Abstract
Of the four genes of the Arabidopsis (Arabidopsis thaliana) INOSITOL TRANSPORTER family (AtINT family) so far only AtINT4 has been described. Here we present the characterization of AtINT2 and AtINT3. cDNA sequencing revealed that the AtINT3 gene is incorrectly spliced and encodes a truncated protein of only 182 amino acids with four transmembrane helices. In contrast, AtINT2 codes for a functional transporter. AtINT2 localization in the plasma membrane was demonstrated by transient expression of an AtINT2-GREEN FLUORESCENT PROTEIN fusion in Arabidopsis and tobacco (Nicotiana tabacum) epidermis cells and in Arabidopsis protoplasts. Its functional and kinetic properties were determined by expression in yeast (Saccharomyces cerevisiae) cells and Xenopus laevis oocytes. Expression of AtINT2 in a Δitr1 (inositol uptake)/Δino1 (inositol biosynthesis) double mutant of bakers' yeast complemented the deficiency of this mutant to grow on low concentrations of myoinositol. In oocytes, AtINT2 mediated the symport of H+ and several inositol epimers, such as myoinositol, scylloinositol, d-chiroinositol, and mucoinositol. The preference for individual epimers differed from that found for AtINT4. Moreover, AtINT2 has a lower affinity for myoinositol (Km = 0.7–1.0 mm) than AtINT4 (Km = 0.24 mm), and the Km is slightly voltage dependent, which was not observed for AtINT4. Organ and tissue specificity of AtINT2 expression was analyzed in AtINT2 promoter/reporter gene plants and showed weak expression in the anther tapetum, the vasculature, and the leaf mesophyll. A T-DNA insertion line (Atint2.1) and an Atint2.1/Atint4.2 double mutant were analyzed under different growth conditions. The physiological roles of AtINT2 are discussed.
Myoinositol and its derivatives are central to numerous metabolic pathways under different physiological conditions. Myoinositol is a precursor in the biosynthesis of UDP-GlcUA, GalUA, Xyl, apiose, and Ara (Loewus and Murthy, 2000; Kanter et al., 2005), it is used for the formation of galactinol, a myoinositol-linked, activated form of Gal that is used for the biosynthesis of raffinose and its derivatives (Kandler and Hopf, 1982), and it is thought to be an initial substrate in the biosynthesis of l-ascorbic acid (Lorence et al., 2004). It may be conjugated to auxins to prevent their biological activity and to allow long-distance transport (Cohen and Bandurski, 1982), and in phospholipids (Lehle, 1990) or glycosylphosphatidylinositol-membrane anchors (Schultz et al., 1998) myoinositol provides the structural basis for membranes and membrane-attached proteins. Inositol-1,4,5-triphosphate plays an important role as second messenger, and myoinositol-1,2,3,4,5,6-hexakisphosphate (phytate) is used for the storage of inositol, phosphorus, and minerals (Shi et al., 2005), or as a structural component of proteins (Macbeth et al., 2005; Tan et al., 2007). Finally, myoinositol may represent a cellular energy currency (Raboy, 2003).
Living cells possess different inositol epimers, the most frequent ones being scylloinositol and d- or l-chiroinositol, and all of these epimers are found in phosphatidyl inositols (Narasimhan et al., 1997). Moreover, monomethylated inositols are found in all plants but are especially common in legumes. In leaves of unstressed soybean (Glycine max), for example, concentrations of pinitol (3-O-methyl-chiroinositol) can be 2-fold higher than the combined concentrations of Suc, all monosaccharides, and myoinositol (Smith and Phillips, 1982; Streeter et al., 2001). This already high concentration is further increased in drought-stressed plants, underlining the important role of unmethylated and methylated inositols as osmoprotectants (Thomas and Bohnert, 1993; Sheveleva et al., 1997; Nelson et al., 1998; Hasegawa et al., 2000; Streeter et al., 2001; Murakeözy et al., 2003).
The first cDNAs of putative plant myoinositol transporters, MITR1 and MITR2, were cloned from ice plants (Mesembryanthemum crystallinum), and MITR1-expressing yeast (Saccharomyces cerevisiae) cells were able to grow on lower extracellular concentrations of myoinositol than control cells (Chauhan et al., 2000). Only recently, Schneider et al. (2006) published a detailed functional characterization of the Arabidopsis (Arabidopsis thaliana) myoinositol transporter AtINT4 (At4g16480). This protein was characterized as a high-affinity, energy-dependent, plasma membrane-localized H+/inositol symporter.
Here we present the molecular cloning of an AtINT2 cDNA (At1g30220), analyses of the tissue-specific expression of the AtINT2 gene, and the functional characterization and subcellular localization of the AtINT2 protein. Like AtINT4, AtINT2 is a plasma membrane-localized H+/inositol symporter. Expression of an AtINT2 cDNA in yeast and in Xenopus oocytes, however, revealed significant differences between the kinetic properties and substrate specificities of these transporters. Moreover, analyses of AtINT2 promoter/GUS and AtINT2 promoter/GFP reporter lines showed an expression pattern that differed from that of AtINT4. Finally, Atint2 mutant lines were studied. The putative physiological roles of AtINT2 are discussed.
RESULTS
Cloning of AtINT2 and AtINT3 cDNAs
The AtINT3 gene (At2g35740) is predicted to encode a protein of 580 amino acids (NP_181117) that is similar to AtINT2 and AtINT4 with 60.4% and 63.4% identical amino acids, respectively. However, we did not succeed in amplifying an AtINT3 cDNA by reverse transcription (RT)-PCR. To overcome this problem we generated AtINT3 overexpressing plant material by infiltration of tobacco (Nicotiana tabacum) leaves with an Agrobacterium tumefaciens strain carrying the plasmid pPU6. The same strain was used for stable transformation of Arabidopsis. pPU6 drives expression of a genomic AtINT3 sequence (from start to stop including two predicted introns) under the control of the 35S promoter. RT-PCR reactions performed with total RNA from infiltrated tobacco leaves or transformed Arabidopsis plants were successful. Unexpectedly, in all cDNAs analyzed the first predicted AtINT3 intron was spliced incorrectly, i.e. 95 bp from the 3′ end of the first predicted exon were removed together with the first intron, and in none of the analyzed sequences the second intron was spliced out.
The obtained AtINT3 cDNAs were 1,742 bp long and carried a stop codon after the first 546 bp. This open reading frame (ORF) encodes a truncated protein with 182 amino acids. Only the first 148 amino acids (covering four of the predicted 12 transmembrane helices of AtINT3) corresponded to the predicted AtINT3 protein sequence. Amino acids 149 to 182 resulted from a frameshift due to the incorrect splicing. The obtained cDNA sequence was deposited in the EMBL database (accession no. AM778029).
Obviously, neither in tobacco nor in Arabidopsis the genomic AtINT3 sequence is spliced as predicted, indicating that AtINT3 is most likely not a functional gene. Therefore, additional expression analyses in yeast and studies of the activity of the AtINT3 promoter were not performed.
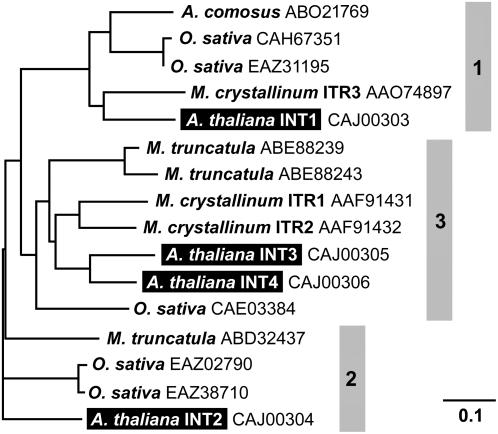
The AtINT2 ORF (At1g30220) encodes a protein of 580 amino acids, with two consensus sequences for N-glycosylation (Asn310 and Asn382) and 12 predicted transmembrane helices (Schneider et al., 2006). Figure 1 presents a phylogenetic tree that is based on the experimentally confirmed protein sequences of AtINT1, AtINT2, and AtINT4 (Schneider et al., 2006), on the predicted sequence of AtINT3, on three confirmed or predicted inositol transporter sequences from ice plant (MITR1, MITR2, and MITR3; Chauhan et al., 2000), and on nine homologous sequences found in publicly accessible data libraries (five from rice [Oryza sativa], three from Medicago truncatula, and one from pineapple [Ananas comosus]). The tree falls into three separate clades (1–3 in Fig. 1) and members of the Arabidopsis family and rice sequences are found in each clade. Interestingly, all members of clades 2 and 3 possess an enlarged loop between the predicted transmembrane helices IX and X with two conserved Cys-X-X-Cys motifs. Neither the enlarged loop nor the conserved motifs are found in any of the proteins of clade 1.
