Abstract
Site-specific integration is an attractive method for the improvement of current transformation technologies aimed at the production of stable transgenic plants. Here, we present a Cre-based targeting strategy in Arabidopsis (Arabidopsis thaliana) using recombinase-mediated cassette exchange (RMCE) of transferred DNA (T-DNA) delivered by Agrobacterium tumefaciens. The rationale for effective RMCE is the precise exchange of a genomic and a replacement cassette both flanked by two heterospecific lox sites that are incompatible with each other to prevent unwanted cassette deletion. We designed a strategy in which the coding region of a loxP/lox5171-flanked bialaphos resistance (bar) gene is exchanged for a loxP/lox5171-flanked T-DNA replacement cassette containing the neomycin phosphotransferase (nptII) coding region via loxP/loxP and lox5171/lox5171 directed recombination. The bar gene is driven by the strong 35S promoter, which is located outside the target cassette. This placement ensures preferential selection of RMCE events and not random integration events by expression of nptII from this same promoter. Using root transformation, during which Cre was provided on a cotransformed T-DNA, 50 kanamycin-resistant calli were selected. Forty-four percent contained a correctly exchanged cassette based on PCR analysis, indicating the stringency of the selection system. This was confirmed for the offspring of five analyzed events by Southern-blot analysis. In four of the five analyzed RMCE events, there were no additional T-DNA insertions or they easily segregated, resulting in high-efficiency single-copy RMCE events. Our approach enables simple and efficient selection of targeting events using the advantages of Agrobacterium-mediated transformation.
Plant transformation is a fundamental technique in plant science research as well as in the production of transgenic crops. Independent of the transformation method that is applied, gene silencing and variable transgene expression are a major problem for the production of stable transgenic plants. Transgene instability and variation in expression levels are mostly caused by transgene integration patterns (multiple insertions often organized as complex loci or rearranged transgene inserts) and genomic location (Peach and Velten, 1991; Kohli et al., 2003; Francis and Spiker, 2005). In addition, transgenes become integrated at random positions in the plant genome (Kim et al., 2007), which may result in unwanted mutations due to insertion in active genes. Site-specific recombination systems have been utilized for the directed and precise single-copy integration at predetermined genomic positions to circumvent the problems mentioned above.
The Cre/lox system of bacteriophage P1 has been developed as a versatile tool due to its simplicity and activity in heterologous systems (Ow, 2002; Gilbertson, 2003). It requires a single recombinase protein, Cre, which does not need additional cofactors to mediate reciprocal recombination between two of its 34-bp loxP DNA recognition sites. Cre monomers bind to loxP sites at two 13-bp inverted repeats that are separated by an 8-bp spacer region, which gives directionality to the target site. DNA flanked by loxP sites positioned in direct orientation will be excised, whereas loxP sites in inverted orientation will result in inversion of the loxP-flanked DNA. The Cre/lox system has been successfully used in plants to remove selectable marker genes that have served their purpose in transformation (Gleave et al., 1999; Corneille et al., 2001; Zuo et al., 2001; Kopertekh et al., 2004) or to resolve complex integrated loci (Srivastava et al., 1999; Srivastava and Ow, 2001, 2002; Zuo et al., 2001). In addition, site-directed integration of DNA molecules at a predetermined site in the genome has been successful in plants, using direct DNA transfer to tobacco (Nicotiana tabacum) protoplasts (Albert et al., 1995; Day et al., 2000), DNA delivered via particle bombardment of rice (Oryza sativa; Srivastava and Ow, 2002; Srivastava et al., 2004), as well as after Agrobacterium-mediated T-DNA delivery to Arabidopsis (Arabidopsis thaliana) and tobacco (Vergunst and Hooykaas, 1998; Vergunst et al., 1998b; Nanto et al., 2005; Nanto and Ebinuma, 2007).
