Abstract
HEXOKINASE1 (HXK1) from Arabidopsis (Arabidopsis thaliana) has dual roles in glucose (Glc) signaling and in Glc phosphorylation. The cellular context, though, for HXK1 function in either process is not well understood. Here we have shown that within normal experimental detection limits, AtHXK1 is localized continuously to mitochondria. Two mitochondrial porin proteins were identified as capable of binding to overexpressed HXK1 protein, both in vivo and in vitro. We also found that AtHXK1 can be associated with its structural homolog, F-actin, based on their coimmunoprecipitation from transgenic plants that overexpress HXK1-FLAG or from transient expression assays, and based on their localization in leaf cells after cryofixation. This association might be functionally important because Glc signaling in protoplast transient expression assays is compromised by disruption of F-actin. We also demonstrate that Glc treatment of Arabidopsis seedlings rapidly and reversibly disrupts fine mesh actin filaments. The possible roles of actin in HXK-dependent Glc signaling are discussed.
In plants, yeast (Saccharomyces cerevisiae), and mammals, Glc regulates many aspects of organismal growth and function through both nutritive metabolic effects and gene regulatory mechanisms. In Arabidopsis (Arabidopsis thaliana), Glc can modulate the expression of almost 1,000 genes (Price et al., 2004; Villadsen and Smith, 2004) and contemporary research has demonstrated many examples of cross talk between Glc and classical plant hormone-signaling pathways (Sheen et al., 1999; Rolland et al., 2002). Independent of nutritional effects, plant Glc signaling interacts with other hormones to promote leaf expansion, vegetative growth, flowering, and senescence (Xiao et al., 2000; Moore et al., 2003). Glc and cytokinin signaling affect cell division activity by inducing cyclin D2 and cyclin D3, respectively (Riou-Khamlichi et al., 2000), whereas Glc and abscisic acid (ABA) signaling can modulate meristem size by inducing a specific protein farnesyl transferase, ERA1 (enhanced response to ABA; Xiao et al., 2000; Ziegelhoffer et al., 2000). Wang et al. (2006) have suggested that Glc signaling in plants might be viewed as initiating growth strategies related to cell proliferation, as well as controlling energy carbon homeostasis.
Hexokinase (HXK) functions in Glc sensing in yeast, plants, and animals (Jang et al., 1997; Moore et al., 2003; Towle, 2005). One of the most intriguing aspects of HXK in different organisms is the different intracellular locations observed for diverse HXK protein family members. In yeast, HXK2 is reported to have dual targeting to both the nucleus and cytosol (Rodríguez et al., 2001) with signaling functions initiated in one or perhaps both compartments (for contrasting views, see Towle, 2005; Rolland et al., 2006). In plants, HXKs are encoded by a modestly large gene family (Claeyssen and Rivoal, 2007) and HXK proteins are reported to occur in the cytosol, mitochondria, plastids, nuclei, and Golgi (Miernyk and Dennis, 1983; Schnarrenberger, 1990; da-Silva et al., 2001; Yanasigawa et al., 2003). For example, in pea (Pisum sativum) leaves, most of the HXK is associated with mitochondria and facilitates respiration (Dry et al., 1983). In spinach (Spinacia oleracea) leaves, SoHXK1 has been reported, based largely on subcellular fractionation studies, to be associated with the external plastid envelope (Wiese et al., 1999). However, a more recent study has shown that SoHXK1 has an N-terminal membrane anchor that is similar to known mitochondrial-associated HXKs and that the GFP fusion protein is expressed only in mitochondria (Damari-Weissler et al., 2007). Available evidence suggests that plastid-localized HXKs might occur only in the stroma and possibly stromules (Kandel-Kfir et al., 2006), and might occur most abundantly in certain sink tissues (Giese et al., 2005). How or whether any of these particular plant HXK protein isovariants might be involved in Glc signaling is mostly a matter of conjecture. In developing castor bean (Ricinus communis) endosperm, mitochondrial HXK is released by sugar phosphate treatment (Miernyk and Dennis, 1983) and could then have an alternate function. In mammals, HXK is known to function in the cellular sensing of Glc metabolism (Towle, 2005). Even so, whereas some mammalian HXKs are bound to mitochondria by hydrophobic N-terminal peptide sequences, the type III isozyme lacks a mitochondrial membrane anchor and has perinuclear localization with unknown functional significance (Wilson, 2003). Both liver HXK IV and plant mitochondrial HXK have been shown to protect respective cells against proapoptotic stimuli (Danial et al., 2003; Kim et al., 2006). It is quite possible that these various localization data represent a broad spectrum of activities of any one complete HXK family.
Among plant HXKs, HXK1 in Arabidopsis (Arabidopsis thaliana; www.arabidopsis.org) is the best-characterized Glc sensor, with separable metabolic and gene regulatory functions (Jang et al., 1997; Moore et al., 2003; Cho et al., 2006). Despite substantial evidence that links Glc signaling and Arabidopsis HXK1 function with the modulation of organ growth, the cellular mechanisms by which this occurs are not well understood. Previous evidence indicates that Arabidopsis HXK1 protein is associated, at least in part, with mitochondria. Heazlewood et al. (2004) reported from proteomic analysis that AtHXK1 is one of 416 identified mitochondrial proteins from a dark-grown Arabidopsis cell culture. AtHXK1 has an N-terminal 24-amino acid peptide that is predicted to function as a targeting domain to mitochondria (Predotar; http://genoplante-info.infobiogen.fr/predotar; see also Olsson et al., 2003; Damari-Weissler et al., 2006). When expressed in tobacco (Nicotiana tabacum) protoplasts, AtHXK1-GFP expression was associated only with mitochondria (Damari-Weissler et al., 2007). On the other hand, based on a protoplast lysis protocol using Triton X-100, Yanasigawa et al. (2003) reported that AtHXK1 occurs at least in part in the nucleus of isolated protoplasts. This has led to a current model of Glc signaling that suggests that HXK-dependent plant Glc signaling occurs in analogous fashion to that observed in yeast (Rolland et al., 2006), in which a nuclear form of HXK acts as a transcriptional regulator. Indeed, Cho et al. (2006) have recently presented evidence that nuclear-localized AtHXK1 can function as a corepressor in a transcriptional complex identified from leaf extracts. However, yeast HXK notably does not substitute for Arabidopsis HXK1 in plant Glc signaling (Jang et al., 1997).
HXK-dependent Glc signaling involves not only transcriptional control, but also translational and posttranslational processes (Ho et al., 2001; Yanasigawa et al., 2003). With this in mind, we have further examined the cellular context for HXK function in Glc signaling. These studies establish that mitochondrial-bound HXK can interact with actin and that a normal functioning actin cytoskeleton is required for some HXK-dependent Glc signaling in transient expression assays. Furthermore, plant Glc treatments result in rapid, extensive alterations in the F-actin network. Our data link actin cytoskeleton functions in plant growth with HXK-dependent Glc signaling.
RESULTS
AtHXK1 Is Associated with Mitochondria
We used several approaches to test whether AtHXK1 is indeed bound to mitochondria and whether it might dissociate and/or move to the nucleus under a range of treatments. First, we examined localization of HXK1-GFP fusion proteins after their transient expression in leaf protoplasts. HXK1-GFP has typical HXK1 functions of abundant Glc phosphorylation activity (data not shown) and normal levels of Glc signaling activity in protoplast transient expression assays (Supplemental Fig. S1). Bioimaging revealed specific mitochondrial association of HXK1-GFP in transfected mesophyll protoplasts from Arabidopsis (Fig. 1, A–C), as well as maize (Zea mays) and pea (data not shown). We showed that HXK1-GFP is expressed at mitochondria by costaining with the organelle-selective reagent, MitoTracker Red (Fig. 1B). The fluorescence patterns from both fluorophores overlap completely and can be merged (Fig. 1C). Moreover, we observed similar fluorescence patterns of expressed HXK1-GFP and mitochondrial porin-GFP (Fig. 1D). However, transfected HXK1 (Fig. 1A) did induce association of mitochondria into larger, less numerous foci than observed for transfected porin-GFP, even with high expression levels of the latter. We also examined whether the localization of HXK1-GFP might change in protoplasts treated with Glc, Suc, auxin, or extended exposure to light or darkness. However, none of these treatments altered the subcellular targeting of HXK1 to the mitochondria (data not shown).