Figure 1.
Phylogenetic tree of the AtINT proteins from Arabidopsis and of related proteins from other plant species. Predicted (AtINT3) and confirmed (AtINT1, AtINT2, AtINT4) amino acid sequences of the four Arabidopsis AtINTs plus sequences of 12 homologous proteins from pineapple, rice, M. truncatula, and ice plant were aligned (ClustalX; Thompson et al., 1997) and a tree was calculated (TreeViewX; Page, 1996). Protein names (if available) and accession numbers are given.
Expression of the AtINT2 cDNA in Yeast
In a first approach we tried to determine the functional properties of AtINT2 in the bakers' yeast mutant D458-1B (Nikawa et al., 1991). This strain carries mutations in the ITR1 (inositol transporter) and INO1 (inositol-1-P synthase) genes and can grow only on high extracellular concentrations of myoinositol. To this end, we cloned the AtINT2 cDNA into the unique NotI site of the yeast expression vector NEV-N-Leu. The resulting plasmids harbored the AtINT2 cDNA in sense (pSS51s) or antisense orientation (pSS51as).
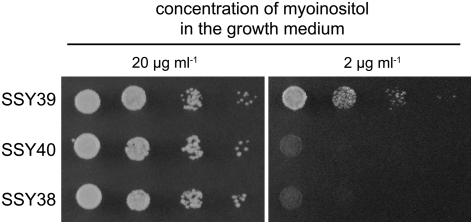
The transformed yeast lines SSY38 (carrying the empty NEV-N-Leu vector), SSY39 (carrying the AtINT2 sense plasmid pSS51s), and SSY40 (antisense AtINT2) were grown on petri plates containing low (2 μg/mL) or high concentrations (20 μg/mL) of myoinositol to visualize a possible complementation of the described mutations. Figure 2 shows that SSY39 cells had, in fact, regained the capacity to grow on low myoinositol. As expected, no growth was seen for the control strains SSY38 and SSY40 on the same medium. After this initial evidence that AtINT2 might encode a myoinositol transporter, we tried to characterize the functional and kinetic properties in detail by transport analyses with 3H-labeled myoinositol in SSY39 cells. However, the uptake rates were too low to yield statistically solid data (data not shown). This suggested that the amount of AtINT2 protein in yeast plasma membranes was sufficient for complementation, but not high enough to allow direct measurements of radiolabeled substrates.
Figure 2.
Complementation of the growth defect of D458-1B. Growth of yeast strains SSY39 (=D458-1B with AtINT2 cDNA in sense), SSY40 (=D458-1B with AtINT2 cDNA in antisense), and SSY38 (=D458-1B with the empty vector) was analyzed on petri plates (minimal medium) supplemented with the indicated concentrations of myoinositol. While all three strains were able to grow on high myoinositol (20 μg mL−1), only the AtINT2-expressing strain was able to grow on low myoinositol (2 μg mL−1).
Expression of the AtINT2 cDNA in Xenopus laevis Oocytes
Xenopus laevis oocytes have been used for the successful expression of AtINT4 (Schneider et al., 2006) and for analyses of several other electrogenic plant plasma membrane transporters (Aoshima et al., 1993; Boorer et al., 1996; Klepek et al., 2005; Reinders et al., 2005). As the yeast complementation data suggested plasma membrane localization also for AtINT2, we decided to analyze AtINT2 in this expression system. To this end, the AtINT2 cDNA was cloned into pDK148 (Jespersen et al., 2002) yielding pSS52s. cRNA was transcribed from the T7 promoter of pSS52s and injected into oocytes that were analyzed for inward currents resulting from the cotransport of cations with potential substrates.
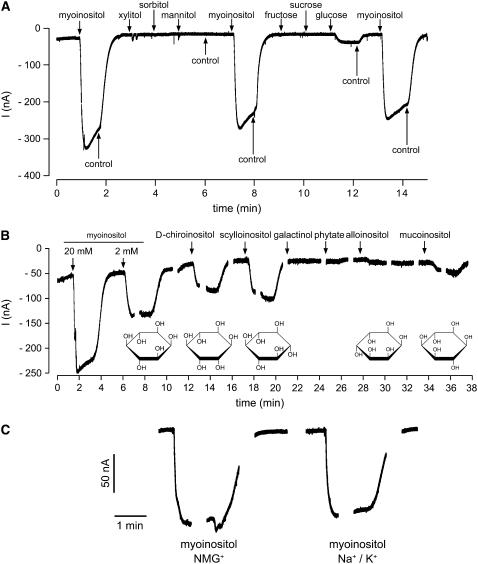
Figure 3A shows a typical recording of inward currents in the presence of various candidate substrates. Analyses were performed with 20-mm solutions of myoinositol, xylitol, mannitol, sorbitol, Glc, Fru, and Suc at an extracellular pH of 5.5. Only application of myoinositol resulted in strong inward currents. Much weaker currents (<10% of myoinositol) were elicited by Glc; no currents were obtained with any of the other compounds (quantitative analyses are shown in Fig. 4A). This demonstrated that, as predicted from the analyses in yeast, AtINT2 does mediate the uptake of myoinositol. Furthermore, the obtained currents confirmed that AtINT2-driven transport is energy dependent, they suggested that a positive charge is likely to be symported with myoinositol, and they demonstrated that AtINT2 is quite specific and does not or hardly transport linear polyols, mono-, or disaccharides.
Figure 3.
Substrate-induced currents of AtINT2-expressing Xenopus oocytes. A, AtINT2 mediates inward currents in response to myoinositol but not to most of the other sugars or sugar alcohols tested (all concentrations were 20 mm). Currents were recorded continuously at pH 5.5 and a membrane potential of −60 mV. B, AtINT2-dependent inward currents elicited by different inositol epimers and derivatives. Currents were continuously recorded at pH 5.5 and a membrane potential of −60 mV, gaps result from voltage pulses applied during these times. Structural formulas of inositol epimers are shown underneath the respective recordings. C, Substrate-induced currents do not result from inward Na+ or K+ currents. Myoinositol-induced currents were recorded in the presence of 112.5 mm N-methylglucamine chloride (left) or 2.5 mm KCl and 110 mm NaCl (right) in the bath solution.
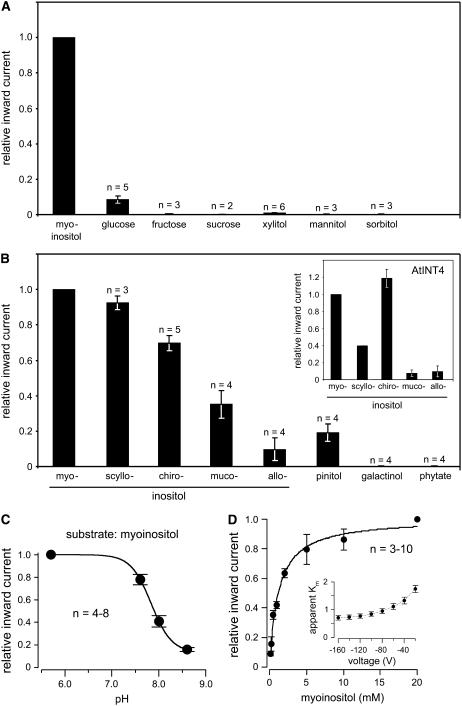
Figure 4.
Quantitative analyses of the substrate specificity, pH dependence, and Km myoinositol of recombinant AtINT2 in Xenopus oocytes. A, Quantitative analysis of inward currents elicited by different potential substrates of AtINT2 showing high specificity for inositol and a marginal capacity to transport Glc. B, Relative inward currents elicited by different inositol epimers and derivatives in AtINT2-expressing or AtINT4-expressing (inset) oocytes. C, pH dependence of myoinositol-elicited inward currents. D, The Km value of AtINT2 for myoinositol was determined by analyzing H+ currents in the presence of different myoinositol concentrations. Normalized currents were plotted against the substrate concentration. Data were fitted with Michaelis-Menten-type kinetics. The voltage dependence of the Km values is shown in the inset. Apparent Km values ranged from 0.7 ± 0.08 at −160 mV to 1.75 ± 0.13 at −20 mV (n = 5). Myoinositol-dependent H+ currents were determined at a membrane potential of −60 mV (A–D) or at voltages indicated (inset in D). Results ± se are presented.