The use of site-specific recombination systems for directed integration requires a two-step procedure. First, a target plant line is produced containing a recombination site, which will subsequently be used as a landing platform for integration of transgenes of interest delivered in a second round of transformation. Several strategies have been employed with the aim of obtaining single-copy, stable, site-specific integrants. Target plants with a single genomic lox target site have been used successfully for the selection of specific integrants. The reversibility of a Cre-mediated integration reaction required methods to control cre expression. These methods include transient expression of Cre (Albert et al., 1995; Vergunst and Hooykaas, 1998) and use of a cre promoter-displacement strategy (Albert et al., 1995; Vergunst et al., 1998b), which can be combined with the use of specific mutant lox sites (the recombination products of such lox sites are no longer able to recombine with each other) to achieve stabilization of the integrated DNA (Albert et al., 1995; Srivastava et al., 2004).
In mammalian systems, efficient recombinase-mediated cassette exchange (RMCE) strategies were developed to overcome the problem of reversible excision. RMCE allows the replacement of a genomic cassette with any desired transgene or DNA construction via a double recombinase-mediated reaction based on the presence of two recombination sites flanking both the genomic and exchange cassettes (Schlake and Bode, 1994; Bouhassira et al., 1997; Baer and Bode, 2001; Lauth et al., 2002; Wallace et al., 2007). A prerequisite for efficient RMCE is that the recombination sites flanking the cassette are not compatible. Otherwise, depending on the orientation of the sites, the cassette might become inverted or deleted from its insertion site in the presence of its cognate recombinase.
For transformation of many plant species, Agrobacterium tumefaciens is the preferred method due to its efficiency and simplicity. This warrants the development of an efficient site-specific integration strategy based on T-DNA, but several caveats have to be addressed. In the bacterium, a part of the large tumor-inducing plasmid, called the T-region, is cleaved at the border sequences by the VirD2 protein, and a single-stranded DNA (ssDNA) copy of the bottom strand (T-strand) is released. VirD2 remains covalently attached to the 5′-end of the T-strand. The T-DNA/VirD2 complex and several other Vir proteins are transported via the bacterial type IV secretion system into host cells. In the plant cell, the ssDNA-binding protein VirE2 binds cooperatively to the T-strand and thereby protects it from degradation (Rossi et al., 1996). Both VirD2 and VirE2 contain nuclear localization signals that facilitate transport of the T-complex into the nucleus (Zupan et al., 1996; Citovsky et al., 1997; Ziemienowicz et al., 2001). Agrobacterium thus delivers T-DNA as a linear ssDNA molecule (Tinland et al., 1994), which is in principle incompatible with site-specific recombination that requires double-stranded DNA. In addition, for an integration event at a single chromosomally introduced recombination site, the DNA molecule needs to be circular to prevent chromosome breakage. Based on several studies, it is known that T-DNA becomes double stranded prior to integration (Offringa et al., 1990; Mozo and Hooykaas, 1992; Narasimhulu et al., 1996; Chilton and Que, 2003; Tzvira et al., 2003). Although circle formation of T-DNA by ligation of the two T-DNA borders has been reported (Bakkeren et al., 1989; Bundock et al., 1995; Zhao et al., 2003), the frequency of occurrence remains unclear. Pansegrau and coworkers (1993) have shown that VirD2 can mediate ligation of the two T-DNA borders in vitro. In vivo, circular T-DNA was only recovered when it was adapted with a replication origin. This was the case when a viral replicon was placed on the T-DNA in maize (Zea mays) cells (Zhao et al., 2003) and turnip (Brassica napus; Bakkeren et al., 1989), or in the presence of the replicator from the yeast (Saccharomyces cerevisiae) 2μ plasmid (Bundock et al., 1995). These data suggest that border ligation may occur, but is not efficient. Previously, in our laboratory, we demonstrated that a Cre/lox integration strategy of T-DNA is feasible using a single loxP site at the target locus. The placement of two loxP sites in the T-DNA replacement vector, allowing Cre-mediated circularization upon entry in the host cell prior to targeting, resulted in the isolation of integrants at workable efficiency (Vergunst et al., 1998b). However, despite the use of methods to control expression of Cre, the integrants were still partially unstable due to the occurrence of the reversible deletion event. In this study, we improve on previously reported Agrobacterium-mediated site-specific integration methods by combining RMCE, heterospecific lox sites, and Cre delivery via a cotransfer method. For Cre technology, many mutant lox sites have been developed. Lee and Saito (1998) demonstrated in an in vitro assay that spacer mutation sites lox5171 recombined with themselves but not with a wild-type loxP site. We designed target and exchange cassettes flanked by loxP and lox5171 in inverted orientation and obtained stable single-copy exchange events in Arabidopsis.