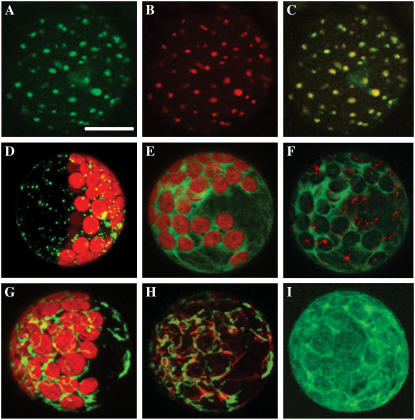
Figure 1.
Intracellular localization of Arabidopsis HXK1-GFP and other constructs in transfected leaf protoplasts. Fluorescence images are maximal projections of Z-stacks, captured with a confocal laser-scanning microscope after 6 to 7 h of expression. A, HXK1-GFP fluorescence, pseudocolored green. Scale bar = 10 μm. B, Stain-specific fluorescence of protoplasts in A incubated with MitoTracker Red, then pseudocolored red. C, Merged images of A and B, showing the resulting yellow fluorescence. D, Mitochondrial porin-GFP (At3g01280), also showing red chlorophyll (Chl) autofluorescence. E, HXK1ΔN-GFP (lacking amino acids 1–27), with Chl autofluorescence. F, HXK1ΔN-GFP and MitoTracker Red, without Chl autofluorescence. G, HXK1ΔC-GFP (lacking amino acids 28–496), with Chl autofluorescence. H, HXK1ΔC-GFP and MitoTracker Red without Chl autofluorescence. I, Yeast HXK2-GFP without Chl autofluorescence.
As further evidence of the mitochondrial targeting of AtHXK1, we showed that the N-terminal, 27-amino acid peptide from HXK1 is both necessary and sufficient for targeting to mitochondria in transfected leaf protoplasts. HXK1ΔN-GFP (lacking amino acids 1–27) was localized to the cytosol (Fig. 1, E and F), whereas HXK1ΔC-GFP (lacking amino acids 28–496) was localized to the mitochondria (Fig. 1, G and H; see also, Kim et al., 2006). Similarly, yeast HXK2-GFP also lacks an N-terminal membrane anchor (Olsson et al., 2003) and is expressed only in the cytosol (Fig. 1I). Neither of the two constructs expressed in the cytosol were observed to localize even partly to the nucleus, or elsewhere, after different protoplast treatments (data not shown).
Second, as a follow-up to the localization experiments using transient expression assays, we also examined protein localization in stable transgenic seedlings that express HXK1-GFP (Fig. 2, A–D). GFP fluorescence occurred in smaller punctate areas of the cells as in transient expression studies, but, in this case, they also occurred in some larger foci. Costaining with MitoTracker Red again showed specific fluorescence of the same smaller foci, plus many of the larger foci. The identity of the larger foci is uncertain. They could represent aggregated mitochondria and/or they could be artifacts from protein overexpression. However, as seen with the protoplast studies, seedling treatments with sugars or extended periods of light or darkness did not affect the mitochondrial location of HXK1-GFP (data not shown).
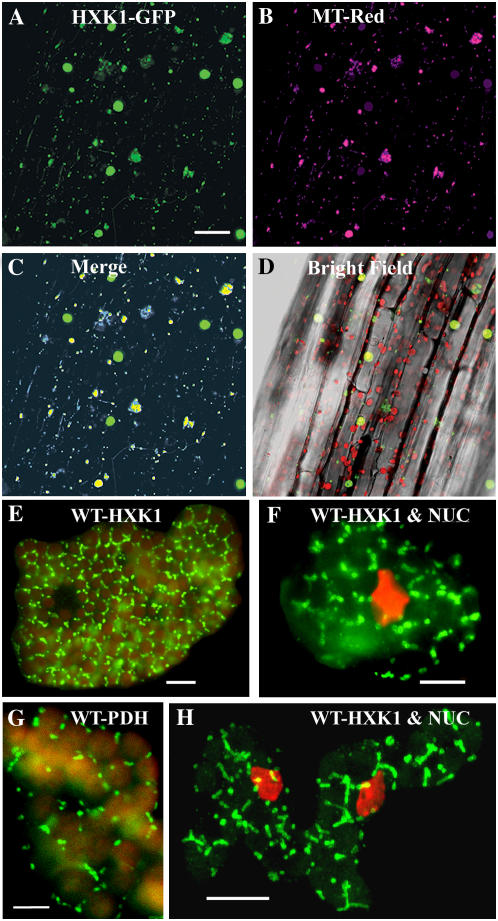
Figure 2.
Intracellular localization of Arabidopsis HXK1 in intact tissues. A to D, HXK1 localization in a seedling hypocotyl by fluorescence imaging of cauliflower mosaic virus 35S:HXK1-GFP expressed in Ler. Glc phosphorylation activity was increased about 18-fold in these seedlings (data not shown). A, Seedling hypocotyl showing punctate GFP fluorescence, occurring as smaller foci as well as larger aggregates. Scale bar = 50 μm. B, Seedling hypocotyl in A stained with MitoTracker Red and pseudocolored magenta. C, Merged images from A and B. Note codistributed blue-yellow fluorescence associated with the smaller foci and some larger foci. D, Hypocotyl images (A–C) merged with a corresponding bright-field image. Red shows Chl autofluorescence. E to H, Cellular immunolocalization of HXK in leaf mesophyll cells of young wild-type seedlings. Following cryofixation and freeze substitution, dissociated cells were labeled with anti-HXK1 polyclonal (E, F, and H) or anti-PDH monoclonal (G) antibodies, and then with secondary antibodies conjugated to FITC. Leaf cells were observed by fluorescence microscopy (E–G) or confocal microscopy (H); scale bars = 10 μm. H, Merged image of several optical sections and the rest (E–G) is each a single optical section. Staining of the nucleus (NUC) with DAPI is pseudocolored red (F and H). E and G, Red shows Chl autofluorescence.
We extended these observations with two different approaches to determine the subcellular localization of native HXK1: immunostaining of seedling leaves after cryofixation and acetone freeze substitution and immunoblotting of purified organelles from mature leaves. In the former case, we used a polyclonal antibody to HXK1, followed by a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Fig. 2, E, F, and H). This antibody reacts well with other HXKs (Moore et al., 2003; B. Moore, unpublished data). As a positive control, we tested a monoclonal antibody to pyruvate dehydrogenase (PDH; Fig. 2G). We observed HXK1 antigen in numerous fusiform shapes that were occasionally in close proximity to chloroplasts and nuclei. We observed that PDH antigen occurred in similarly shaped structures.
