | |||
| 6263 | DNA-dependent DNA replication | 1.62E-06 | 7/238 2.9% | 33/20,763 0.1% | |||
| 74 | Regulation of progression through cell cycle | 3.24E-06 | 8/238 3.3% | 54/20,763 0.2% | |||
| 9653 | Morphogenesis | 1.84E-05 | 13/238 5.4% | 212/20,763 1.0% | |||
| 7169 | Transmembrane receptor protein Tyr kinase signaling pathway | 9.86E-05 | 9/238 3.7% | 115/20,763 0.5% | |||
| MF | 3777 | Microtubule motor activity | 1.76E-13 | 14/244 5.7% | 56/22,033 0.2% | ||
| 3677 | DNA binding | 3.17E-07 | 45/244 18.4% | 1,600/22,033 7.2% | |||
| 4674 | Protein Ser/Thr kinase activity | 1.39E-06 | 15/244 6.1% | 232/22,033 1.0% | |||
| CC | 5875 | Microtubule-associated complex | 1.17E-12 | 13/242 5.3% | 52/21,632 0.2% | ||
| 786 | Nucleosome | 2.98E-10 | 11/242 4.5% | 54/21,632 0.2% | |||
| 31225 | Anchored to membrane | 5.46E-08 | 15/242 6.1% | 189/21,632 0.8% | |||
| 5634 | Nucleus | 2.49E-07 | 41/242 16.9% | 1,427/21,632 6.5% | |||
| 43232 | Intracellular non-membrane-bound organelle | 1.82E-05 | 22/242 9.0% | 621/21,632 2.8% | |||
| CYCD4;1 | 217 (≥0.6) | BP | 6416 | Protein biosynthesis | 2.60E-13 | 40/163 24.5% | 1,164/20,763 5.6% |
| 42254 | Ribosome biogenesis and assembly | 2.98E-08 | 11/163 6.7% | 107/20,763 0.5% | |||
| 19538 | Protein metabolism | 6.09E-06 | 49/163 30.0% | 3,005/20,763 14 4% | |||
| MF | 3740 | Structural constituent of ribosome | 1.81E-25 | 33/176 18.7% | 309/22,033 1.4% | ||
| CC | 5840 | Ribosome | 1.53E-27 | 36/178 20.2% | 339/21,632 1.5% | ||
| 5830 | Cytosolic ribosome (sensu Eukaryota) | 7.41E-11 | 13/178 7.3% | 107/21,632 0.4% | |||
| 5843 | Cytosolic small ribosomal subunit (sensu Eukaryota) | 3.95E-08 | 8/178 4.4% | 46/21,632 0.2% | |||
| CYCD4;2 | 4 (≥0.5) | None | |||||
| CYCD5;1 | 139 (≥0.5) | BP | 6260 | DNA replication | 2.01E-06 | 7/104 6.7% | 63/20,763 0.3% |
| 74 | Regulation of progression through cell cycle | 1.24E-05 | 6/104 5.7% | 54/20,763 0.2% | |||
| 46472 | GPI anchor metabolism | 7.35E-05 | 3/104 2.8% | 6/20,763 0.0% | |||
| CYCD6;1 | 79 (≥0.7) | BP | 74 | Regulation of progression through cell cycle | 9.11E-07 | 6/64 9.3% | 54/20,763 0.2% |
| 6260 | DNA replication | 1.76E-06 | 6/64 9.3% | 63/20,763 0.3% | |||
| MF | 3753 | Cyclin-dependent protein kinase regulator activity | 1.10E-03 | 3/66 4.5% | 16/22,033 0.0% | ||
| CC | 31225 | Anchored to membrane | 1.46E-06 | 8/59 13.5% | 189/21,632 0.8% |
To analyze further the function of genes whose expression correlates with each CYCD subgroup, the overrepresentation of specific Gene Ontology (GO) categories was analyzed. Top correlators of each CYCD gene were classified into functional subgroups using BiNGO, an algorithm for GO significance analysis (Maere et al., 2005; Table III). The GO category consisted of biological process (BP), molecular function (MF), and cellular component (CC). GO terms in Table III were first selected for their highest significance based on the P value and second by the position in the resulting network graphic, as nodes furthest down the hierarchy are probably the most relevant (Maere et al., 2005). GO categories overrepresented in correlators to the three CYCD3 genes are linked to cytoskeleton organization and biogenesis (BP), microtubule motor activity (MF), and microtubule-associated complex (CC), whereas GO categories of the top correlators to CYCD4;1 are overrepresented in protein biosynthesis (BP), structural constituent of ribosome (MF), and ribosome (CC). The most significant GO categories overrepresented in CYCD1;1 correlators are linked to cell surface and transmembrane receptor kinase signaling pathway (BP), protein Ser/Thr kinase activity (MF), and membrane-anchored proteins (CC). Although CYCD6;1 correlators are also overrepresented in the same GO category for the CC as anchored to membrane, other overrepresented GO categories are linked to cell cycle progression, as suggested by the global correlation with mitotic activity (regulation of progression through cell cycle [BP], cyclin-dependent protein kinase regulator activity [MF]). CYCD5;1 correlators appear to be linked more to DNA replication than mitotic activity.