Myoinositol represents only one of nine possible inositol epimers. Seven of these epimers (allo-, d-chiro-, l-chiro-, muco-, myo-, neo-, and scylloinositol) are found in living cells, the others (cis- and epiinositol) possibly exist only as synthetically prepared compounds. Especially d-chiroinositol (altered orientation of the OH group at position 1 of myoinositol) and scylloinositol (altered orientation of the OH group at position 2 of myoinositol) were also found in plant phospholipids (Chien et al., 1996; Narasimhan et al., 1997; Carstensen et al., 1999). We compared the ability of AtINT2 to transport different epimers of inositol, including the frequently found epimers myo-, d-chiro-, and scylloinositol and two epimers that were not or hardly detected in plant cells (alloinositol [altered orientation of the OH group at position 6 of myoinositol] and mucoinositol [altered orientations of the OH groups at positions 1 and 6 of myoinositol]). Figures 3B (original recording) and 4B (quantitative analysis) demonstrate that in addition to the already shown transport of myoinositol (Figs. 3A and 4A) AtINT2 does also catalyze the transport of scyllo- and d-chiroinositol with only slightly reduced rates (90% and 70%, respectively). Lower current amplitudes (35% and 10%) were obtained in the presence of muco- and alloinositol. We also analyzed the myoinositol derivatives pinitol (monomethylated chiroinositol), phytate (6-fold phosphorylated), and galactinol (inositol attached to Gal). While pinitol was transported by AtINT2 (20% of the rate of myoinositol), no currents were elicited by phytate and galactinol (Figs. 3B and 4B).
The epimer specificity has so far neither been determined for the previously characterized AtINT4 transporter (Schneider et al., 2006) nor for any other plant myoinositol transporter. To observe possible differences in the substrate specificities of AtINT2 and AtINT4 we studied the transport of different inositol epimers also in Xenopus oocytes injected with AtINT4 cRNA. The insert in Figure 4B demonstrates that AtINT2 and AtINT4 show, in fact, clearly different specificities.
The identity of the cotransported ion was determined in the presence of different cations in the extracellular solution. There was no difference between the substrate-induced currents elicited in a buffer containing 110 mm NaCl and 2.5 mm KCl or in a buffer that replaced these salts by N-methylglucamine chloride (112.5 mm; Fig. 3C; n = 6). This demonstrates that neither K+ nor Na+ ions are symported, and that AtINT2 is most likely an inositol/H+ symporter.
Figure 4C shows the pH dependence of inward H+ currents elicited by myoinositol in AtINT2-expressing Xenopus oocytes. AtINT2 shows highest transport rates at physiological and acidic pH values. At more alkaline values, myoinositol-induced currents decrease rapidly, which can be taken as additional, though circumstantial evidence that protons are the cosubstrate of AtINT2.
Finally, the Km value of AtINT2 was determined for myoinositol. Figure 4D shows Michaelis-Menten kinetics determined at a membrane potential (Δψ) of −60 mV and at an extracellular pH of 5.5. Comparison of the Km value calculated from this and similar analyses at different membrane potentials (insert in Fig. 4D) revealed that the Km of AtINT2 is slightly voltage dependent (higher membrane potentials resulted in higher substrate affinity). However, a stronger decrease in the affinity of AtINT2 for myoinositol was observed only at unphysiologically low values of Δψ. At −80 mV, a Km of 0.95 ± 0.09 mm (se; n = 5) could be determined. This Km is significantly lower than that determined for AtPLT5 in Xenopus oocytes (3.5 ± 0.3 mm; Klepek et al., 2005), but about 4-fold higher than the Km for myoinositol of AtINT4 (0.24 ± 0.01; Schneider et al., 2006), indicating that AtINT2 is an inositol transporter with medium affinity.
Analysis of AtINT2 Expression in AtINT2 Promoter/GUS Plants and in AtINT2 Promoter/GFP Plants
For analyses of the tissue specificity of AtINT2 expression we generated and analyzed AtINT2 promoter/GUS plants and AtINT2 promoter/GFP plants. To this end a 1,448-bp promoter fragment was used to drive expression of GUS or GFP in plants that had been selected for BASTA resistance after transformation with the plasmids pLEX111 (=AtINT2 promoter/GUS) or pLEX106 (=AtINT2 promoter/GFP).
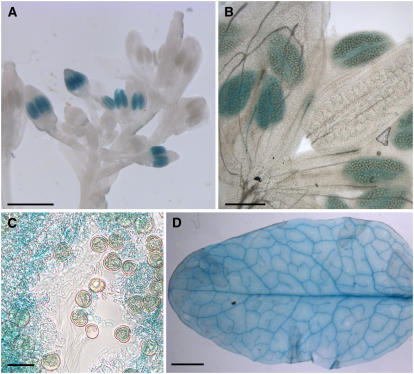
We obtained numerous transformants with both constructs and analyzed 30 independent AtINT2 promoter/GUS lines and 24 independent AtINT2 promoter-GFP lines. In none of the AtINT2 promoter/GFP lines we were able to detect GFP fluorescence, suggesting that the activity of the AtINT2 promoter is rather weak. This was confirmed by analyses of AtINT2 promoter/GUS plants. Although all plants showed the same tissue specificity of the AtINT2 promoter, the obtained GUS staining was weak and in many plants it was detected only after prolonged staining. The results are summarized in Figure 5. Strongest GUS staining was detected in anthers (Fig. 5A). Analyses at higher magnification (Fig. 5B) and in destroyed anther tissue (Fig. 5C) revealed that this anther-specific GUS staining is not localized in pollen, but rather in a cell layer of the anther wall, most likely the tapetum. This is deduced from two observations: (1) the outermost cell layer of intact anthers shown in Figure 5B were not stained, and (2) individual pollen grains in squashed anther tissue did not show any GUS staining. Interestingly, the previously reported GUS staining in anthers of AtINT4 promoter/GUS plants (Schneider et al., 2006) was pollen specific.
Figure 5.
Analysis of AtINT2 promoter/GUS plants. A, GUS histochemical staining of an inflorescence showing GUS staining only in anthers. Notably GUS staining is missing from very young and from fully developed anthers. B, A higher magnification of one of the developing flowers shown in A, suggesting that GUS staining is not in the pollen, but rather in the surrounding tissue, potentially in the tapetum. C, Squashed, GUS-stained anther confirming that the GUS activity is absent from the pollen grains. D, GUS histochemical staining in the vascular strands of a rosette leaf. Weaker staining is seen all over the leaf. Bars are 20 μm in C, 100 μm in B, 1 mm in A, and 2 mm in D.
AtINT2 promoter-dependent GUS activity was also seen in Arabidopsis leaves (Fig. 5D) and very weakly in roots (data not shown). Clearly, stronger staining was obtained in leaf vascular tissue of AtINT2 promoter-GUS plants, however, staining was also seen in all other parts of the leaves.
Immunohistochemical Analyses of AtINT2 Localization
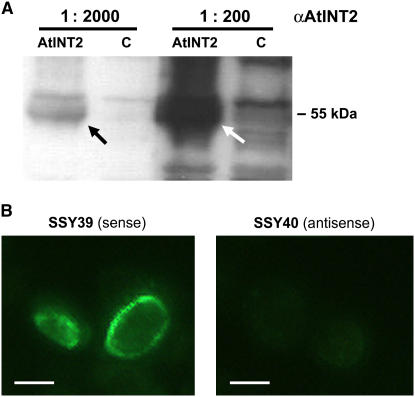
Antisera were raised in one guinea pig and two rabbits against a 26-amino acid peptide from the very C terminus of AtINT2. This sequence is not found in any other Arabidopsis protein and does not show significant homology to the C termini of the other AtINT proteins. The quality of the obtained sera was tested on detergent extracts from total membranes isolated from yeast strains ScLEX41 (SEY2102 [Emr et al., 1983] that harbors the NEV-N vector [Sauer and Stolz, 2000] with AtINT2 cDNA in sense orientation) and SSY9 (SEY2102 that harbors the empty NEV-N vector). Figure 6A shows a western-blot of these extracts after electrophoretic separation and incubation with the unpurified guinea pig antiserum (αAtINT2-GP; similar results were obtained with the rabbit-derived antisera). A band with an apparent molecular mass of about 55 kD was detected only in extracts from AtINT2-expressing cells. This band was absent from extracts from SSY9 control cells showing that the labeled band in ScLEX41 cells represents the AtINT2 protein.