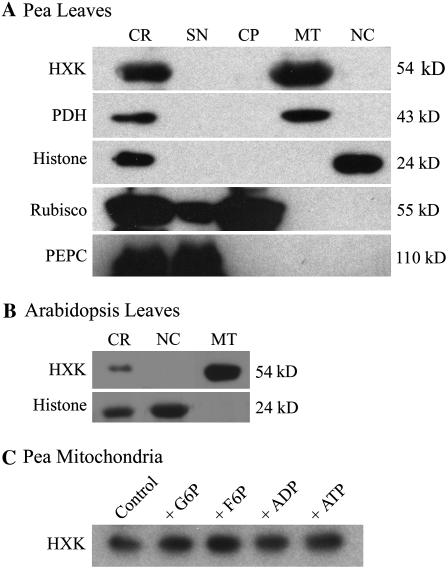
We also examined by western blot the distribution of AtHXK1 antigen in different organelle fractions of pea and Arabidopsis leaves (Fig. 3, A and B). As reported elsewhere (Dry et al., 1983), pea protein that cross-reacts with anti-HXK1 antibody occurred only in the isolated mitochondrial fractions and was not present in the cytosolic, nuclear, or chloroplast fractions. Proteins from isolated Arabidopsis leaf mitochondria also cross-reacted with anti-HXK1 antibody under these conditions, whereas the nuclear protein fraction did not. Thus, AtHXK1 did not occur at readily detectable levels in isolated nuclei from pea or Arabidopsis. Because isolated mitochondria from castor bean endosperm will release bound HXK following treatment with hexose phosphates (Miernyk and Dennis, 1983), we also tested whether different phosphorylated metabolites might induce the release of HXK from isolated pea mitochondria (Fig. 3C). However, treatment of the mitochondrial fraction with a variety of sugar phosphates or adenylates did not result in any apparent release of HXK1 antigen. These results extend similar recent observations that treatments of maize and rice (Oryza sativa) mitochondria with a variety of sugars and related metabolites failed to solubilize bound HXK (Rezende et al., 2006).
Figure 3.
HXK localization in isolated leaf organelles. A, Western blot of Percoll-purified pea leaf organelles, showing corresponding marker antigens and molecular weights after SDS-PAGE. CR, Crude homogenate; SN, postmitochondrial supernatant; MT, mitochondria; NC, nucleus. B, Western blot of isolated mitochondria and nuclei from Arabidopsis Ler leaves. Leaf organelle fractions also were monitored by fluorescence staining or Chl autofluorescence. These confirmed the purity of pea leaf organelles and also indicated that the Arabidopsis mitochondrial fraction contained low amounts of chloroplasts, whereas the Arabidopsis nuclear fraction was free of mitochondria and chloroplasts (images not shown). C, Western blot for HXK from a washed pea mitochondrial fraction, following overnight incubation at 4°C with 10 mm solutions of indicated metabolites.
AtHXK1 Can Interact with Porin and Actin Proteins
To identify proteins that might interact with HXK1, we first used a proteomics approach to immunocapture HXK1-FLAG from leaf extracts of a constitutive overexpression line made in gin2-1, a HXK1 null mutant (Moore et al., 2003). HXK1-FLAG was bound to primary mouse anti-FLAG and then partially purified using magnetic beads coated with a secondary antibody. This isolation procedure avoided pelleting by centrifugation. After washing the beads that had bound proteins, FLAG peptide was used to release HXK1 by competitive binding interactions to the primary antibody. About 25% of bound HXK1 was typically released by this process and about 40 proteins were identified by mass spectroscopy as coeluting with HXK1 (data not shown). Among identified proteins were seven other mitochondrial proteins, including the two most abundant Arabidopsis porin isoforms (At5g515090 and At3g01280; Clausen et al., 2004). In mammals, mitochondrial porins are the docking proteins in the outer membrane for some HXKs (Wilson, 1997).
We further examined the possible interaction of HXK1 with the two mitochondrial porins by testing with coimmunoprecipitation assays of transiently expressed proteins after labeling with 35S-Met (Fig. 4A). Only when HXK1-FLAG was coexpressed with either porin or hemagglutinin (HA) clones was the anti-FLAG antibody also able to pull down porin proteins and the anti-HA antibody able to pull down HXK1-FLAG. As one pair of controls, anti-FLAG antibody did not capture porin-HA protein, and anti-HA antibodies were not able to capture HXK1-FLAG protein. These data demonstrate that both porins can interact with AtHXK1, apparently as docking proteins. As an additional control, we showed that HXK1 interactions with cellular proteins are not promiscuous in this assay because a cytosolic calcium-dependent protein kinase could not be coimmunoprecipitated (Fig. 4B).
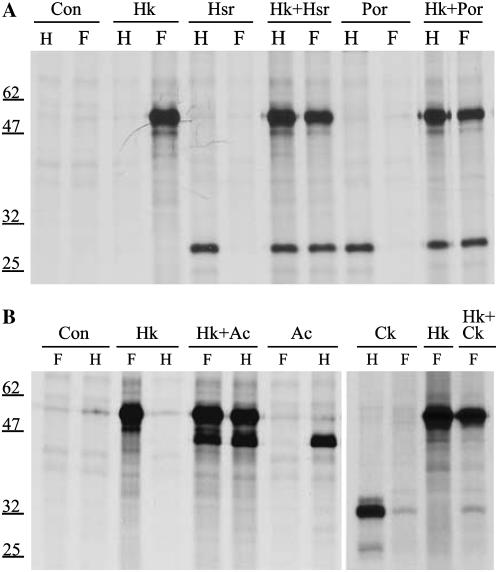
Figure 4.
Interaction assays between HXK1 and mitochondrial porin proteins and between HXK1 and actin proteins after their transient expression in maize leaf protoplasts. A, Protoplasts were transfected with HXK1-FLAG and/or porin-HA, labeled with [35S]Met, then pull-down assays were done using anti-FLAG antibody (F) or anti-HA antibody (H). HK, HXK1; Hsr, VDAC (At5g515090); por, porin (At3g01280). B, Protoplasts were transfected with HXK1-FLAG, ACT2-HA, and/or truncated calcium-dependent protein kinase (Ck)-HA. Comparable pull-down assays were done as in A.
Among the coeluting proteins from the FLAG proteomic experiment, we also identified vegetative actins (ACT2 and ACT8). Notably, ACT2 and ACT8 proteins differ by only one amino acid. This was interesting because certain mammalian HXKs can bind to actin (Murata et al., 1997; Wagner et al., 1999). To test whether the corresponding Arabidopsis proteins can interact, we carried out coimmunoprecipitation assays between HXK1-FLAG and ACT2-HA (Fig. 4B) and ACT8-HA (data not shown). In both cases, positive interaction was observed, with appropriate controls. Because we do not know that actin-HA was incorporated into actin filaments, these data do not discriminate whether HXK1 can bind to monomeric actin (G-actin) or to actin filaments (F-actin). However, when ACT2-GFP was expressed in leaf protoplasts, the fluorescent tag was present only in microfilaments (see below; Fig. 6B). Thus, these data suggest that HXK1 can physically interact with F-actin.
Figure 6.
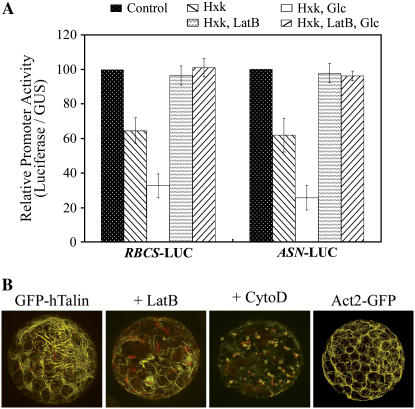
Influence of actin manipulations on Glc signaling in transient expression assays using Arabidopsis leaf protoplasts. A, Influence of 2 μm LatB on repression of pea RBCS-LUC or Arabidopsis ASN1-Luc without effector, +HXK1, and +HXK1 with 2 mm Glc. Error bars are sd values for at least three replications. Comparable results were obtained also using 10 μm CytoD (data not shown). B, Influence of 2 μm LatB or 10 μm CytoD on F-actin structure in protoplasts of GFP-hTalin seedlings. [See online article for color version of this figure.]