Conserved Motifs in CYCD Gene Promoters
The striking conservation of CYCD subgroups across the angiosperms suggests that important distinct functions are likely. We therefore examined CYCD promoters up to 1,000 bp from the predicted start codon for potential conserved regulatory motifs using WeederH (Pavesi et al., 2007), an algorithm that identifies conserved transcription factor binding sites in a given sequence by comparing it to one or more homologs. This approach does not need or compute an alignment of the sequences investigated and so is suited for identifying short conserved motifs that may be located at different positions within the sequence examined. Promoter sequences were compared between the two dicot species Arabidopsis and poplar, using either the closest poplar orthologous gene or all genes in the subgroup in the case of CYCD3. Results are shown in Table IV and identify putative binding sites for a number of defined transcription factors as well as novel motifs. The number of occurrences of each motif within the Arabidopsis genome was also determined as an indication of specific regulatory significance. Notable is the presence of conserved E2F binding sites in only CYCD3 and CYCD5 subgroup promoters, as well as a conserved MSA (Ito et al., 2001) sequence in CYCD5. The presence of an E2F binding site in the Arath;CYCD3;1 has been previously reported (Huntley et al., 1998). E2F activity is positively regulated by CYCD-mediated phosphorylation of RBR, suggesting that CYCD3 and CYCD5 may be involved in a positive feedback mechanism promoting the onset of S phase, as is the case for cyclin E in mammals (Koff et al., 1991). Indeed, CYCD3;1 and CYCD5;1 genes increase in expression in the G1 or G1 to S phases of the cell cycle (Menges et al., 2005). A number of new potential regulatory sequences were also identified, and a conserved 24-bp sequence was found to be present in both poplar and Arabidopsis CYCD7 genes. The initial part of this region (8 bp) was also conserved in rice. In general, comparisons to rice sequences showed much lower levels of conservation and fewer conserved motifs, although motifs found in the Arabidopsis-poplar comparisons could also be located in rice promoters, but the much higher number of substitutions found in rice promoters suggests that further investigation is required before claiming that these represent equivalent functional sequences.
Table IV.
Conserved motifs in promoters of Arabidopsis and poplar CYCD genes
Comparisons were made using WeederH as described in the text and “Materials and Methods.” Scores and positions of the sequences relative to the ATG start codon in Arabidopsis CYCD are shown, as well as putative matches to known transcription factor binding sites. The prevalence of the sequence within the Arabidopsis genome is also given.