RESULTS
Experimental Design
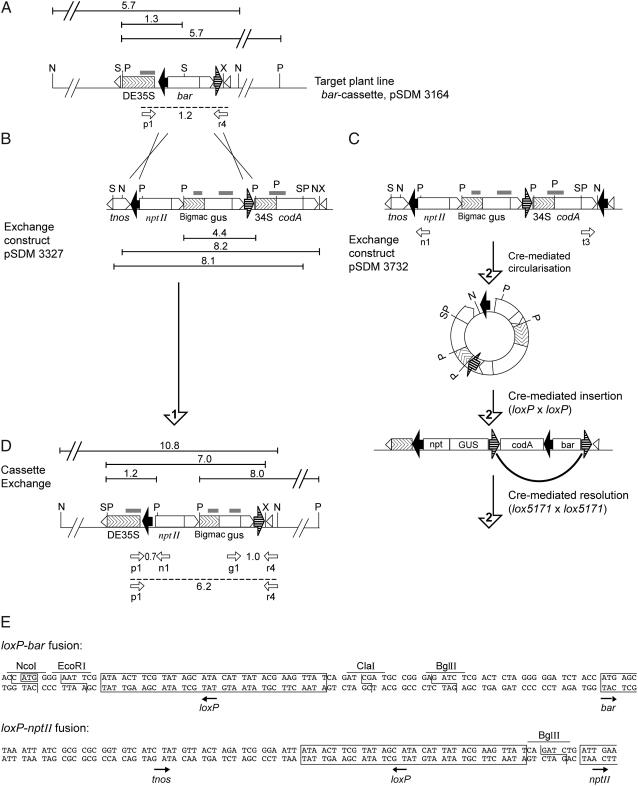
The rationale for our RMCE strategy is depicted in Figure 1. In a first step, the target construct was introduced in the plant genome by Agrobacterium-mediated transformation. This construct contains the bialaphos resistance (bar) coding region, flanked by loxP and the mutant lox5171 site in an inverted orientation (Fig. 1A). The loxP-bar coding region (Fig. 1E) is expressed from the strong cauliflower mosaic virus (CaMV) 35S promoter adjacent to the lox-flanked exchange cassette, providing resistance to phosphinothricin (PPT). The replacement T-DNA contains an exchange cassette with a promoterless neomycin phosphotransferase (nptII) gene and a β-glucuronidase (gus) gene allowing visualization and analysis of gene expression levels. Upon RMCE, the bar gene will be replaced by the nptII gene, effectively exchanging bar gene expression for nptII gene expression and allowing preferential selection for recombinants on medium containing kanamycin. Outside the exchange cassette, near the right T-DNA border repeat, the conditional negative selectable marker codA is inserted to allow selection of RMCE events in the absence of random insertions expressing codA.
Figure 1.