Figure 6.
Immunohistochemical detection of recombinant AtINT2 protein in western blots of yeast total membranes and in thin sections of AtINT2-expressing yeast cells. A, Unpurified αAtINT2 (diluted 1:200 or 1:2,000) that had been raised against 26 amino acids from the AtINT2 C terminus labeled a 55-kD band in detergent extracts from yeast total membranes after gel electrophoresis and blotting to nitrocellulose filters (10 μg lane−1; AtINT2 = AtINT2-expressing cells; C = control cells). B, Incubation of thin sections with affinity-purified αAtINT2 and fluorescence-tagged second antibody yielded fluorescence only in AtINT2-expressing cells (SSY39) but not in control cells (SSY40). Bars are 2 μm. [See online article for color version of this figure.]
The difference between the apparent molecular mass (55 kD) and the molecular mass predicted from the DNA sequence (63.4 kD) was not unexpected. Typically, lipophilic proteins run at lower apparent molecular masses on SDS gels. This unusual running behavior results from excess binding of SDS of highly lipophilic proteins (Beyreuther et al., 1980; Gahrtz et al., 1994; Barth et al., 2003; Schneider et al., 2006).
Next we tested the quality of the antisera on thin sections of yeast cells that expressed AtINT2 either in sense (SSY39) or in antisense orientation (SSY40; see above). Sections of these cells were prepared using the identical protocol usually applied for the fixation and embedding of plant material (Meyer et al., 2004). Figure 6B demonstrates that affinity-purified αAtINT2-R2 antiserum (from rabbit 2) does recognize the AtINT2 protein in sections of SSY39 cells (sense), but not in SSY40 cells (antisense). This result demonstrated that the antigenic epitope recognized by αAtINT2-R2 was not destroyed during fixation and suggested that αAtINT2-R2 should also detect AtINT2 in plant tissue sections.
Therefore, affinity-purified αAtINT2-R2 was used to study the localization of AtINT2 protein in sections of Arabidopsis leaves and flowers. Antibody binding was visualized with an anti-rabbit IgG-fluorescein isothiocyanate-isomer 1 conjugate. Unfortunately, in none of the tested Arabidopsis tissues αAtINT2-R2-dependent fluorescence was detected. Based on the data from AtINT2-expressing yeast cells (Fig. 6B) we could exclude that this results from a destruction of the antigenic epitope during fixation and/or embedding. The complete lack of immunosignals in plant sections rather suggested that the concentration of the antigen is too low for immunodetection of AtINT2 with our antisera. This interpretation was supported by the lack of GFP fluorescence in AtINT2 promoter/GFP plants and by the observed weak GUS histochemical staining.
Analysis of the Subcellular Localization of AtINT2 by Transient Expression of AtINT2-GFP
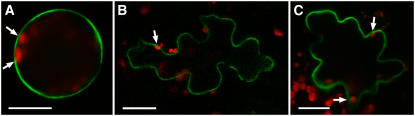
The subcellular localization of AtINT2 in planta was analyzed using an AtINT2 protein with GFP fused to its C terminus. To this end, the plasmid pMG002 that drives expression of the AtINT2-GFP fusion under the control of an enhanced 35S promoter was used for transient expression in Arabidopsis protoplasts (Figs. 7A) or in particle bombarded epidermis cells of Arabidopsis (Fig. 7B) or tobacco (Fig. 7C). Transformed cells and protoplasts were analyzed by confocal microscopy. In all analyses the red autofluorescence of the chloroplasts was localized inside the GFP-labeled structure (arrows in Fig. 7). No GFP fluorescence was found in any other structure inside the transformed cells, indicating that in both plant expression systems (Arabidopsis and tobacco) the AtINT2-GFP fusion protein is located in the plasma membrane.
Figure 7.
Subcellular localization of AtINT2 in AtINT2-GFP-expressing cells. A, Single optical section of an Arabidopsis protoplast. B, Single optical section from an Arabidopsis epidermis cell after particle bombardment of a detached leaf. C, Transient expression in a tobacco epidermis cell after particle bombardment of a detached tobacco leaf. Images were taken on a confocal laser-scanning microscope; red fluorescence in A to C shows chlorophyll autofluorescence. Arrows show localization of chloroplasts inside the GFP-labeled structure. Scale bars are 20 μm.
Analysis of an Atint2 Mutant Line
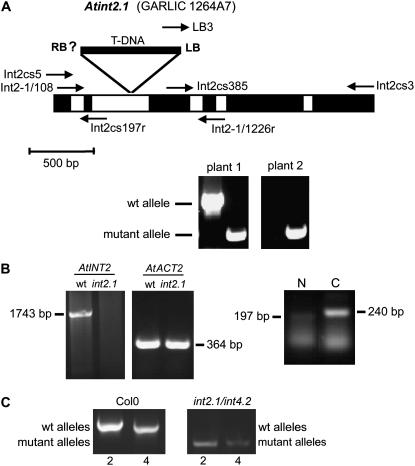
Screening of publicly accessible libraries identified a mutant line (GARLIC_1264A7 = Atint2.1) with a T-DNA insertion in the second intron of the AtINT2 gene, 644 bp after the start ATG (Fig. 8A). We performed PCR reactions to identify homozygous Atint2.1 plants (e.g. plant 2 in the PCR shown in Fig. 8A) and used these plants for further analyses. Homozygous mutant plants showed a complete loss of intact AtINT2 mRNA (1,742-bp band in Fig. 8B). Partial AtINT2 mRNAs, however, representing sequences flanking the T-DNA insertion could be amplified also from Atint2.1 RNA preparations. The 197-bp fragment (N in Fig. 8B) flanking the predicted right border of the T-DNA insertion (RB in Fig. 8A) is likely to result from AtINT2 promoter activity. The 240-bp fragment (C in Fig. 8B) flanking the identified left border (Fig. 8A) of the T-DNA insertion most likely results from promoter activity within the T-DNA insertion. Growth analyses of homozygous Atint2.1 plants under standard conditions (on soil in the growth chamber: 21°C, 60% relative humidity, long day [16 h light/8 h dark] or short day [8 h light/16 h dark]), on different concentrations of NaCl (10–100 mm), in the presence of mannitol, or on different concentrations of myoinositol in the growth medium (0–100 mm) revealed no visible or metabolic (e.g. altered sugar, polyol, or cyclitol concentrations; data not shown) differences between the T-DNA insertion line and the isogenic wild type. Also the fertility of the plants was not affected by the T-DNA insertion in AtINT2.
Figure 8.
Characterization of GARLIC line 1264A7 (=Atint2.1). A, Schematic drawing of the AtINT2 genomic sequence from start to stop. Exons are shown in black, the five introns in white. The position of the T-DNA insertion and of the characterized left border (LB) are indicated. Arrows show the positions and directions of primers used for PCR reactions in A and B. Example PCRs for two progeny plants of GARLIC_1264A7 seeds identifying wild-type (primers: Int2-1/108 and Int2-1/1226r) and mutant (primers: LB3 and Int2-1/1226r) alleles are presented. Plant 1 was characterized as heterozygous, plant 2 as homozygous. B, RT-PCR-derived bands showing the complete AtINT2 ORF (primers: Int2cs5 and Int2cs3) and the partial AtACT2 ORF (primers: ACT2-846f and ACT2-1295r) from RNA isolated from wild-type or Atint2.1 plants. While an AtINT2 product was obtained only from wild-type plants, the ACT2 fragment was amplified both from wild-type and mutant plants. cDNA fragments were amplified from truncated mRNAs upstream and downstream of the insertion site (N: primers Int2cs5 and Int2cs197r; C: primers Int2cs385 and Int2-1/1226r). C, Characterization of an Atint2.1/Atint4.2 double mutant. Only in Col-0 but not in the double mutant the wild-type alleles of AtINT2 and AtINT4 were identified (primers: Int2-1/108 and Int2-1/1226r for AtINT2; int4-802 and int4-1826r for AtINT4; Schneider et al. [2006]). In contrast, mutant alleles were amplified only in the double mutant (primers: LB3 and Int2-1/1226r for Atint2.1; int4-802 and LBb1 for Atint4.2; Schneider et al. [2006]).