| CYCD Subgroup | Comparison | Motif | Score | Position | No. of Hits (The Arabidopsis Information Resource) | Putative Binding Site (Plant Care) |
|---|---|---|---|---|---|---|
| CYCD1 | Arabidopsis versus best poplar | TAACCGTCTCTC | 32.4704 | −135 | 5 | Not found |
| GCAACAACACCA | 18.7287 | −113 | 15 | Not found | ||
| ATAAATAAAAAC | 17.525 | −96 | 157 | TATA box element | ||
| CYCD2 | Arabidopsis versus all poplar | TTACTTTG | 15.832 | −948 | 2,478 | DNA-binding protein of sweet potato (Ipomoea batatas) that binds to the SP8a |
| (ACTGTGTA) and SP8b (TACTATT) sequences of sporamin and β-amylase genes | ||||||
| GGTGGTAG | 12.2088 | −645 | 431 | Zinc-finger protein in alfalfa (Medicago sativa) roots, regulates salt tolerance | ||
| ACGTTACC | 11.5886 | −440 | 381 | Trihelix DNA-binding factor GT-3a | ||
| AGGTGTTTAAGA | 11.0741 | −930 | 7 | Not found | ||
| TAATTAACTTAA | 10.8628 | −795 | 45 | SBF-1 | ||
| TGACAAAATATT | 10.1763 | −780 | 45 | Not found | ||
| CYCD3 | Arabidopsis versus all poplar | TTTCACGC | <0.001 | (Consensus) | 465 | E2F class I sites |
| TTTCTCGCGC | <0.001 | (Consensus) | 13 | E2F class I sites | ||
| CYCD5 | Arabidopsis versus best poplar | TTTGGCGGCACC | 56.5282 | −189 | 3 | E2F complex |
| TAACCGCT | 28.5116 | −229 | 362 | MSA, mitosis-specific activator | ||
| CAAATAAAACCT | 15.346 | −278 | 15 | ccaat/tata box like | ||
| AACTAAATGATT | 15.0417 | −926 | 24 | Storekeeper, plant-specific DNA-binding protein important for tuber- specific and Suc- inducible gene expression | ||
| CYCD6 | Arabidopsis versus best poplar | TTAAATAC | 25.571 | −417 | 2,595 | L1-specific homeodomain protein Arabidopsis MERISTEM LAYER1 |
| AAACAACT | 17.3557 | −622 | 2,849 | CA-rich element | ||
| TCTAATAAAAAC | 16.0271 | −665 | 24 | Not found | ||
| TTATGAATATTT | 13.199 | −569 | 54 | Not found | ||
| TTATAAAAACAC | 11.5132 | −493 | 31 | TATA box element | ||
| CYCD7 | Arabidopsis versus poplar | ATTTACCGCGTTTCA TGTGGGACG | 34.1076 | −467 | 1 | Tobacco GT1/rice GT2 AT CAMTA3 AT RAP1 |
| CGACGTCT | 19.0557 | −445 | 257 | OCS-like elements; bZIP transcription factor from Antirrhinum majus | ||
| TTAAAGCT | 15.3937 | −931 | 1,736 | Dof2, single zinc finger transcription factor | ||
| CGTGGACTCACG | 12.6834 | −569 | 2 | Not found | ||
| CACCAGTTTCAT | 11.3275 | −1,000 | 7 | Not found | ||
| CYCD3/6 | With coexpressed genes | TTTCCCGCGC | <0.001 | (Consensus) | E2F binding site |
We further analyzed the promoters of the CYCD3 and CYCD6 genes, which showed some degree of coexpression (Fig. 7), together with 29 other cell cycle genes clustered together (Fig. 7; from KRP7 to CYCA2;4). We employed the motif-finding tool Weeder (Pavesi et al., 2004) that identifies conserved sequence motifs in promoter sequences from coregulated genes likely to represent binding sites for the common factors regulating the genes. The presence of an E2F site in the promoters of the CYCD3 genes was confirmed, with Weeder identifying an E2F-like conserved motif (TTTCCCGCGC). However, no reliable occurrence of the motif could be detected in the CYCD6;1 promoter (and in a few other genes), suggesting, not unexpectedly, that the similar expression profile of the genes investigated results from the action of several different transcription factors. The presence of E2F sites in the promoters of genes of this cluster was further verified by applying pscan, a recently developed tool (Pavesi and Zambelli, 2007) that employs descriptors of the binding specificity of known factors to identify overrepresented sites in a set of promoter sequences from coregulated genes. The program, by using the binding matrixes available in the TRANSFAC database (Matys et al., 2006) for plant transcription factors, predicted E2F as a common regulator for most of the genes (P value < 5 × 10−4). Also in this case, however, no reliable E2F site was predicted in the CYCD6;1 promoter. Nevertheless, E2F binding sites are clearly a common feature among this coregulated group, supporting the methodology used for global correlation expression analysis.