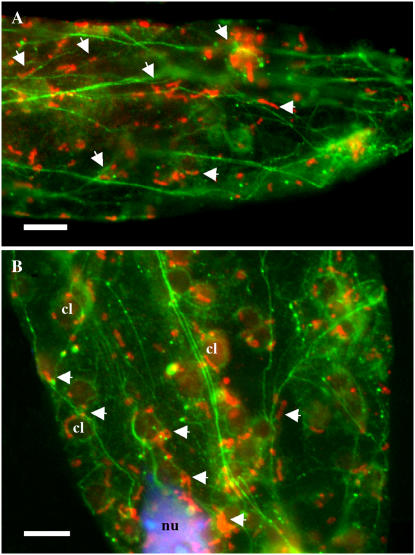
To further clarify whether this interaction can occur in mesophyll cells, we examined the dual immunolocalization of HXK and ACT proteins following cryofixation and freeze substitution of seedling leaf tissue (Fig. 5). As is typical of mesophyll cells, F-actin was observed to form a network of filaments and bundles, including baskets around the chloroplasts and nucleus. However, many of the expected fine mesh filaments are not so readily seen with the epifluorescence microscope. We did observe that numerous fusiform HXK fluorescent structures are associated with or immediately adjacent to actin filaments. In many cases, these fusiform structures were seen along filaments that encircle the chloroplasts and nuclei (Fig. 5B). Thus, most of the visible foci for HXK fluorescence in the single optical sections were associated with F-actin.
Figure 5.
Association of HXK1-localized mitochondria with F-actin. Following cryofixation and freeze substitution, leaf cells of wild-type (Ler) plants were double labeled with anti-HXK1 polyclonal antibody and antiactin monoclonal antibody. Texas Red- and FITC-conjugated secondary antibodies were used to detect HXK1 and actin, respectively. A and B, Fluorescence images of single optical sections of two representative cells are shown. DNA staining with DAPI is shown in blue (B). Arrowheads show the association of some of the HXK1-labeled mitochondria (red) with actin filaments (green) throughout both fields and with the actin basket surrounding the nucleus (nu) and chloroplasts (cl). Chloroplasts in B show slight autofluorescence. Scale bars = 10 μm.
HXK1 and Glc Interactions with F-Actin Might Be Functionally Important
Given our results that HXK1 and actin possibly interact both in vitro and in vivo, we examined whether this putative interaction might have functional significance. First, we tested known reagents that disrupt actin filaments for their possible effects on HXK1-dependent Glc signaling in transient expression assays, using Arabidopsis leaf protoplasts (Fig. 6). In these assays, activities of two different HXK1 reporters were evaluated as a function of expressed luciferase (LUC) activity from promoter-LUC fusion constructs. Activities were normalized for small variances in transfection efficiency by assaying a control promoter construct, UBQ10-GUS. Notably, expression of the control promoter was not affected by cotransfected HXK1, by Glc addition, or by drug treatment (data not shown). In these assay conditions, cotransfection of HXK1 plus treatment with 2 mm Glc resulted in a 3-fold reduction in reporter activities (Fig. 6). Interestingly, the addition of latrunculin-B (LatB) did not affect expression of either promoter in the absence of HXK1 and Glc, but it did completely block effector-dependent repression of both reporter activities. To validate the influence of LatB, we also showed that its treatment extensively disrupted the finer actin filaments of leaf protoplasts from transgenic plants that express the cytoskeleton marker protein, GFP-hTalin (Fig. 6B). Substitution of cytochalasin D (CytoD) for LatB also blocked Glc signaling (data not shown) and even more extensively disrupted the F-actin (Fig. 6B). However, Glc signaling was not affected by treatment with 20 μm oryzalin to disrupt the organization of microtubules (data not shown).
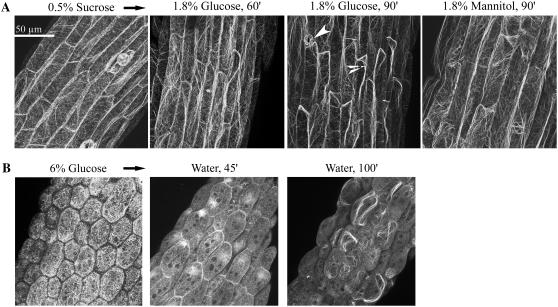
Recognizing that many stimuli are reported to affect F-actin structure (e.g. auxin, Ca, ABA; Dröbak et al., 2004; Gu et al., 2004), we used a GFP-hTalin reporter of F-actin to test both short- and long-term effects of Glc treatment on F-actin organization in seedlings. These seedlings had a normal phenotype when grown on agar plates with 0.5% Suc and had readily visualized actin filament structures. In hypocotyl cells, we observed a few prominent longitudinal fibers located toward the periphery of columnar-shaped cells, with a larger number of transverse and finer mesh filaments (Fig. 7A). Transfer of GFP-hTalin seedlings from Suc plates to a solution of 1.8% (0.1 m) Glc resulted in the rapid loss of many transverse and fine mesh filaments, with increased bundling of the longitudinal F-actin cables (Fig. 7A). This response was specific to Glc and not mannitol. Additionally, seedling transfer instead to 0.18% (10 mm) Glc, but not mannitol, had the same effect on the cytoskeleton after longer treatment time of 10 to 12 h (data not shown). We also tested a transgenic line expressing GFP-Fimbrin ACTIN-BINDING DOMAIN2 (GFP-fABD2; Ketelaar et al., 2004), but in our hands the fine mesh filaments were not so readily visualized (see “Discussion”).
Figure 7.
Glc disrupts F-actin organization as visualized in hypocotyls of GFP-hTalin seedlings. A, Influence of short-term Glc treatment on F-actin organization after seedling transfer to solutions of 1.8% (0.1 m) Glc or mannitol. Prior to treatment, seedlings were grown on agar plates with 0.5% Suc (5 d). Note the loss of many fine filaments after 60 and 90 min of Glc treatment. Arrowheads point to an open stomate and an actin cable. Scale bar = 50 μm. Gain and amplifier offset on the confocal microscope were kept constant throughout the acquisition time courses. Mannitol treatment had no apparent influence on F-actin organization throughout the 90-min treatment time course (earlier images not shown). B, Visualization of actin organization in seedlings developmentally repressed by Glc and the recovery of F-actin after seedling transfer to water. Seedlings were grown on 6% Glc (7 d), then transferred to water for indicated times. These seedlings had arrested growth, with pale white cotyledons. Images are of different seedling hypocotyls. Note the initial absence of fine mesh filaments before transfer and the beginning reorganization of F-actin after transfer to water. Seedlings transferred from 6% mannitol to water showed no apparent changes in their filaments over several hours (observations).
We next tested how Glc treatment might affect F-actin organization after long-term growth at elevated Glc levels. One well-established Glc bioassay is a developmental repression phenotype observed in wild-type seedlings grown on plates with 6% Glc (Jang et al., 1997). In Glc-arrested seedlings, cell morphology was altered as shown by reduced length and increased width of hypocotyl cells (Fig. 7B). Also in these seedlings, the actin cytoskeleton had decreased numbers of fine transverse filaments, with increased numbers of shorter longitudinal fibers that appeared to lack directional organization. These also had increased diffuse fluorescence, perhaps due to some actin molecules not being incorporated into filaments. Staining of hypocotyls with MitoTracker Red indicated that mitochondria still remained associated with the altered actin filaments in repressed seedlings (data not shown). As long as the Glc-repressed seedlings were maintained in solutions of 6% Glc, the actin network underwent no visible structural changes. However, upon transfer of repressed GFP-hTalin seedlings to a solution of water or 0.5% Suc, we observed rapid reorganization of F-actin (Fig. 7B). This included an apparent polar aggregation of actin filaments, followed by formation of longitudinal cables. With additional treatment time, fine mesh filaments also form in transferred seedlings (observations). Growth of GFP-hTalin seedlings on 6% mannitol was normal, as was their F-actin structure before and after transfer to water (observations). These results collectively indicate that Glc treatments have profound and rapid effects on the organization of the actin cytoskeleton. The possible significance of these data to HXK1-dependent Glc signaling will be considered below.