Analysis of an Atint2.1/Atint4.2 Double Mutant
A possible reason for the lack of a detectable phenotype in the Atint2.1 mutant might be a functional complementation of the defective Atint2.1 allele by AtINT4. AtINT4 promoter/GUS and AtINT4 promoter/GFP analyses (Schneider et al., 2006) and the AtINT2 promoter/GUS data presented in Figure 6 showed a partial overlap of AtINT2 and AtINT4 expression patterns in the vasculature. Therefore, we crossed the Atint2.1 mutant and the previously described Atint4.2 mutant (Schneider et al., 2006).
For the identification of Atint2.1/Atint4.2 double mutants, comparative PCRs were performed with genomic DNA isolated from Columbia-0 (Col-0) plants or from potential double mutants and a set of three primers that allowed simultaneous amplification of fragments from wild-type and mutant alleles in a single PCR reaction. Figure 8C shows the characterization of a homozygous Atint2.1/Atint4.2 double mutant that yielded only PCR fragments of the two mutant alleles. These fragments were not amplified from Col-0 DNA that showed the expected wild-type fragments for both genes. Like the Atint2.1 single mutant, the Atint2.1/Atint4.2 double mutant neither developed a growth phenotype under different growth conditions nor did we observe differences in the carbohydrate compositions of leaf extracts (myoinositol, Glc, Fru, Suc; data not shown).
DISCUSSION
This article presents a detailed characterization of AtINT2 as a plasma membrane-localized H+ symporter with medium affinity for various inositol epimers, and of AtINT3 as a gene that very likely encodes a truncated protein. AtINT2 and AtINT3 represent two of four predicted inositol transporter genes (AtINT1 to AtINT4) that form a subfamily within the MST-like superfamily of Arabidopsis that was named after the AtSTP family of plasma membrane-localized monosaccharide transporters (Sauer et al., 1990; Büttner and Sauer, 2000; Büttner, 2007).
So far, less than 50% of the members of the MST-like superfamily (53 genes in Arabidopsis) have been characterized on a functional basis. This includes most members of the STP subfamily (Büttner, 2007), one member of the PLT subfamily (AtPLT5; Klepek et al., 2005), one member of the TMT subfamily of tonoplast-localized monosaccharide transporters (AtTMT1; Wormit et al., 2006), one member of the VGT subfamily of vacuolar Glc transporters (AtVGT1; Aluri and Büttner, 2007), and one member of the AtINT subfamily (AtINT4; Schneider et al., 2006). Individual members of other subfamilies were studied (AtERD6 [At1g08930], Kiyosue et al., 1998; pGlcT [At5g16150], Weber et al., 2000; AtSFP1 [At5g27350], Quirino et al., 2001), but not functionally characterized.
AtINT3 Seems to Be a Pseudogene
Our attempts to obtain an AtINT3 cDNA via RT-PCR from total RNA from Arabidopsis rosettes failed repeatedly. Moreover, publicly accessible libraries did not contain AtINT3 cDNA clones. Finally, no correctly spliced AtINT3 cDNAs could be isolated from plant tissues (Arabidopsis or tobacco) that expressed genomic AtINT3 sequences under the control of the 35S promoter. All obtained cDNAs were incorrectly spliced and encoded only a truncated protein. As can be seen from the phylogenetic tree shown in Figure 1, AtINT3 is closely related to AtINT4, suggesting that the corresponding genes may have duplicated in the more recent past of Arabidopsis evolution. Obviously one of the duplicated genes, AtINT3, accumulated mutations that eventually caused a loss of function. Similar observations have been reported for members of the Arabidopsis Suc transporter family (Sauer et al., 2004).
AtINT2 Is a Plasma Membrane-Localized H+ Symporter That Differs in Its Substrate Specificities from AtINT4
The successful functional analyses in yeast (complementation of a defect in the plasma membrane myoinositol transporter Itr1p) and Xenopus suggested that AtINT2 might be a plasma membrane-localized protein in plant cells. This was confirmed by transient expression analyses of an AtINT2-GFP construct that resulted in GFP fluorescence exclusively in the plasma membrane of transformed cells (Fig. 7). Moreover, the Xenopus data characterized AtINT2 as an H+/inositol symporter with medium affinity to myoinositol. In contrast to AtINT4 that has a 4-fold lower, Δψ-independent Km for myoinositol (Schneider et al., 2006) the affinity of AtINT2 increases with increasing Δψ.
Another difference between AtINT2 and AtINT4 was observed, when the currents elicited by different inositol epimers were compared (Figs. 3B and 4B). At most likely saturating substrate concentrations (20 mm), AtINT2 catalyzed the uptake of myoinositol and scylloinositol with similar rates. d-chiro-, muco-, and alloinositol were also transported, although with decreasing uptake rates. Under identical conditions, however, AtINT4 preferred myoinositol and d-chiroinositol (d-chiroinositol even slightly better), transported scylloinositol with significantly lower rates, and muco- and alloinositol turned out to be only poor substrates for AtINT4. This demonstrates that AtINT2 and AtINT4 respond differently to altered orientations of the OH groups at position 1 of myoinositol (as in chiroinositol; best substrate for AtINT4) and position 2 (as in scylloinositol; very good substrate for AtINT2). In contrast, an altered orientation of the OH group at position 6 of myoinositol (as in muco- and alloinositol) resulted in reduced transport by both proteins.
AtINT2 and AtINT4 transported pinitol, the monomethylated derivative of chiroinositol, only with reduced rates (about 20%–30% of chiroinositol; Fig. 4B; Schneider et al., 2006), suggesting that unmethylated inositols are the preferred substrates in Arabidopsis. This is supported by results of Miyazaki et al. (2004) obtained with yeast cells expressing inositol transporter cDNAs from ice plants (named McITR1 or MITR1; Chauhan et al., 2000) or Arabidopsis (named AtITR1 or AITR1; identical to AtINT4; Schneider et al., 2006). In these analyses, even a 600-fold excess of ononitol (4-O-methyl-myoinositol) inhibited myoinositol uptake only by 75%.
Although data on tissue or cellular concentrations of scyllo- and chiroinositol are not available for Arabidopsis, the finding of these inositol epimers in numerous other plant species (Kinnard et al., 1995; Narasimhan et al., 1997; Ichimura et al., 2000) makes it quite likely that Arabidopsis also contains different inositol epimers, and that AtINT2 and AtINT4 may, in fact, see different potential substrates.
Substrate specificities of inositol transporters have also been studied in several nonplant organisms, and for some of these transport systems they are similar to those described for AtINT2 and AtINT4. For the ciliate Tetrahymena vorax, for example, it was found that the most frequently occurring epimers myo-, scyllo-, and d-chiroinositol were transported across the plasma membrane (Kersting and Ryals, 2004). Similarly, it was shown for human HepG2 liver cells that myoinositol and d-chiroinositol are transported by the same protein (Ostlund et al., 1996). Interestingly, however, in the yeast Candida albicans (Jin and Seyfang, 2003) and in the protozoan parasite Leishmania donovani (Mongan et al., 2004) transporters were identified that are highly specific for myoinositol, because no or only marginal competition by scyllo- or d-chiroinositol was seen. Moreover, for both inositol transport systems a 50% inhibition of myoinositol transport by phytate was observed. This compound elicited no current in AtINT2-expressing Xenopus oocytes (Figs. 3B and 4B).
AtINT2 Has a Highly Conserved, Cys-Rich Sequence in a Predicted Extracellular Loop
Analyses of the protein sequences of the (putative) inositol transporters shown in the phylogenetic tree in Figure 1 revealed that the loops between the predicted transmembrane helices IX and X of all transporters in clades 2 and 3 are about 80 amino acids longer than the respective loops of the proteins in clade 1. These additional 80 amino acids possess an unusually large number of highly conserved Cys residues. Four of these residues are part of two Cys-X-X-Cys motifs typically separated by only few amino acids (e.g. 399-CMTCLKASSPSCGYC-413 in AtINT2 and 406-CMKCLRSECGFC-417 in AtINT4). Since AtINT2 and AtINT4 were both localized to the plasma membrane, their Cys-X-X-Cys-containing loops are predicted to face the extracellular space and the lumen of the endoplasmic reticulum during biosynthesis and targeting.
Possibly, these highly conserved Cys residues are substrates for endoplasmic protein disulphide isomerases. Alternatively, the Cys-X-X-Cys motifs may provide some yet uncharacterized function to the extracellular loop or confer special redox sensitivity to the protein. Thioredoxins, for example, and bacterial disulphide isomerases have quite similar Cys-X-X-Cys motifs (Elton et al., 2005) and most of these proteins were shown to catalyze redox reactions.