Other than known E2F sites, Weeder (and WeederH) identified several other conserved sites not directly comparable to known elements. A more in-depth analysis of promoter sequences and regulatory elements for CYCD and coexpressed genes and the experimental validation of predicted elements will be the subject of future research.
CONCLUSION
The identification of the CYCD genes of poplar and their analysis, together with those of Arabidopsis and rice, shows the strong conservation of six CYCD subgroups across the angiosperms. Despite the low sequence identity shared with plant CYCD proteins, certain gene features are nevertheless conserved with vertebrates. Moss contains a single CYCD gene that appears to represent an ancestral form, whereas green algae do not show gene conservation with higher plants. This analysis strongly supports conserved and differential functions for six of the seven defined CYCD subgroups and further suggests that the CYCD2 and CYCD4 subgroups cannot be distinguished in an evolutionary analysis.
MATERIALS AND METHODS
Clustering and Bootstrap Analysis
Information of all 10 Arabidopsis (Arabidopsis thaliana), 14 rice (Oryza sativa), and 22 poplar (Populus trichocarpa) CYCD sequences used for the analysis here has been downloaded from The Institute for Genomic Research (TIGR; Arabidopsis: http://www.tigr.org/tdb/e2k1/ath1/, TIGR annotation version 5.0, June 2007; rice: http://www.tigr.org/tdb/e2k1/osa1/, TIGR rice annotation release 5, June 2007) and the poplar genome project (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html., Populus genome release 1.1, June 2007).
For multiple sequence alignment, full protein sequences were analyzed with ClustalW v.1.83 (http://www.ebi.ac.uk/clustalw/index.html; Thompson et al., 1994). To estimate large phylogenies by maximum likelihood, the resulting multiple sequence alignment file in phylip format was imported into PHYML (http://atgc.lirmm.fr/phyml/; Guindon and Gascuel, 2003) and analyzed further by using the default WAG amino acid substitution model and, as a starting tree, the default BIONJ distance-based tree. Bootstrap values (n = 100) were calculated and presented at each node of the resulting unrooted trees.
Global Expression Correlation Analysis
The Nottingham Arabidopsis Stock Centre's (NASC) microarray database (NASCArrays, http://affy.arabidopsis.info/narrays/experimentbrowse.pl) was the source of data for the generation of scatterplots and the calculation of the relative correlation value. All Affymetrix ATH1 GeneChip array data deposited at NASC are normalized using the MASuite 5.0 Scaling Protocol Algorithm to exclude the top 2% and bottom 2% of signal intensities before the mean is calculated. All signal values from each individual slide are scaled such that the mean is made equal to 100. The superbulk gene file was downloaded in June, 2005 from http://affy.arabidopsis.info/narrays/help/usefulfiles.html. The file consisted of nearly 1,800 hybridizations (Supplemental Table S3), each with expression level measurements for over 22,500 genes. The arrays are derived from varied experiments, tissues, conditions, treatments, and genetic backgrounds, providing the diversity for expression correlation analysis. A cutoff value of 1 (all values <1 were discarded) was applied to the data before performing the analysis. A few slides (<50 from three different experiments) that used RNA from species other than Arabidopsis or that involved preamplification of the RNA used as the source for the hybridization were not included. All GeneChip arrays used for our calculation are listed in Supplemental Table S3, providing a NASCArrays experiment reference number, a short description of the experiment, and a hyperlink to the NASC Web site providing detailed information on each experiment, such as conditions and number of replicate slides used. This name is the same as in the superbulk gene file. For the correlation analysis, no further array normalization or processing of replicates was performed. The correlation analysis was performed essentially as described by Toufighi et al. (2005) by calculating the Pearson correlation coefficient for each gene pair from a two-gene scatterplot in the linear space using standard linear regression analysis. Briefly, between two sets of expression values (where X = [X1, X2,…, XN] and Y = [Y1, Y2,…, YN]), the Pearson correlation coefficient is defined as
 |
and ranges from 1, for perfect correlation, to −1 for perfect anticorrelation (Toufighi et al., 2005). A detailed description of the program employed for automation of the correlation calculation will be published elsewhere (P. Morandini and G. Pavesi, unpublished data).