DISCUSSION
In this study, we have shown a number of novel cellular aspects of HXK and Glc functions. First, AtHXK1 can bind to the two most abundant Arabidopsis porin molecules in the outer mitochondrial membrane (Fig. 4; proteomic data not shown). HXK is metabolically active with other glycolytic enzymes located at the mitochondria (Giegé et al., 2003). However, its exclusively noncytosolic localization is distinct because only a subset of other glycolytic enzymes is apparently bound to mitochondria. Plant porin molecules have been suggested to act as scaffolds for organizing a number of mitochondrial-associated proteins, based on their binding of Fru-1,6-bisP aldolase and Suc synthase isoforms (Holtgräwe et al., 2005; Subbaiah et al., 2006). We were not able to confirm those interactions because we did not detect any other glycolytic enzymes or Suc synthase among proteins interacting with AtHXK1-FLAG (data not shown). Our experiments did show that HXK1 is not solubilized following treatment of mitochondria with various adenylates and sugar phosphates (Fig. 3). Similar results were recently shown using maize and rice mitochondrial bound HXKs (Rezende et al., 2006). In that study, even treatment with the antifungal derivative clotrimazole failed to solubilize plant mitochondrial HXKs under conditions in which mammalian mitochondrial HXKs were solubilized. Thus, it is likely that binding of plant versus mammalian HXKs to respective mitochondrial porins occurs by different interactions.
A second novel finding in this study is that AtHXK1 might be an actin-associated protein based on their immunoprecipitation from leaf extracts and from protoplasts after their transient expression and based on their immunolocalization in flash-frozen leaf cells (Figs. 4 and 5). This is not so surprising because a recent two-hybrid study identified several plant glycolytic enzymes as actin-binding proteins (Holtgräwe et al., 2005). These include Fru-1,6-bisP aldolase, glyceraldehyde 3-P dehydrogenase, and enolase. Also, as part of the actin superfamily, HXK shares a common evolutionary origin based on similar tertiary structures that contain the actin fold with the nucleotide-binding pocket (Bork et al., 1992; van den Ent et al., 2001). Actin can self polymerize and we have shown in this study by using GFP fusion proteins that AtHXK1 also has a tendency to self aggregate (Fig. 1). There are reports in mammals of HXK interacting either with actin-binding proteins or with F-actin. Liver HXK4 occurs in a mitochondrial complex with WAVE-1, a multifunctional protein that regulates actin polymerization or that can be a protein kinase A anchoring protein (Danial et al., 2003). Liver glucokinase expressed in COS-7 cells was shown to colocalize with actin filaments (Murata et al., 1997). Also, HXK from bovine heart acts possibly as a capping protein in vitro when present during polymerization of G-actin (Wagner et al., 1999).
A third novel finding in this study is that Glc treatment of seedlings that express a cytoskeletal marker protein caused both short- and long-term effects (Fig. 7). The use of cytoskeleton-GFP marker proteins has come under increased scrutiny due to reporter-specific secondary effects on the organizational state of the cytoskeleton or with regard to particular elements that are labeled. Both transgenic reporter lines that we have used, GFP-hTalin and GFP-fABD2, can have F-actin bundling activity and can alter cytoskeleton dynamics when highly expressed (Ketelaar et al., 2004; Wilsen et al., 2006). Whereas caution is needed in their use to study detailed functions, such as myosin-dependent organelle movement and fine level analyses of F-actin structure in relation to growth mechanisms, these lines are still the best tools available for live cell studies. Furthermore, the GFP-hTalin line used primarily in this study has a modest transgene expression level, with no overt phenotype under normal growth conditions (Takemoto et al., 2003; personal observations). In our hands, the GFP-fABD2 reporter is not as bright as the GFP-hTalin reporter and fine mesh filaments were not as distinctly visualized.
Nonetheless, we have shown that short-term Glc treatment resulted in a loss of fine transverse filaments, with increased bundling into longitudinal cables, as seen in Arabidopsis hypocotyl cells (Fig. 7). This pattern of filament rearrangement is different from that following treatment of etiolated coleoptiles with auxin or light, which also were visualized with a GFP-talin construct (Holweg et al., 2004). Whereas both light and Glc treatments cause a loss of fine mesh filaments, light treatment in the study by Holweg et al. (2004) resulted in the realignment of actin into dispersed longitudinal strands that lack the extensive bundling that we observed following short-term Glc treatment. We further have shown that long-term Glc treatment that causes arrested seedling development resulted in further alterations in F-actin organization. The repressed seedlings lacked many finer filaments as well as peripheral longitudinal strands. This pattern is consistent with the absence of an active growth axis. Furthermore, upon transfer from Glc to water or 0.5% Suc, much of the F-actin rapidly adopted a polarized organization followed by formation of longitudinal strands. The different visualized effects of short-term versus long-term Glc treatments further support the validity of these observations. One remaining question is whether an association of AtHXK1 with actin might facilitate the Glc-dependent rearrangement of F-actin. We hypothesize that this is true because gin2-1 seedlings are Glc tolerant and have largely normal seedling growth on 6% Glc plates (Moore et al., 2003).
A current model of plant HXK signaling is that the sensor protein can translocate to the nucleus and thereby control transcription of target genes (Rolland et al., 2006). These data, however, do not specifically support this model. Even though it is always difficult to exclude that a given protein does not occur at, or move to, an alternate subcellular site, our data indicate that AtHXK1 is continuously bound to mitochondria (Figs. 1–3). One could argue that the relatively large size of the HXK1-GFP fusion protein (>80 kD) might limit its uptake into the nucleus. However, we have shown that this construct does have both catalytic and signaling activities (Supplemental Fig. S1; data not shown). Recently, Cho et al. (2006) used the same HXK1-FLAG transgenic line as we used herein and provided evidence for the presence of HXK1 in Arabidopsis nuclei bound to the CAB2 promoter through interactions with unconventional proteins (VHA-B1 and RPT5B). Using the same polyclonal antibody as we used, those authors estimated that approximately 1% of leaf HXK1 protein is present in nuclei. However, this estimate is inconsistent with our organelle fractionation studies (Fig. 3) for unknown reasons. Cho et al. (2006) did process more than 100-fold greater amounts of leaf tissue than we did, specifically to identify possible nuclear proteins that interact with HXK1. If the reported interactions by Cho et al. (2006) are indeed rare, then our lack of also identifying those two proteins might be explained on that basis or if they remained bound to the separation resin under our very gentle isolation conditions. Nonetheless, our protein immunolocalization following rapid freeze fixation also does not corroborate a nuclear location for any HXKs in Arabidopsis mesophyll cells. Even with this caveat of conflicting data to their model, it is not certain what might be the function of nuclear HXK1. Conceivably, the reported interactions could modulate transcriptional control in a less abundant, perhaps nonphotosynthetic, leaf cell type.
One observation that cannot be readily explained by the model for transcriptional control by nuclear HXK is that the disruption of F-actin by treatment with LatB or CytoD blocked HXK1-dependent signaling in transient expression assays (Fig. 6). The plant actin cytoskeleton forms a diverse system of 200+ proteins that provides structural support to the cell, is modulated by a variety of cellular signals, and transduces certain environmental stimuli (Meagher and Fechheimer, 2003; Dröbak et al., 2004). We envision either of two mechanisms by which actin might have a role in Glc signaling. First, 1-h Glc treatment causes reversible loss and reconfiguration of fine filaments that precedes in time the typical outputs of Glc signaling that are monitored in microarray studies (e.g. Price et al., 2004; Bläsing et al., 2005). These studies generally use a longer Glc pulse of 3 to 4 h at comparable concentrations to produce changes in gene expression. Therefore, monomeric G-actin or an associated protein such as actin-depolymerizing factor might be activated by Glc treatment and have a direct role mediating Glc-dependent transcriptional control of gene expression. In plants, there are a number of actin-related proteins and actin-binding proteins that localize to the nucleus, such as nuclear ARP4 to ARP9 (Kandasamy et al., 2004), profilins (Baluska et al., 2001; Kandasamy et al., 2002b), and actin-depolymerizing factor proteins (Dong et al., 2001). In animals, monomeric actin is part of several chromatin-remodeling complexes (de Lanerolle and Cole, 2002). In human muscles, G-actin acts downstream of a Rho GTPase to repress a coactivator of the serum response transcription factor, which regulates expression of many muscle-specific genes (Posern and Treisman, 2004). In that case, treatment with LatB or CytoD blocks F-actin treadmilling and promotes target gene expression. Our attempts to titrate LatB to possibly attenuate the repression of seedling development on 6% Glc plates, however, were not successful (X. Xia and B. Moore, unpublished data).