Strategy to obtain Cre-mediated cassette exchange. A, Chromosomally introduced target cassette (T-DNA pSDM3164). The bar gene is flanked by the loxP site (large black arrow) and the lox5171 site (large dashed arrow) in inverted orientation, with the strong CaMV 35S promoter and double enhancer sequence (DE35S) adjacent to the cassette providing resistance to PPT. B, Exchange T-DNA pSDM3327 contains the nptII coding sequence and gus expression unit driven by the BigMac promoter, flanked by lox sites in similar configuration as the bar cassette. The nos transcription terminator, tnos, and three translation stop signals in the different reading frames are included to prevent nptII expression from neighboring sequences upon random integration. The conditional negative selectable marker codA with the 34S promoter is placed outside the cassette. Arrow 1 indicates direct cassette exchange via a double crossover reaction loxP × loxP and lox5171 × lox5171, mediated by Cre that is provided via a cotransforming T-DNA. C, Exchange T-DNA pSDM3732 contains an additional loxP site near the right-border repeat. Arrows marked with a 2 indicate cassette exchange via a Cre-mediated circularization step of the exchange T-DNA prior to insertion at the target site (A) and resolution of the inserted exchange cassette. Circle formation can occur prior to or after random integration of pSDM3732. One of the two hypothetical exchange reactions is drawn for circularized exchange T-DNA pSDM3732 via insertion at the loxP site, followed by resolution at the lox5171 site. D, Result of precise cassette exchange (with either exchange construct pSDM3327 [B] or pSDM3732 [C]). The restored nptII selectable marker allows selection for correct cassette exchange using kanamycin. Large black arrows, loxP sites; large dashed arrows, mutant lox sites, lox5171; the direction of arrowheads in the boxes indicates promoter regions; small white triangles, A. tumefaciens left- and right-border repeats. For the Southern strategy, probes are drawn as gray boxes above the constructs; N, NsiI; P, PstI; S, SalI; X, XhoI; the expected fragment length (kb) is indicated above the lines. Beneath the constructs, white arrows indicate primer binding sites, the expected size of the PCR fragments is given, with the RMCE-PCR product from primers p1 and r4 indicated with a dashed line. E, Nucleotide sequence of the DE35S-loxP-bar fusion of the target cassette and the loxP-nptII fusion of the exchange cassette. Upon cassette exchange, nptII expression is initiated at the transcription and translation signals of the DE35S promoter, located outside the cassette. Restriction endonuclease recognition sites used for cloning are indicated. loxP sites are framed and their orientation is indicated with an arrow. The translational start codon ATG at the NcoI site for bar and nptII is framed.
We tested two versions of the exchange T-DNA. Plasmid pSDM3327 contains a T-DNA with a simple RMCE cassette (Fig. 1B) flanked by a loxP and a lox5171 site. Using this T-DNA vector, RMCE will be the result of simultaneous loxP/loxP and lox5171/lox5171 recombination between the target locus and exchange T-DNA. We anticipated that simultaneous recombination would be an infrequent event, resulting in chromosome breakage or translocation events, respectively. Therefore, we inserted an additional loxP site in direct orientation with the loxP site near the right-border repeat, resulting in plasmid pSDM3732 (Fig. 1C). This would allow efficient Cre-mediated circularization of the T-DNA, providing a circular lox substrate that may first integrate at the target site by a single loxP/loxP or lox5171/lox5171 recombination step (one of two possible intermediates is depicted in Fig. 1C). In a second step, Cre may resolve the integrated product, resulting in a precise selectable exchange (Fig. 1D). In the case of T-DNA with the simple RMCE cassette, T-DNA border ligation may result in a similar circular substrate, but probably this does not occur frequently (Bundock et al., 1995; Vergunst and Hooykaas, 1998; Zhao et al., 2003). Here, we used a cotransformation approach to introduce Cre recombinase in the same cell as the exchange T-DNAs by means of an Agrobacterium strain harboring a T-DNA vector containing a strong expression unit for the cre gene.
Target Plant Line
Arabidopsis ecotype C24 root explants were cocultivated with LBA1100 (pSDM3164). Several target lines were selected and the transgenic plants were screened for the presence of single-copy nontruncated inserts using the outermost XhoI site close to the right-border repeat of the T-DNA for identification. One single-copy line containing the full T-DNA insertion with the XhoI site was chosen for further study. Thermal asymmetric interlaced (TAIL)-PCR was performed and sequence analysis showed that the T-DNA was inserted in the twelfth intron of the annotated gene At5g49570.1. The left-border repeat had remained intact, except for the outermost two G-residues. Of the right-border repeat, only two G-residues remained. Based on the sequence data, we designed primer r4, which is complementary to a short stretch of plant DNA sequence, the T-DNA junction, and the XhoI site of the T-DNA for later analysis of RMCE events (Fig. 1, A and D).