AtINT2 Is Expressed in Anthers and in the Vasculature
Expression of the AtINT2 gene was observed in anthers (Fig. 5, A–C), in the vascular tissue (Fig. 5D), and to a lesser extent also in the leaf mesophyll (Fig. 5D). The low intensity of the obtained GUS staining and the lack of GFP fluorescence suggested low expression levels for AtINT2, and, in fact, we were not able to immunolocalize AtINT2 protein in plant tissue, although the antiserum was shown to work on western blots (Fig. 6A) and in thin sections of AtINT2-expressing yeast cells.
At first sight, the expression patterns of AtINT2 (Fig. 5) and AtINT4 (Schneider et al., 2006) look similar. The promoters of both genes are active in anthers and in the vasculature. Detailed comparisons show, however, that the anther-specific activity of the AtINT2 promoter is restricted to cells of the anther wall, most likely the tapetum, a nutrient-supplying cell layer, whereas the anther-specific activity of the AtINT4 promoter is restricted to the pollen grains. Moreover, the AtINT2 promoter is active only during a very limited period of anther development (Fig. 5A), whereas AtINT4 drives GUS expression preferentially in the pollen of fully developed anthers (Schneider et al., 2006). These differences in the expression pattern suggest also different physiological roles for the two transporters and make functional redundancy of AtINT2 and AtINT4 rather unlikely. In contrast to AtINT4, which is likely to mediate inositol import into germinating and growing pollen tubes (Schneider et al., 2006), the function of AtINT2 may involve the release of inositols from the tapetum to the developing pollen. It is known that 6-fold phosphorylated myoinositol, phytate, is a major constituent of pollen grains (1%–5% of dry weight; Loewus, 1990; Raboy, 1990) and AtINT2 may play a role in feeding myoinositol to the developing pollen. Given a sufficient substrate accumulation inside the cells of the tapetum, even under physiological pH gradients a release function for AtINT2 appears likely, as other plasma membrane H+ symporters work in either direction (Carpaneto et al., 2005).
More detailed analyses of the GUS data from AtINT2 and AtINT4 promoter/GUS plants revealed also differences in the expression patterns in leaves. Whereas the AtINT2 promoter is active in the vasculature and to a lesser extent also in all other parts of the leaf (Fig. 5D), the activity of the AtINT4 promoter is strictly confined to the vasculature.
The Physiological Role of AtINT2 Remains to Be Elucidated
Due to the absence of a phenotypic difference between Atint2.1 single or Atint2.1/Atint4.2 double mutants and wild-type plants, a more detailed prediction of the physiological role(s) of AtINT2 is difficult. In the vasculature it may be involved in the supply of inositol for galactinol and thus for raffinose biosynthesis. The concentrations of raffinose in Arabidopsis, however, are very low (Haritatos et al., 2000), and a partial or complete lack of raffinose would not be expected to strongly influence the development of Arabidopsis plants. The low activity of the AtINT2 promoter in all other leaf cells might indicate that the function of AtINT2 in the leaf mesophyll is simply the retrieval of inositols that diffused out of the cells. This has been postulated for numerous other transporters as well, and, in fact, keeping the cell wall concentrations of free polyols, sugars, amino acids, and other potential nitrogen and carbon sources low (cell wall hygiene) is certainly a very effective mechanism to reduce growth and development of extracellular pathogens.
One may speculate that triple mutants that include also a T-DNA insertion in the AtINT1 gene might provide additional information. However, only recently AtINT1 has been characterized as a tonoplast-localized inositol transporter, and Atint1 mutants show increased cellular inositol concentrations and reduced root development (S. Schneider and N. Sauer, unpublished data). The construction of an Atint1/Atint2/Atint4 triple mutant is on the way, however, with the background of the observed Atint1-based phenotypic differences additional effects of Atint2 or Atint4 mutations will be difficult to identify.
MATERIALS AND METHODS
Strains and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Col wild type) plants were grown in growth chambers on potting soil under a 16 h light/8 h dark regime (22°C, 60% relative humidity) or in the greenhouse under ambient conditions. For expression of AtINT2 cDNAs in yeast (Saccharomyces cerevisiae) we used strains D458-1B (Nikawa et al., 1991) or SEY2102 (Emr et al., 1983). Escherichia coli DH5α (Hanahan, 1983) was used for all cloning steps, fusion proteins for antisera were made in E. coli Rosetta (Novagen). Transformation of Arabidopsis was performed with Agrobacterium tumefaciens GV3101 (Holsters et al., 1980).
cDNA Cloning of AtINT3
AtINT3 cDNA was amplified from tobacco (Nicotiana tabacum) plants infiltrated with Agrobacteria carrying plasmid pPU6 or from Arabidopsis plants stably transformed (Clough and Bent, 1998) with this plasmid. pPU6 is derived from pEARLEYGATE 100 (Earley et al., 2006) and was designed for expression of genomic AtINT3 sequences (start to stop including two predicted introns) under the control of the 35S promoter. For pPU6 construction, genomic AtINT3 DNA was amplified from Arabidopsis (AtINT3g + 1f: 5′-CACCATGGTGGAAGAAGCATCGAAATCAG-3′; AtINT3g + 1926r: 5′-CTAAGGCGTTTCCACTTGATTTTCCTTGGTCG-3′), inserted into pENTR/D-TOPO (Invitrogen), sequenced, and cloned into pEARLEYGATE 100.
After isolation of total RNA (RNeasy plant mini kit; Qiagen) from infiltrated tobacco leaves (24 h after infiltration) or from rosette leaves of BASTA-selected Arabidopsis plants, AtINT3 cDNA was amplified using the primers AtINT3g + 1f and AtINT3g + 1926r, cloned into pCR-Blunt-II-Topo (Invitrogen), and sequenced.
AtINT2 cDNA Cloning and Constructs for Yeast Expression
PCR-based cloning of the AtINT2 cDNA was described (Schneider et al., 2006). During this PCR, NotI restriction sites were added to both ends of the cDNA. For yeast expression, these sites were cut with NotI, and the cDNA was cloned into the yeast expression vector NEV-N-Leu, a modification of the expression vector NEV-N, where the Ura2 selection marker had been replaced by Leu-3 (Sauer and Stolz, 1994). Plasmids containing the AtINT2 cDNA in sense (pSS51s) or antisense orientation (pSS51as) plus the empty NEV-N-Leu vector were used for transformation (Gietz et al., 1992) of D458-1B, yielding strains SSY39, SSY40, and SSY38, respectively. If not otherwise indicated, uptake experiments with myo-[1,2-3H(N)]-inositol (American Radiolabelled Chemicals Inc.) were performed at a concentration of 1 mm (final specific activity: 0.2 μCi μmol−1) in 50-mm sodium phosphate buffer (pH 5.0) as described (Sauer et al., 1990).
Synthesis and Injection of cRNA and Two-Electrode Voltage Clamp
Oocytes were injected (General Valve Picospritzer III, Parker Hannifin Corp.) with cRNA from AtINT2 or AtINT4 or with RNase-free water and stored at 16°C in ND96 solution (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, adjusted to pH 7.4 with NaOH) supplemented with 1% penicillin-streptomycin (Sigma) or Barth's solution [88 mm NaCl, 1 mm KCl, 0.33 mm Ca(NO3)2, 0.41 mm CaCl2, 82 mm MgCl2, 2.4 mm NaHCO3, 10 mm HEPES, adjusted to pH 7.6 with NaOH] supplemented with 0.1% gentamycin (Sigma). Oocyte currents were studied 4 to 6 d after injection with the two-electrode voltage-clamp technique (Hedrich et al., 1995) using a Turbo Tec-10Cx amplifier (NPI electronic GmbH). During two-electrode voltage-clamp measurements, oocytes were constantly superfused with modified Ringer solution (110 mm NaCl, 2.5 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2) at room temperature. The solution was buffered using 10 mm MES (pH 5.5, adjusted with TRIS) or TRIS (pH 7.4, 8.0, and 8.5, adjusted with MES). Sugars and sugar alcohols were added to the indicated concentrations. For continuous whole-cell current recordings, oocytes were clamped at a holding potential of −60 mV. Starting from this holding potential, voltage-step protocols were applied to test potentials ranging from +60 mV up to −160 mV in 20 mV decrements.