A second possible mechanism for the role of actin in supporting Glc signaling could involve targeted modulation of translational control. The association of some cellular polysomes and other translational components with the cytoskeleton has been recognized for a number of years in plants as well as animals (Davies et al., 1998). For example, the loss of fine mesh actin filaments in potato (Solanum tuberosum) tubers is associated with the loss of wound-induced message translation activity under conditions of low O2 (Morelli et al., 1998). Researchers have been able to isolate four defined populations of polysomes: free, membrane bound, actin bound, and actin membrane bound polysomes. In one study, ABA treatment of germinating embryos from triticale caryopses is reported to greatly increase the abundance of the cytoskeleton-associated polysomes (Weidner et al., 2000). In this study, disruption of F-actin by LatB or CytoD did not affect expression of RBCS-LUC or ASN1-LUC in the absence of HXK1 and Glc, but only blocked the HXK1 plus Glc-dependent repression of LUC expression (Fig. 6). It is possible that HXK-dependent repression of LUC protein expression in this case might be due to blocking translation rather than transcription of the introduced promoter construct. By this model, Glc treatment might increase the level of actin-associated polysomes and therefore shift the translation of LUC transcript to a LatB-sensitive population.
Possible functional interactions between mitochondria, HXK, and actin are intriguing to consider. Because mitochondria traffic on F-actin (Van Gestel et al., 2002), they can be viewed as linking two major processes that consume ATP, cytoplasmic streaming, and Glc metabolism. The binding of mitochondria to F-actin is likely mediated by a number of proteins, including myosin motors and microfilament-binding proteins. Because HXK1 overexpression can lead to mitochondrial clumping (Fig. 1), HXK might affect the trafficking of mitochondria on specific microfilaments. Furthermore, Glc-dependent rearrangement of F-actin could prove important to the distribution of mitochondria among dynamic cellular populations that vary in their mobility (Van Gestel et al., 2002). Mitochondria could have even direct effects on Glc signaling. For example, in skeletal muscle, the transcription factors MondoA and Mlx shuttle between the mitochondria and nucleus (Sans et al., 2006). Nuclear MondoA-Mlx heterodimers bind to carbohydrate response elements in promoters of several glycolytic and lipogenic genes. However, we are not aware of any plant transcription factors that shuttle from mitochondria to the nucleus.
CONCLUSION
In summary, results of this study indicate that a major component of HXK-dependent Glc signaling might require a previously unrecognized interaction with the actin cytoskeleton. This observation provides a novel gateway to understand an important interface between Glc signaling and plant growth. Rop GTPases often mediate signaling events that modulate actin organization and affect certain types of plant growth (Gu et al., 2004). Whether any of the Rop GTPases have a role in Glc signaling remains to be investigated.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana L. Heynh.) ecotype Columbia-0 and ecotype Landsberg erecta (Ler) mutants were obtained from the Arabidopsis Biological Resource Center; gin2-1, and transgenic gin2-1, which express HXK1-FLAG, were as previously described (Moore et al., 2003); seeds of transgenic Arabidopsis expressing hTalin-GFP were generously provided by Dr. Adrienne Hardham (Australian National University); seeds of Arabidopsis that express GFP-fABD2 were a gift from Dr. David McCurdy (University of Newcastle); and seeds of maize (Zea mays; Seed Genetics) and pea (Pisum sativum var. Little Marvel; Gurneys Seed and Nursery Co.) were purchased. Plants were chamber grown with a 12-h photoperiod at 125 μmol m−2 s−1 and a 23°C day/20°C night thermoperiod. Seeds were germinated and grown on agar plates after 2 d at 4°C as in Jang et al. (1997). Greening maize seedlings (Jang and Sheen, 1994) were used as a source of protoplasts for protein expression and immunoprecipitation.
Plasmid and Transgenic Constructs
Two Glc-responsive promoters driving firefly LUC, pea RBCS-LUC (Schäffner and Sheen, 1991), and Arabidopsis ASN1-LUC (Dr. Li Zhou; see also Price et al., 2004), and an internal control reporter, UBQ10-GUS, were cloned in the plant expression vector previously described (Kovtun et al., 1998). ACT2 cDNA (At3g18780) was amplified from a previously described plasmid (Kandasamy et al., 2002a) and subcloned as a C-terminal GFP fusion (Chiu et al., 1996) in the plant expression vector. Porin genes At3g01280 (por301) and At5g15090 (hsr2) were amplified from cDNA stocks U12693 and U18357 (Arabidopsis Biological Resource Center), respectively, using the following primers: por301, 5′-GGGATCCCATGGTGAAAGGTCCCGGT and 5′-GAAGGCCTAGGCTTGAGTGCGAGAGCCAATCC; and hsr2, 5′-GTCAAGGCCTGGGCTTGAGAGCGAGAGCAATC and 5′-GGGATCCATGGTTAAAGGTCCAGGACTCTACACT. These were subcloned as C-terminal GFP and double HA constructs in the expression vector. HXK1-HA and HXK1-FLAG were as described previously (Moore et al., 2003) and HXK was subcloned as a C-terminal GFP fusion. HXK1ΔC (amino acids 1–27) was made by inserting a StuI site after amino acid 27 of HXK1-GFP, using Quick Change (Stratagene) and the following primers: 5′-GGTTGTTCGACGACGGAGGCCTATGCAGAGCTCAGGG and 5′-CCCTGAGCTCTGCATAGGCCTCCGTCGTCGAACAACC. HXK1ΔN (amino acids 28–496) was made by deletion using Quick Change (Stratagene) and the following primers: 5′-CCTTGCTCCGTGGATCCCGATGCAGAGCTCAGGGAAG and 5′-CTTCCCTGAGCTCTGCATCGGGATCCACGGAGCAAGG. HXK1-GFP was subcloned into the minibinary vector pCB302 (Xiang et al., 1999) with the bar selection marker and plants of Arabidopsis Ler were transformed by Agrobacterium-mediated floral dip (Clough and Bent, 1998). Homozygous lines with a single insert were selected for characterization. All PCR-based cloning was confirmed by DNA sequencing.
Protoplast Transient Expression Assays
Arabidopsis leaf protoplasts were isolated as described previously and transfected using polyethylene glycol (Hwang and Sheen, 2001). Transfection efficiencies were >60%. Protoplasts were transfected with RBCS-LUC (4 μg) or ASN1-LUC (6 μg) ± HXK1-HA (6 μg) as an effector + UBQ10-GUS (2 μg) as an internal control, and empty vector to make equal concentrations of cDNA. Following transfection, protoplasts were incubated in the light at 35 mmol m−2 s−1, then treated with ±2 mm Glc after 90 min, and harvested after 6 to 8 h. The protoplast pellet was resuspended in lysis buffer and GUS and LUC activity was measured as previously (Jefferson, 1987; Sheen, 1996). Promoter activity is expressed as LUC/GUS values and normalized to untreated controls.
Coimmunoprecipitation of 35S-Labeled Proteins
[35S]Met (25 μCi; Perkin-Elmer) was added to maize protoplasts 90 min after polyethylene glycol transfection. Protoplasts were harvested after 10 h and resuspended in 200 μL immunoprecipitation buffer (150 mm NaCl, 50 mm Tris chloride, pH 7.5, 5 mm EDTA, 1% [v/v] Triton X-100, 1 mm dithiothreitol, 1 mm NaF, 1 mm NaVO3, complete protease inhibitors [Roche Molecular Biochemicals], and 0.075% [v/v] SDS). Anti-HA (Roche) or anti-FLAGM2 (Sigma-Aldrich) antibodies were used with protein A agarose (Roche) for immunoprecipitation. Proteins were solubilized, electrophoresed on 10% SDS gels, and visualized by fluorography.