Exchange T-DNA as a Circular Substrate for Cre
Single-stranded T-DNA can become double stranded extrachromosomally to undergo RMCE with the target locus directly. However, it is also possible that T-DNA integrates randomly prior to RMCE with the target locus. In both cases, simultaneous loxP/loxP and lox1571/lox1571 recombination events are required. We were interested in whether circularization could increase RMCE efficiency. To test whether circularization of exchange T-DNA pSDM3732 occurred in planta, the following experiment was performed. Arabidopsis C24 roots were transformed with A. tumefaciens carrying exchange T-DNA pSDM3732 in the presence of a cre-delivering strain. After 1 d of cocultivation, roots were harvested and chromosomal DNA was isolated. Cre-mediated recombination at the loxP sites in pSDM3732 will result in a circular molecule that can be detected using primers n1 and t3 (Fig. 1C). A 0.93-kb fragment was detected and directly sequenced. The fragment contained the expected sequences for precisely recombined loxP sites (and not border fusion or tandem T-DNA structures), showing that indeed Cre-mediated circle formation occurs.
Stability of the Exchange DNA in the Presence of Cre
The exchange cassette contains the loxP site and the heterospecific mutant lox site 5171 in inverted orientation. Initially, Lee and Saito (1998) reported that there was no detectable cross-recombination between these two sites. Later, a low level of recombination between these sites was observed using a sensitive assay in Escherichia coli (Siegel et al., 2001). We observed some instability of the exchange T-DNA when present in A. tumefaciens containing a cre expression plasmid (data not shown). To avoid unsolicited recombination of the exchange vectors in Agrobacterium prior to T-DNA transfer, we performed cocultivations with target plant material using two Agrobacterium strains: one providing the exchange cassette and one providing the cre expression unit.
RMCE Experiments
Roots from homozygous (BB) or hemizygous (B−) target plants were cocultivated with two Agrobacterium strains: the first carrying one of the two exchange T-DNAs and the second harboring binary vectors with a cre cassette driven either by the nopaline synthase (nos) promoter or the BigMac promoter as indicated in Table I. Control experiments were performed with the exchange T-DNA-providing strain only.
Table I.
Efficiency of RMCE after transformation of Arabidopsis target plant 3164 with Cre and exchange T-DNA constructs
| Experiment | Target Plant bar Locusa | Cre Constructb | Exchange T-DNA Construct | No. of Root Explantsc | No. of Kanamycin- Resistant Calli | RMCE Eventsd | RMCE per No. of Kanamycin-Resistant Callie |
|---|---|---|---|---|---|---|---|
| 1 | BB | nos-cre | 3327 | 1,557 | 6 | 3 | 0.50 |
| 2 | BB | nos-cre | 3327 | 1,363 | 4 | 1 | 0.25 |
| 3 | BB | nos-cre | 3327 | 847 | 5 | 3 | 0.60 |
| bm-cre | 3327 | 860 | 3 | 2 | 0.67 | ||
| 4 | B− | nos-cre | 3327 | 664 | 5 | 2 | 0.40 |
| bm-cre | 3327 | 755 | 1 | 1 | 1.00 | ||
| 5 | B− | nos-cre | 3327 | 1,725 | 8 | 4 | 0.50 |
| nos-cre | 3732 | 1,705 | 8 | 3 | 0.38 | ||
| 6 | B− | nos-cre | 3327 | 1,080 | 6 | 2 | 0.33 |
| bm-cre | 3327 | 977 | 2 | 0 | 0 | ||
| nos-cre | 3732 | 1,030 | 2 | 1 | 0.50 | ||
| bm-cre | 3732 | 1,021 | 0 | 0 | 0 |
Hemizygous (B−) or homozygous (BB) plant material.
bm, BigMac promoter.
Roots were cut into explants of 2 to 3 mm in size; up to 500 explants were plated per petri dish.
RMCE events determined by PCR using primers p1 and r4.
RMCE events per no. of kanamycin-resistant calli.
In six independent RMCE experiments (Table I), a total of 50 kanamycin-resistant calli were obtained (ranging from 1/132 to 1/755 calli/root explant). When Cre was not provided during cocultivation, similar numbers of kanamycin-resistant calli were, however, obtained (data not shown). This suggests that translational fusions of the nptII gene with endogenous plant DNA sequences were formed, even though in the exchange T-DNA the nptII coding region is preceded by translational stop codons in all frames and the nos terminator signal to avoid such fusions. These fusions might have been the result of infrequent integration of exchange T-DNAs in which the left border had been truncated, resulting in loss of the stop codons. Plants that were regenerated from these control calli, however, had progeny that grew very poorly on kanamycin-containing medium, suggesting that the nptII expression level was low or cell type specific. In contrast, progeny of putative recombinant plants grew well on kanamycin-containing medium.