AtINT2 Promoter/GUS and AtINT2 Promoter/GFP Constructs and Plant Transformation
A 1,448-bp AtINT2 promoter fragment was PCR amplified from genomic Arabidopsis DNA. The primers (INT2-p5: 5′-ATAACTTTAAGCTTTTTGTTAGAA-3′; INT2-p3: 5′-TCCTCCCTCCATGGTTTTTTGGGTTAAAAGAGTTAGAA-3′) introduced an N-terminal HindIII and a C-terminal NcoI site that were used to clone the fragment in front of the ORF of GFP and a transcriptional terminator in a pUC19-based plasmid (pEPS1/pUC19; Imlau et al., 1999). The fragment was sequenced and the AtINT2 promoter/GFP/terminator box was cloned into pGPTV-BAR (Becker et al., 1992) yielding the plasmid pLEX163 that was used for transformation of Arabidopsis (Clough and Bent, 1998). Plasmid pLEX111 (AtINT2 promoter/GUS) was also generated in pGPTV-BAR using the same promoter fragment.
Transient Expression of AtINT2-GFP
The AtINT2 coding sequence was PCR amplified using the primers AtINT2-5-BspH (5′-TCATGAAGGGAGGAATAATACATG-3′) and AtINT2-5-bspH (5′-TCATGACTGCACTCTGGTTTTGTTTCTC-3′). These primers introduced BspHI sites at the start and at the very end of the AtINT2 ORF, thereby replacing the stop codon of the original AtINT2 sequence. This modified AtINT2 ORF was inserted into the unique NcoI cloning site representing the start ATG of the GFP ORF in the pSO35e plasmid (Klepek et al., 2005). The continuous ORF was confirmed by sequencing. The resulting plasmid was named pMG002.
pMG002 was used for transient expression of AtINT2 in Arabidopsis protoplasts (polyethylene glycol transformation; modified after Abel and Theologis, 1994) or in tobacco or Arabidopsis epidermis cells (particle bombardment; Klepek et al., 2005).
Analysis of the T-DNA Insertion Line Atint2.1
The T-DNA insertion line Atint2.1 (GARLIC_1264A7 = SAIL_1264_A07) was identified using the Salk Institute T-DNA Express gene-mapping tool (Alonso et al., 2003). Homozygous plants were identified in PCR reactions on genomic DNA with the primers Int2-1/108 (5′-ATCGGTGGTCTTCTCTTTGGT-3′) and Int2-1/1226r (5′-CCGGGAGTGTAAACATTAAGACA-3′) in combination with the primer LB3 (5′-TAGCATCTGAATTTCATAACCAATCTC-3′) that binds near the left border within the T-DNA insertion.
Primers Int2cs5 (5′-ATATCTCTGCGGCCGCAAAAATGGAGGGAGGAATAATACAT-3′), Int2cs3 (5′-ATATCTCTGCGGCCGCTCATGCACTCTGGTTTTGTTTCTCA-3′), Int2-1/108, and Int2-1/1226r were used for RT-PCRs with total RNA from wild-type and mutant Arabidopsis leaves. Primers AtACT2g + 846f (5′-ATTCAGATGCCCAGAAGTCTTGTT-3′) and AtACT2g + 1295r (5′-GAAACATTTTCTGTGAACGATTCCT-3′) were used to amplify the AtACT2 mRNA.
Immunohistochemical Techniques and Western-Blot Analyses
For αAtINT2 production, two oligonucleotides were annealed and cloned into the EcoRI/HindIII-digested vector pMAL-c2 (New England Biolabs) yielding pSS11-AK2. The oligonucleotides encoded the C-terminal 26 amino acids of AtINT2. pSS11-AK2 was used to express a fusion of this peptide to the maltose-binding protein in E. coli Rosetta (Novagen). Expression of the protein was induced with isopropyl thiogalactoside, solubilized proteins were separated on polyacrylamide gels (Laemmli, 1970), bands were excised, and proteins extracted and lyophilized. Antisera were generated by Pineda-Antikörper-Service.
Binding of αAtINT2 to microtome sections was visualized by treatment with anti-rabbit IgG-fluorescein isothiocyanate-isomer 1 conjugate (Sigma-Aldrich). Microscopic slides were mounted in antifading medium (ProLong Antifade kit; Molecular Probes) and viewed under appropriate excitation light.
Protein extracts of total membrane fractions from bakers' yeast were prepared as described (Sauer and Stolz, 2000), separated on polyacrylamide gels (Laemmli, 1970), and transferred to nitrocellulose filters (Dunn, 1986). Filters were treated with anti-rabbit or anti-guinea pig IgG-peroxidase conjugate (diluted 1:4,000) followed by incubation with Lumi-Light western Blotting Substrate (Roche Diagnostics GmbH).
Ion Chromatography
Concentrations of sugars and sugar alcohols were determined with an ICS-3000 system (Dionex) with a gradient pump (ICS-3000 SP), a degaser module, an autosampler (ICS-3000 AS), and a pulsed amperometric detector (ICS-3000 DC). Anionic compounds were separated on a CarboPack MA1 column (4 × 250 mm) connected to a guard column of the same material (4 × 10 mm) and an ATC-1 anion trap column that was placed between the eluent and separation columns to remove anionic contaminants in the eluents. As eluent, 612 mm sodium hydroxide made from purest water (Millipore) and 50% NaOH (Fluka) was used. The column was equilibrated at a flow rate of 0.4 mL min−1. The duration of the run was 60 min. Calibration and quantitative calculation of carbohydrates was performed with the Dionex chromeleon software 6.7.
Microscopy and Detection of GFP Fluorescence
Images of GFP fluorescence were made with an epifluorescence microscope (Zeiss Axioskop, Carl Zeiss Jena GmbH; excitation wavelength 460–500 nm) or with a confocal laser-scanning microscope (Leica TCS SPII, Leica Microsystems). Emitted fluorescence was monitored at detection wavelengths longer than 510 nm. Confocal images were processed using the Leica Confocal Software 2.5 (Leica Microsystems).
GUS plants were analyzed using a stereomicroscope (Leica MZFLIII; Leica Microsystems) or a microscope (Zeiss Axioskop, Carl Zeiss Jena GmbH). Images were processed using analySIS Doku 3.2 Software (Soft Imaging System).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AM778029 for AtINT3.