Organelle Isolation
Organelles from leaves of 10-d-old pea or 4-week-old Arabidopsis were isolated following low-speed blending. Mitochondria were isolated on Percoll gradients as described in Millar et al. (2001). Chloroplasts (Fitzpatrick and Keegstra, 2001) and nuclei (Watson and Thompson, 1986) were isolated from the initial low-speed pellet. Fraction purities were initially monitored by fluorescence microscopy (see below). For western blots, organelle proteins were electrophoresed by SDS-PAGE, transferred to Immobilon-P (Millipore), and probed with antibodies: rabbit polyclonal anti-HXK1 (Zhou et al., 1998), polyclonal anti-RuBP carboxylase/oxygenase (Dr. Rowan Sage), mouse monoclonal anti-PDH (Dr. Jan Miernyk), polyclonal antihistone (Talbert et al., 2002), and polyclonal phosphoenolpyruvate carboxylase (Dr. Ray Chollet).
Fluorescence Microscopy
Protoplasts and seedlings were monitored using an Axiovert 200 m fluorescence microscope with Apotome (Carl Zeiss) and band-pass filters (Chroma), with most reported images acquired using an LSM 510 confocal laser-scanning microscope (Carl Zeiss). Mitochondria were visualized by staining with MitoTracker Red (Molecular Probes) and nuclei by staining with Hoechst 33342 (Molecular Probes). During organelle purification, fractions were routinely visualized for cross contamination, including also chlorophyll fluorescence for chloroplasts. For visualizing actin disruption in protoplasts, samples were treated 30 min with 2 μm LatB, 10 μm CytoD, or 20 μm oryzalin (Molecular Probes). To visualize actin disruption in vivo, transgenic plants that express GFP-hTalin were grown on agar plates with 0.5% Suc medium for 7 d, then transferred to deep well slides with either 1.8% (0.1 m) Glc, 1.8% mannitol, or 0.5% Suc. To visualize actin organization in Glc-repressed seedlings, transgenic plants of either GFP-hTalin or fABD2-GFP were grown on 6% Glc plates for 6 d. In one experiment, seedlings were then placed in water or 0.5% Suc to visualize F-actin reorganization.
Cellular Immunolocalization
Immunolocalization of HXK1 and F-actin was done using methods previously described (Baskin et al., 1996; Kandasamy et al., 2002a), following leaf cryofixation and acetone freeze substitution of young seedlings. Because anti-HXK1 is a polyclonal antibody and anti-ACT (MAbGPa) is a monoclonal antibody, we labeled cells with corresponding secondary antibodies conjugated to FITC. Double labeling of actin and HXK1 was done by incubating the cells in a mixture of anti-actin and anti-HXK1 primary antibodies for 12 h and the FITC-conjugated anti-mouse and Texas Red-conjugated anti-rabbit secondary antibodies for 4 h. DNA was stained with 4′,6-diamino-phenylindole (DAPI; Sigma-Aldrich) and 0.1 μg/mL phosphate-buffered saline. For control, mitochondria were labeled with anti-PDH monoclonal antibody. Cells were visualized with a fluorescence microscope (Carl Zeiss) or a Leica scanning confocal microscope (TCS-SP2).
HXK1-FLAG Protein Isolation and Analysis
Leaves of greenhouse-grown plants of line HXK1-FLAG29 were collected into liquid N2. About 0.35 g fresh weight were extracted using a mortar and pestle with 5 volumes of buffer (100 mm Tris-HCl, pH 7.8, 10% glycerol, 5 mm MgCl2, 15 mm KCl, 1 mm EDTA, 2 mm dithiothreitol, 0.5% CHAPS, and 1× protease inhibitors [Roche]). Extracts were microfuged at 12,000g for 10 min at 4°C. The supernatant was kept on ice, whereas the pellet was reextracted with 0.5 mL of grinding buffer plus 0.31 m KCl by flopping for 30 min at 4°C. Following microfuging, both extracts were assayed for glucokinase activity (Doehlert, 1989) and protein content (Bio-Rad Laboratories). Extracts were combined to give 1 to 1.2 mL with 12 to 15 units of activity at 0.15 m KCl, then incubated with 4 μg of anti-FLAG M2 antibody (Sigma-Aldrich) for 60 min on a flopper at room temperature.
Extracts were transferred to washed anti-mouse magnetic beads (MagnaBind Beads; Pierce) and flopped for 60 min at room temperature. Beads were separated and washed three times with buffer (50 mm Tris-HCl, pH 7.8, 150 mm KCl, 10% glycerol), then incubated for 30 min at room temperature with FLAG peptide elution medium (50 mm Tris-HCl, pH 7.8, 150 mm KCl, 5% glycerol, 2% CHAPS, 125 μg/mL FLAG peptide). FLAG peptide (Massachusetts General Hospital Core Facility) was quantified by A274 (ɛ = 1,420 m−1 cm−1). The eluted fraction generally had 10% to 15% of initial glucokinase activity. Protein was concentrated for electrophoresis using the Pierce PAGE prep kit (catalog no. 26800). Protein samples were electrophoresed on 10% SDS-PAGE gels. Gels were stained using the Colloidal Blue staining kit (Invitrogen), then the gel lane was excised into four sections and submitted for liquid chromatography-tandem mass spectrometry analysis (Harvard Microchemistry and Proteomics Analysis Facility).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_119057, NM_112764, NM_110994, and NM_121513.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Glc signaling activity of HXK1-GFP in protoplast transient expression assays.
Supplementary Material
Acknowledgments
We are very grateful to the late Dr. Shu-Hua Cheng for procedural advice and useful comments on the manuscript and also to Dr. Jen Sheen for sharing expression vectors and DNA constructs.
This work was supported by the U.S. Department of Agriculture-National Research Initiative Plant Biochemistry Program (grant no. 2001–035318) and the South Carolina Agricultural Experiment Station (technical contribution no. 5272 of the Clemson University Experiment Station). This material is based upon work supported by the Cooperative State Research, Education and Extension Service/U.S. Department of Agriculture (project no. SC–170090).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Brandon d. Moore (moore8@clemson.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baluska F, von Witsch M, Peters M, Hlavacka A, Volkmann D (2001) Mastoparan alters subcellular distribution of profiling and remodels F-actin cytoskeleton in cells of maize root apices. Plant Cell Physiol 42 912–922 [DOI] [PubMed] [Google Scholar]
- Baskin T, Miller DD, Vos JW, Wilson JE, Hepler PK (1996) Cryofixing single cells and multicellular specimens enhances structure and immunocytochemistry for light microscopy. J Microsc 182 149–161 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 89 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6 325–330 [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127 579–589 [DOI] [PubMed] [Google Scholar]
- Claeyssen E, Rivoal J (2007) Isozymes of plant hexokinase: occurrence, properties and functions. Phytochemistry 68 709–731 [DOI] [PubMed] [Google Scholar]
- Clausen C, Ilkavets I, Thomson R, Philippar K, Vojta A, Mohlmann T, Neuhaus E, Fulgosi H, Soll J (2004) Intracellular localization of VDAC proteins in plants. Planta 220 30–37 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- da-Silva WS, Rezende GL, Galina A (2001) Subcellular distribution and kinetic properties of cytosolic and non-cytosolic hexokinases in maize seedling roots: implications for hexose phosphorylation. J Exp Bot 52 1191–1201 [PubMed] [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang C-Y, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, et al (2003) BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424 952–956 [DOI] [PubMed] [Google Scholar]
- Damari-Weissler H, Ginzburg A, Gidoni D, Mett A, Krassovskaya I, Weber APM, Belausov E, Granot D (2007) Spinach SoHXK1 is a mitochondria-associated hexokinase. Planta 226 1053–1058 [DOI] [PubMed] [Google Scholar]
- Damari-Weissler H, Kandel-Kfir M, Gidoni D, Mett A, Belausov E, Granot D (2006) Evidence for intracellular spatial separation of hexokinases and fructokinases in tomato plants. Planta 224 1495–1502 [DOI] [PubMed] [Google Scholar]
- Davies E, Abe S, Larkins BA, Clore AM, Quatrano RS, Weidner S (1998) The role of the cytoskeleton in plant protein synthesis. In J Bailey-Serres, DR Gallie, eds, A Look Beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Biologists, Rockville, MD, pp 115–124
- de Lanerolle P, Cole AB (2002) Cytoskeletal proteins and gene regulation: form, function, and signal transduction in the nucleus. Sci STKE 139: PE30 [DOI] [PubMed]
- Doehlert DC (1989) Separation and characterization of four hexose kinases from developing maize kernels. Plant Physiol 89 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C-H, Kost B, Xia G, Chua N-H (2001) ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Mol Biol 45 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröbak BK, Franklin-Tong VE, Staiger CJ (2004) The role of the actin cytoskeleton in plant cell signaling. New Phytol 163 13–30 [DOI] [PubMed] [Google Scholar]