First, the top correlators to each of the nine CYCD genes represented by a probe on the ATH1 GeneChip array (CYCD7;1 is not represented) were identified (cutoff values between ≥0.5 and ≥0.75) and used further to analyze the significant overrepresentation of GO categories using BiNGO software (Maere et al., 2005). Details of selected top correlators are summarized in Supplemental Table S6.
Next, calculations were done for all 86 core cell cycle genes against the whole list of 21,692 genes uniquely represented by a probe on Affymetrix's ATH1 GeneChip array. For each gene pair, the resulting value of the linear Pearson correlation coefficient was imported into GeneMaths (version 2.01) to visualize values in a colored representation. To identify the relationship of core cell cycle regulators and to further group these genes based on correlation of expression across a wide range of random experiments, hierarchical clustering analysis was performed and a new matrix was calculated using as clustering algorithm the unweighted pair group method using arithmetic averages (large N/p; Eisen et al., 1998). A detailed picture of the resulting heatmap after correlation analysis of 21,692 genes to 86 cell cycle genes is provided in Supplemental Figure S2, whereas the picture shown in Figure 7 represents a version of the heatmap compressed on the x axis.
Promoter Analysis
For all sequence sets, with the exception of the three CYCD3 genes, we compared the Arabidopsis promoter sequence to the one derived from the closest (according to sequence similarity) poplar orthologous gene. Each of the sequence pairs was analyzed by using the WeederH algorithm (Pavesi et al., 2007) that looks for motifs and regions significantly conserved in sequences from orthologous genes. The program was run with default settings and Arabidopsis background oligo frequencies. Extending the comparison to all the orthologous poplar genes did not yield significant results.
The CYCD3 set was analyzed with the motif finder tool Weeder (Pavesi et al., 2004). Two sequence sets were built with: (1) the promoters of the three Arabidopsis CYCD3 genes and the orthologous genes in poplar; and (2) the promoters of the genes belonging to a cluster of coexpressed cell-cycle genes, comprising the CYCD3 genes and CYCD6;1 (Fig. 7, from KRP7 to CYCA2;4). Weeder was run in double-strand mode, default (large) search, with Arabidopsis background oligo frequencies. On the first set, motifs reported had to appear in 75% of the sequences of the set (thus allowing motifs to be absent from one sequence of the set), while on the second, they had to appear in at least one-half of the sequences. Motifs reported in this work are those listed as interesting by Weeder on both sets (redundant motifs were filtered out), with Weeder score greater than one. The second sequence set was also submitted to the analysis of pscan (Pavesi and Zambelli, 2007), with default parameters, using the set of binding site matrixes available for plant transcription factors in Transfac professional version 10.4 (Matys et al., 2006).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Table I.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Protein sequence of CYCD in mammals and plants.
Supplemental Figure S2. Expanded version of Figure 7.
Supplemental Table S1. CYCD genes of Arabidopsis, poplar, and rice showing gene ID, exon number, exon length, genomic sequence, cDNA, and protein length.
Supplemental Table S2. CYCD genes of Arabidopsis, poplar, and rice showing gene ID, LxCxE motif sequence, cyclin signature, and position of cyclin_N and cyclin_C domains.
Supplemental Table S3. The superbulk gene file used for the correlation analysis presented here was downloaded in June, 2005 from the NASC Web site (http://affy.arabidopsis.info/narrays/help/usefulfiles.html).
Supplemental Table S4. Gene expression analysis using MPSS (Brenner et al., 2000) in various Arabidopsis tissues or after treatment were downloaded from the Arabidopsis MPSS database (http://mpss.udel.edu/at/; Meyers et al., 2004).