Acknowledgments
We thank Ruth Stadler for experimental help and Angelika Wolf for growing the Arabidopsis plants. We are grateful to Christoph Korbmacher for the frequent and generous supply with Xenopus laevis oocytes.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Arabidopsis Functional Genomics Network; grant no. Sa 382/13–1 to N.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Norbert Sauer (nsauer@biologie.uni-erlangen.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5 421–427 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Aluri S, Büttner M (2007) Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc Natl Acad Sci USA 104 2537–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshima H, Yamada M, Sauer N, Komor E, Schobert C (1993) Heterologous expression of the H+/hexose cotransporter from Chlorella in Xenopus oocytes and its characterization with respect to sugar specificity, pH and membrane potential. J Plant Physiol 141 293–297 [Google Scholar]
- Barth I, Meyer S, Sauer N (2003) PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20 1195–1197 [DOI] [PubMed] [Google Scholar]
- Beyreuther K, Bieseler B, Ehring R, Griesser HW, Mieschendahl M, Müller-Hill B, Triesch I (1980) Investigation of structure and function of lactose permease of Escherichia coli. Biochem Soc Trans 8 675–676 [DOI] [PubMed] [Google Scholar]
- Boorer KJ, Loo DD, Frommer WB, Wright EM (1996) Transport mechanism of the cloned potato H+/sucrose cotransporter StSUT1. J Biol Chem 271 25139–25144 [DOI] [PubMed] [Google Scholar]
- Büttner M (2007) The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett 581 2318–2324 [DOI] [PubMed] [Google Scholar]
- Büttner M, Sauer N (2000) Monosaccharide transporters in plants: structure, function and physiology. Biochim Biophys Acta 1465 263–274 [DOI] [PubMed] [Google Scholar]
- Carpaneto A, Geiger D, Bamberg E, Sauer N, Fromm J, Hedrich R (2005) Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under control of sucrose gradient and pmf. J Biol Chem 280 21437–21443 [DOI] [PubMed] [Google Scholar]
- Carstensen S, Pliska-Matyshak G, Bhuvarahamurthy N, Robbins KM, Murthy PP (1999) Biosynthesis and localization of phosphatidyl-scyllo-inositol in barley aleurone cells. Lipids 34 67–73 [DOI] [PubMed] [Google Scholar]
- Chauhan S, Forsthoefel N, Ran Y, Quigley F, Nelson DE, Bohnert HJ (2000) Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J 24 511–522 [DOI] [PubMed] [Google Scholar]
- Chien CT, Lin TP, Juo CG, Her GR (1996) Occurance of a novel galactopinitol and its changes with other non-reducing sugars during development of Leucaena laucocephala seeds. Plant Cell Physiol 37 539–544 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cohen JD, Bandurski RS (1982) Chemistry and physiology of the bound auxins. Annu Rev Plant Physiol 33 403–430 [Google Scholar]
- Dunn SD (1986) Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on western blots by monoclonal antibodies. Anal Biochem 157 144–153 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Elton TC, Holland SJ, Frost LS, Hazes B (2005) F-like type IV secretion systems encode proteins with thioredoxin folds that are putative DsbC homologues. J Bacteriol 187 8267–8277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr SD, Scheckman R, Flessel MC, Thorner J (1983) An MFα1-SUC2 (σ-factor-invertase) gene fusion for study of protein localisation and gene expression in yeast. Proc Natl Acad Sci USA 80 7080–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz M, Stolz J, Sauer N (1994) A phloem specific sucrose-H+ symporter from Plantago major L. supports the model of apoplastic phloem loading. Plant J 6 697–706 [DOI] [PubMed] [Google Scholar]
- Gietz D, Jean WS, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of E. coli with plasmids. J Mol Biol 166 557–580 [DOI] [PubMed] [Google Scholar]
- Haritatos E, Ayre BG, Turgeon R (2000) Identification of phloem involved in assimilate loading in leaves by activity of the galactinol synthase promoter. Plant Physiol 123 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert H (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51 463–499 [DOI] [PubMed] [Google Scholar]
- Hedrich R, Moran O, Conti F, Busch H, Becker D, Gambale F, Dreyer I, Kuch A, Neuwinger K, Palme K (1995) Inward rectifier potassium channels in plants differ from their animal counterparts in response to voltage and channel modulators. Eur Biophys J 24 107–115 [DOI] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inze D, Engler G, Villarroel R, et al (1980) The functional organization of the nopaline Agrobacterium tumefaciens plasmid pTiC58. Plasmid 3 212–230 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Kohata K, Yamaguchi Y, Douzono M, Ikeda H, Koketsu M (2000) Identification of L-inositol and scyllitol and their distribution in various organs in Chrysanthemum. Biosci Biotechnol Biochem 64 865–868 [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen T, Grunnet M, Angelo K, Klaerke DA, Olesen SP (2002) Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques 32 536–540 [DOI] [PubMed] [Google Scholar]
- Jin JH, Seyfang A (2003) High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology 149 3371–3381 [DOI] [PubMed] [Google Scholar]
- Kandler O, Hopf H (1982) Oligosaccharides based on sucrose (sucrosyl oligosaccharides). In FA Loewus, W Tanner, eds, Plant Carbohydrates 1. Encyclopedia of Plant Physiology: Plant Carbohydrates I, Intracellular Carbohydrates, New Series, Vol 13 A. Springer-Verlag, Berlin, pp 348–383
- Kanter U, Usadel B, Guerineau F, Li Y, Pauly M, Tenhaken R (2005) The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221 243–254 [DOI] [PubMed] [Google Scholar]
- Kersting M, Ryals PE (2004) Sodium-dependent transport of [3H](1D)chiro-inositol by Tetrahymena. J Eukaryot Microbiol 51 307–311 [DOI] [PubMed] [Google Scholar]
- Kinnard RL, Narasimhan B, Pliska-Matyshak G, Murthy PP (1995) Characterization of scyllo-inositol-containing phosphatidylinositol in plant cells. Biochem Biophys Res Commun 210 549–555 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Abe H, Yamaguchi-Shinozaki K, Shinozaki K (1998) ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochim Biophys Acta 1370 187–191 [DOI] [PubMed] [Google Scholar]
- Klepek YS, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N (2005) Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell 17 204–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lehle L (1990) Phosphatidyl inositol metabolism and its role in signal transduction in growing plants. Plant Mol Biol 15 647–658 [DOI] [PubMed] [Google Scholar]
- Loewus FA (1990) Structure and occurrence of inositols in plants. In DJ Morré, WF Boss, FA Loewus, eds, Inositol Metabolism in Plants. Wiley-Liss Inc, New York, pp 1–11
- Loewus FA, Murthy PPN (2000) myo-Inositol metabolism in plants. Plant Sci 150 1–19 [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, VanDemark AP, Lingam AT, Hill CP, Bass BL (2005) Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N (2004) Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol 134 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S, Rice M, Quigley F, Bohnert HJ (2004) Expression of plant inositol transporters in yeast. Plant Sci 166 245–252 [Google Scholar]
- Mongan TP, Ganapasam S, Hobbs SB, Seyfang A (2004) Substrate specificity of the Leishmania donovani myo-inositol transporter: critical role of inositol C-2, C-3 and C-5 hydroxyl groups. Mol Biochem Parasitol 135 133–141 [DOI] [PubMed] [Google Scholar]
- Murakeözy EP, Nagy Z, Duhazé C, Bouchereau A, Tuba Z (2003) Seasonal changes in the levels of compatible osmolytes in three halophytic species of inland saline vegetation in Hungary. J Plant Physiol 160 395–401 [DOI] [PubMed] [Google Scholar]
- Narasimhan B, Pliska-Matyshak G, Kinnard R, Carstensen S, Ritter MA, van Weymarn L, Murthy PN (1997) Novel phosphoinositides in barley aleurone cells. Plant Physiol 113 1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Rammesmayer G, Bohnert HJ (1998) Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell 10 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Tskugoshi Y, Yamashita S (1991) Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J Biol Chem 266 11184–11191 [PubMed] [Google Scholar]
- Ostlund RE Jr, Seemayer R, Gupta S, Kimmel R, Ostlund EL, Sherman WR (1996) A stereospecific myo-inositol/D-chiro-inositol transporter in HepG2 liver cells. J Biol Chem 271 10073–10078 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Reiter WD, Amasino RD (2001) One of two tandem Arabidopsis genes homologous to monosaccharide transporters is senescence-associated. Plant Mol Biol 46 447–457 [DOI] [PubMed] [Google Scholar]
- Raboy V (1990) Biochemistry and genetics of phytic acid biosynthesis. In DJ Morré, WF Boss, FA Loewus, eds, Inositol Metabolism in Plants. Wiley-Liss Inc, New York, pp 55–76
- Raboy V (2003) myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64 1033–1043 [DOI] [PubMed] [Google Scholar]
- Reinders A, Panshyshyn JA, Ward JM (2005) Analysis of transport activity of Arabidopsis sugar alcohol permease homolog AtPLT5. J Biol Chem 280 1594–1602 [DOI] [PubMed] [Google Scholar]
- Sauer N, Friedländer K, Gräml-Wicke U (1990) Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J 9 3045–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N, Ludwig A, Knoblauch A, Rothe P, Gahrtz M, Klebl F (2004) AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J 40 120–130 [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J (1994) SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine tagged protein. Plant J 6 67–77 [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J (2000) Expression of foreign transport proteins in yeast. In SA Baldwin, ed, Practical Approach Series. Oxford University Press, Oxford, pp 79–105
- Schneider S, Schneidereit A, Konrad KR, Hajirezaei M-R, Gramann M, Hedrich R, Sauer N (2006) Arabidopsis thaliana INOSITOL TRANSPORTER 4 mediates high affinity H+-transport of myoinositol across the plasma membrane. Plant Physiol 141 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A (1998) GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci 3 426–431 [Google Scholar]
- Sheveleva E, Chmara W, Bohnert HJ, Jensen RG (1997) Increased salt and drought tolerance by D-ononitol production in transgenic Nicotiana tabacum L. Plant Physiol 115 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T (2005) The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J 42 708–719 [DOI] [PubMed] [Google Scholar]
- Smith AE, Phillips EV (1982) The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Physiol Plant 54 31–33 [DOI] [PubMed] [Google Scholar]
- Streeter JG, Lohnes DG, Fioritto RJ (2001) Patterns of pinitol accumulation in soybean plants and relationships to drought tolerance. Plant Cell Environ 24 429–438 [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645 [DOI] [PubMed] [Google Scholar]
- Thomas JC, Bohnert HJ (1993) Salt stress perception and plant growth regulators in the halophyte Mesembryanthemum crystallinum. Plant Physiol 103 1299–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Servaites JC, Geiger DR, Kofler H, Hille D, Groner F, Hebbeker U, Flügge UI (2000) Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, Meyer S, Schmidt U, Martinoia E, Neuhaus HE (2006) Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18 3476–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]