- Dry IB, Nash D, Wiskich JT (1983) The mitochondrial localization of hexokinase in pea leaves. Planta 158 152–156 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick LM, Keegstra K (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J 27 59–65 [DOI] [PubMed] [Google Scholar]
- Giegé P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ (2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese J-O, Herbers K, Hoffmann M, Klösgen RB, Sonnewald U (2005) Isolation and functional characterization of a novel plastidic hexokinase from Nicotiana tabacum. FEBS Lett 579 827–831 [DOI] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7 527–536 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Fillippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SL, Chao YC, Tong WF, Yu SM (2001) Sugar coordinately and differentially regulates growth- and stress-related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol 125 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtgräwe D, Scholz A, Altmann B, Scheibe R (2005) Cytoskeleton-associated, carbohydrate-metabolizing enzymes in maize identified by yeast two-hybrid screening. Physiol Plant 125 141–156 [Google Scholar]
- Holweg C, Sublin C, Nick P (2004) Arabidopsis myosin XI mutant is defective in organelle movement and polar auxin transport. Plant Cell Physiol 45 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413 383–389 [DOI] [PubMed] [Google Scholar]
- Jang JC, León P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Sheen J (1994) Sugar sensing in higher plants. Plant Cell 6 1665–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5 387–405 [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB (2004) Plant actin-related proteins. Trends Plant Sci 9 196–202 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2002. a) Functional nonequivalency of actin isovariants in Arabidopsis. Mol Biol Cell 13 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2002. b) Plant profiling isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil Cytoskeleton 52 22–32 [DOI] [PubMed] [Google Scholar]
- Kandel-Kfir M, Damari-Weissler H, German MA, Gidoni D, Mett A, Belausov E, Petreikov M, Adir N, Granot D (2006) Two newly identified membrane-associated and plastidic tomato HXKs: characteristics, predicted structure and intracellular localization. Planta 224 1341–1352 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Anthony RG, Hussey PJ (2004) Green fluorescent protein-mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor's actin depolymerizing activity in vitro. Plant Physiol 136 3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lim JH, Ahn CS, Park K, Kim GT, Kim WT, Pai HS (2006) Mitochondria-associated hexokinases lay a role in the control of programmed cell death in Nicotiana benthamaiana. Plant Cell 18 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu W-L, Zeng W, Sheen J (1998) Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395 716–720 [DOI] [PubMed] [Google Scholar]
- Meagher RB, Fechheimer M (2003) The Arabidopsis cytoskeletal genome. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–26 [DOI] [PMC free article] [PubMed]
- Miernyk JA, Dennis DT (1983) Mitochondrial, plastid and cytosolic isozymes of hexokinase from developing endosperm of Ricinus communis. Arch Biochem Biophys 226 458–468 [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Lever CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336 [DOI] [PubMed] [Google Scholar]
- Morelli JK, Zhou W, Yu J, Lu C, Vayda ME (1998) Actin depolymerization affects stress-induced translational activity of potato tuber tissue. Plant Physiol 116 1227–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Katagiri H, Ishihara H, Shibasaki Y, Asano T, Toyoda Y, Pekiner B, Pekiner C, Miwa I, Oka Y (1997) Co-localization of glucokinase with actin filaments. FEBS Lett 406 109–113 [DOI] [PubMed] [Google Scholar]
- Olsson T, Thelander M, Ronne H (2003) A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens. J Biol Chem 278 4439–4447 [DOI] [PubMed] [Google Scholar]
- Posern G, Treisman R (2004) Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 16 588–596 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende GL, Logullo C, Meyer L, Machado LB, Oliveira-Carvalho AL, Zingali RB, Cifuentes D, Galina A (2006) Partial purification of tightly bound mitochondrial hexokinase from maize (Zea mays L.) root membranes. Braz J Med Biol Res 39 1159–1169 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JMS, Murray JAH (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A, De La Cera T, Herrero P, Moreno F (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 355 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14 S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE (2006) MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol 26 4863–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffner AR, Sheen J (1991) Maize rbcS promoter activity depends on sequence elements not found in dicot rbcS promoters. Plant Cell 3 997–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnarrenberger C (1990) Characterization and compartmentation, in green leaves, of hexokinases with different specificities for glucose, fructose, and mannose and for nucleoside triphosphates. Planta 181 249–255 [DOI] [PubMed] [Google Scholar]
- Sheen J (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274 1900–1902 [DOI] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2 410–418 [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Palaniappan A, Duncan K, Rhoads DM, Huber SC, Sachs MM (2006) Mitochondrial localization and putative signaling function of sucrose synthase in maize. J Biol Chem 281 15625–15635 [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR (2003) GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J 33 775–792 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Maseulli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towle HC (2005) Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab 16 489–494 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Löwe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413 39–44 [DOI] [PubMed] [Google Scholar]
- Van Gestel K, Köhler RH, Verbelen J-P (2002) Plant mitochondria move on F-actin, but their positioning in the cortical cytoplasm depends on both F-actin and microtubules. J Exp Bot 53 659–667 [DOI] [PubMed] [Google Scholar]
- Villadsen D, Smith SM (2004) Identification of more than 200 glucose-responsive Arabidopsis genes none of which responds to 3-O-methylglucose or 6-deoxyglucose. Plant Mol Biol 55 467–477 [DOI] [PubMed] [Google Scholar]
- Wagner O, Zinke J, Dancker P, Grill W, Bereiter-Hahn J (1999) Viscoelastic properties of f-actin, microtubules, f-actin/α-actinin, and f-actin/hexokinase determined in microliter volumes with a novel nondestructive method. Biophys J 76 2784–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM (2006) A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol Biol Cell 17 4257–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JC, Thompson WF (1986) Purification and restriction endonuclease analysis of plant nuclear DNA. Methods Enzymol 118 57–75 [Google Scholar]
- Weidner S, Lukaszewicz D, Amarowicz R (2000) Significant role for polysomes associated with the cytoskeleton in the control of protein synthesis during germination of triticale caryopses in the presence of abscisic acid. Acta Physiol Plant 22 185–193 [Google Scholar]
- Wiese A, Gröner F, Sonnewald U, Deppner H, Lerchl J, Hebbeker U, Flügge UI, Weber A (1999) Spinach hexokinase 1 is located in the outer envelope membrane of plastids. FEBS Lett 461 13–18 [DOI] [PubMed] [Google Scholar]
- Wilsen KL, Lovy-Wheeler A, Voigt B, Menzel D, Kunkel JG, Hepler PK (2006) Imaging the actin cytoskeleton in growing pollen tubes. Sex Plant Reprod 19 51–62 [Google Scholar]
- Wilson JE (1997) Homologous and heterologous interactions between hexokinase and mitochondrial porin: evolutionary implications. J Bioenerg Biomembr 29 97–102 [DOI] [PubMed] [Google Scholar]
- Wilson JE (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206 2049–2057 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44 451–461 [DOI] [PubMed] [Google Scholar]
- Yanasigawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature 425 521–525 [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer EC, Medrano LJ, Meyerowitz EM (2000) Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development. Proc Natl Acad Sci USA 97 7633–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.