Supplemental Table S5. Results of MPSS gene expression analysis using various rice tissues or various treatments were downloaded from the rice MPSS database (http://mpss.udel.edu/rice/; Nakano et al., 2006).
Supplemental Table S6. Top gene correlators to CYCD genes in Arabidopsis.
Supplementary Material
Acknowledgments
Thanks to Luca Mizzi for software development and to Klaus Herbermann for help with bioinformatics analysis.
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (grant no. BBS/B/13268).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James A.H. Murray (j.murray@biotech.cam.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bisova K, Krylov DM, Umen JG (2005) Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol 137 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28 341–350 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58 424–441 [DOI] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JMS, Murray JAH (2000) Cyclin D control of growth rate in plants. Nature 405 575–579 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Murray JAH (2003) The plant cell cycle. Annu Rev Plant Biol 54 235–264 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, Murray JAH (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Cheng MG, Roussel MF, Sherr CJ (1998) Glycogen synthase kinase 3 beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 12 3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Sherr CJ (1997) A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol 17 7362–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ (1997) Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquintin-proteasome pathway. Genes Dev 11 957–972 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato JY, Livingston DM (1993) Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73 487–497 [DOI] [PubMed] [Google Scholar]
- Germain D, Russell A, Thompson A, Hendley J (2000) Ubiquitination of free cyclin D1 is independent of phosphorylation on threonine 286. J Biol Chem 275 12074–12079 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52 696–704 [DOI] [PubMed] [Google Scholar]
- Guo J, Song J, Wang F, Zhang XS (2007) Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol Biol 64 349–360 [DOI] [PubMed] [Google Scholar]
- Healy JMS, Menges M, Doonan JH, Murray JAH (2001) The Arabidopsis D-type cyclins CycD2 and CycD3 both interact in vivo with the PSTAIRE cyclin-dependent kinase Cdc2a but are differentially controlled. J Biol Chem 276 7041–7047 [DOI] [PubMed] [Google Scholar]
- Huntley R, Healy JMS, Freeman D, Lavender P, de Jager SM, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, et al (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol 37 155–169 [DOI] [PubMed] [Google Scholar]
- Huntley RP, Murray JAH (1999) The plant cell cycle. Curr Opin Plant Biol 2 440–446 [DOI] [PubMed] [Google Scholar]
- Inze D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40 77–105 [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP (1995) Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376 313–320 [DOI] [PubMed] [Google Scholar]
- Kawamura K, Murray JAH, Shinmyo A, Sekine M (2006) Cell cycle regulated D3-type cyclins form active complexes with plant-specific B-type cyclin-dependent kinase in vitro. Plant Mol Biol 61 311–327 [DOI] [PubMed] [Google Scholar]
- Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, Philippe M, Roberts JM (1991) Human cyclin-E, a new cyclin that interacts with 2 members of the Cdc2 gene family. Cell 66 1217–1228 [DOI] [PubMed] [Google Scholar]
- Kono A, Ohno R, Umeda-Hara C, Uchimiya H, Umeda M (2006) A distinct type of cyclin D, CYCD4;2, involved in the activation of cell division in Arabidopsis. Plant Cell Rep 25 540–545 [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson M, Parinyapong P, Sakvarelidze L, Leader D, Shaw P, Doonan JH (2004) CYCD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell 16 2364–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La HG, Li J, Ji ZD, Cheng YJ, Li XL, Jiang SY, Venkatesh PN, Ramachandran S (2006) Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol Genet Genomics 275 374–386 [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jurgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84 61–71 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 21 3448–3449 [DOI] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JAH (2005) D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA 102 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34 D108–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, De Jager SM, Gruissem W, Murray JAH (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41 546–566 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JAH (2007) Plant D-type cyclins: structure, roles and functions. In J Bryant, D Francis, eds, Eukaryotic Cell Cycle, Society for Experimental Biology Seminar Series 59. Taylor and Francis, Abingdon, UK, pp 1–28 [PubMed]