For comparison, random integration of a pDE35S-loxP-nptII control T-DNA after cocultivation with LBA1100 (pSDM3066) was obtained with an efficiency varying from 0.5 to 1 callus per root explant (data not shown).
PCR Analysis Indicates a High Percentage of RMCE
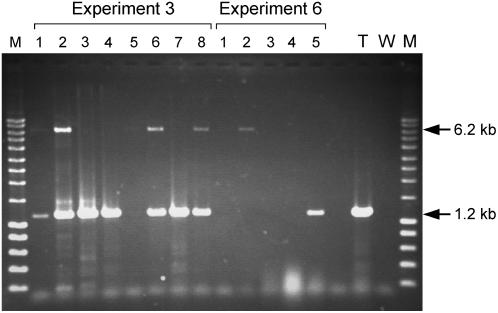
To identify loxP crossover events, chromosomal DNA of kanamycin-resistant calli was analyzed by PCR using primer set p1 (annealing to the DE35S promoter sequence of the target site) and n1 (annealing to the nptII sequence of the exchange cassette; Fig. 1D). Kanamycin-resistant calli derived from cocultivation experiments with the exchange vector in the absence of Cre did not contain the DE35S-loxP-nptII junction, but 37 (74%) of the kanamycin-resistant calli derived from cocultivations in the presence of Cre amplified a DE35S-loxP-nptII-specific junction indicative of site-specific recombination (data not shown). Subsequently, DNA samples in which a DE35S-loxP-nptII junction had been detected were screened for the presence of the lox5171 junction using primer g1 (annealing to the nptII-gus exchange cassette) and primer r4 (the right-border junction of the target DNA; Fig. 1D). Thirty-three of 37 (89%) samples were positive for the lox5171 junction (data not shown), suggesting correct RMCE had occurred in these calli. To confirm that both recombinant lox junctions were physically linked, a PCR reaction with primer set p1/r4 was conducted. PCR conditions were optimized with respect to primers, polymerase, Mg2+ concentration, and amount of template to detect the 6.2-kb fragment indicative of RMCE. The 6.2-kb RMCE-PCR product was detected in 22 (67%) of 33 calli for which both loxP and lox5171 crossover sites were detected. Difficulties with RMCE-PCR may have resulted in underestimation of the actual number of positive reactions. Figure 2 shows a gel of a typical PCR experiment in which several samples scored negative for RMCE-PCR (experiment 3, lane 5; experiment 6, lanes 1 and 4), but were confirmed positive using different DNA concentrations in later PCR reactions. In experiments that used homozygous target plants, a 1.2-kb PCR fragment is also expected if only one of the two alleles has undergone RMCE (Fig. 2, experiment 3). For most of the calli derived from hemizygous starting material, both a 1.2- and a 6.2-kb fragment were, however, also observed. This indicated that the DNA material used for PCR was likely obtained from chimeric callus that was derived from both target and recombinant cells. Summarizing, our PCR data suggest that in 44% (22 of 50) of the kanamycin-resistant calli identified, an RMCE event at the target locus had occurred.
Figure 2.
PCR analysis to detect correct RMCE. Shown is the ethidium bromide-stained gel from a typical RMCE-PCR analysis with primers p1 and r4, detecting the bar-target cassette (1.2 kb) and the exchanged cassette (6.2 kb) in chromosomal DNA of kanamycin-resistant calli of experiments 3 and 6. M, Molecular weight marker (1 kb; Smartladder, Eurogentec); T, callus of target plant; W, water control. Sample 7 is derived from cocultivation in the absence of Cre.