- Menges M, Samland AK, Planchais S, Murray JAH (2006) The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Matvienko M, Ghazal H, Agrawal V, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis by massively parallel signature sequencing. Nat Biotechnol 22 1006–1011 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13 261–291 [DOI] [PubMed] [Google Scholar]
- Moses AM, Heriche JK, Durbin R (2007) Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol 8 R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18 243–252 [DOI] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC (2006) Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res 34 D731–D735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414 514–521 [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Menges M, Murray JAH (2007) The plant cyclins. In D Inze, ed, Cell Cycle Control and Plant Development. Blackwell Publishing, Oxford, pp 31–61
- Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, et al (2002) Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109 87–99 [DOI] [PubMed] [Google Scholar]
- Nugent JHA, Alfa CE, Young T, Hyams JS (1991) Conserved structural motifs in cyclins identified by sequence-analysis. J Cell Sci 99 669–674 [DOI] [PubMed] [Google Scholar]
- Oakenfull EA, Riou-Khamlichi C, Murray JAH (2002) Plant D-type cyclins and the control of G1 progression. Philos Trans R Soc Lond B Biol Sci 357 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G (2004) Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res 32 W199–W203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Zambelli F (2007) Prediction of over represented transcription factor binding sites in co-regulated genes using whole genome matching statistics. In Application of Fuzzy Sets Theory, Proceedings of WILF 2007, Lecture Notes in Artificial Intelligence 4578. Springer, Berlin, pp 651–658
- Pavesi G, Zambelli F, Pesole G (2007) WeederH: an algorithm for finding conserved regulatory motifs and regions in homologous sequences. BMC Bioinformatics 8 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S, Samland AK, Murray JAH (2004) Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J 38 616–625 [DOI] [PubMed] [Google Scholar]
- Qi R, John PC (2007) Expression of genomic AtCYCD2;1 in Arabidopsis induces cell division at smaller cell sizes: implications for the control of plant growth. Plant Physiol 144 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) Pest sequences and regulation by proteolysis. Trends Biochem Sci 21 267–271 [PubMed] [Google Scholar]
- Renaudin JP, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inze D, Jacobs T, Kouchi H, Rouze P, Sauter M, et al (1996) Plant cyclins: a unified nomenclature for plant A-, B- and D- type cyclins based on sequence organization. Plant Mol Biol 32 1003–1018 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Rombauts S, Van de Peer Y, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7 535–538 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544 [DOI] [PubMed] [Google Scholar]
- Robbens S, Khadaroo B, Camasses A, Derelle E, Ferraz C, Inze D, Van De Peer Y, Moreau H (2005) Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus Tauri. Mol Biol Evol 22 589–597 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (1993) Mammalian G1 cyclins. Cell 73 1059–1065 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (1995) D-type cyclins. Trends Biochem Sci 20 187–190 [DOI] [PubMed] [Google Scholar]
- Singh M, Krajewski M, Mikolajka A, Holak TA (2005) Molecular determinants for the complex formation between the retinoblastoma protein and LxCxE sequences. J Biol Chem 280 37868–37876 [DOI] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Shah ZH, Murray JAH (1995) A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jurgens G, Mayer U (2002) The Arabidopsis Hinkel gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol 12 153–158 [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Yang Y, Grotz N, Campisi L, Jack T (2000) An enhancer trap line associated with a D-class cyclin gene in Arabidopsis. Plant Physiol 124 1658–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Acosta JAT, Engler JD, Raes J, Magyar Z, De Groodt R, Inze D, De Veylder L (2004) Molecular characterization of Arabidopsis Pho80-like proteins, a novel class of CDKA;1-interacting cyclins. Cell Mol Life Sci 61 1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43 153–163 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase CDK1. Nature 425 859–864 [DOI] [PubMed] [Google Scholar]
- Uemukai K, Iwakawa H, Kosugi S, De Jager S, Kato K, Kondorosi E, Murray JAH, Ito M, Shinmyo A, Sekine M (2005) Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Mol Biol 57 83–100 [DOI] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Kong HZ, Sun YJ, Zhang XH, Zhang W, Altman N, Depamphilis CW, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.