RMCE Events Detected by Southern-Blot Analysis
The majority of kanamycin-resistant calli regenerated plants (R1). Offspring of six R1 plants obtained from independent calli were further subjected to extensive Southern-blot analysis. These R1 plants include the numbers 51N2, 51N3, 51N5, 52N1, 52N3, and 41B, in which the first identifier refers to the experiment, the second identifier indicates the used exchange cassette (1 = pSDM3327; 2 = pSDM3732), N indicates the use of nos-cre, B indicates cotransformation with BigMac-cre, and, when more than one callus was identified, the last identifier indicates the callus number from which the plants originated. All six R1 plants originated from transformation experiments using hemizygous target plants so as not to further complicate Southern analysis. PCR had indicated that an RMCE event had occurred as demonstrated by the 6.2-kb fragment amplified with primers p1 and r4, except for 52N1, which showed the presence of both loxP and lox5171 junctions, but failed to show the correct RMCE fragment. We included this line to be able to confirm our PCR data. Seeds from the R1 plants were germinated on medium containing kanamycin. Their progeny segregated in a ratio of 3:1 (resistant:sensitive) as expected. Progeny plants were diploid, as determined by flow cytometry. Kanamycin-resistant R1 progeny were grown further and analyzed independently by Southern-blot analysis to be able to detect segregation of possibly integrated random exchange cassette or cre DNA sequences.
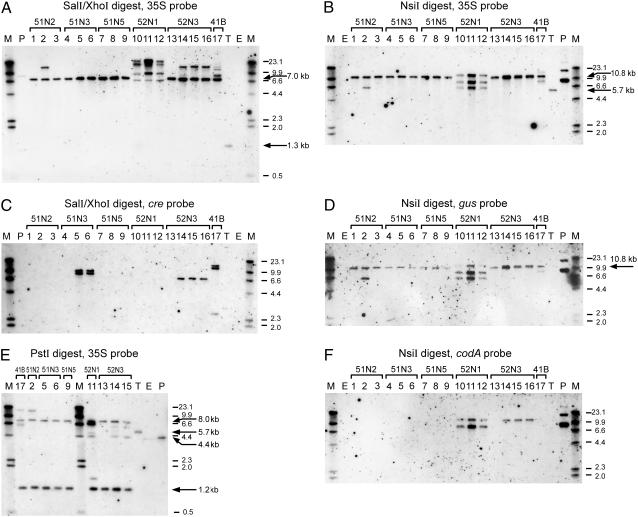
Correct RMCE is detected by a 7.0-kb SalI-XhoI fragment (Fig. 1D) hybridizing with the 35S probe. This product was found for all analyzed offspring of five lines (Fig. 3A), but not for 52N1 as expected from the PCR results. For the target cassette, this SalI-XhoI digest yielded the expected 1.3-kb SalI fragment (Figs. 1A and 3A, lane T). RMCE was further confirmed with an NsiI digest hybridized with the 35S probe (Fig. 3B). Digestion of target plant DNA with NsiI yields a 5.7-kb fragment (Figs. 1 and 3B, lane T). Accurate RMCE will thus yield a 10.8-kb NsiI fragment (Fig. 1D), which was indeed detected for all lines except 52N1. Hybridization of the NsiI blot with the gus probe (Fig. 3D) cemented our conclusion that correct RMCE had occurred in the five lines that were preselected as RMCE events based on PCR; similar banding patterns were observed as in the NsiI/35S blot (Fig. 3B), except for the target line (lane T) in which the 5.7-kb 35S fragment did not hybridize with the gus probe as expected, and in lane 17 (41B) in which the largest fragment detected in the NsiI/35S blot did not hybridize with the gus probe (Fig. 3D). This larger fragment originates from a random insertion of the pBigMac-cre construct pSDM3088 in 41B, which is detected using a 35S probe due to cross-hybridization with the BigMac promoter. In all other lines, the nos-cre construct was used during transformation and not detected using a 35S probe.
Figure 3.
Southern-blot analyses of putative RMCE lines. Chromosomal DNA of target plant (T), untransformed control ecotype C24 (E), and plants of putative RMCE lines derived from experiments 4 and 5 was either digested with NsiI (B, D, and F) or PstI (E) or double digested with SalI and XhoI (A and C), and hybridized with 35S (A, B, and E), cre (C), gus (D), or codA (F) probes, respectively. Codes above the horizontal brackets indicate the name of the parent plant line of which the individuals in the lanes are derived. Numbering of the individuals is consistently used for all blots. The name of the line indicates the following. First identifier, Experiment number; second identifier, 1 for exchange T-DNA 3327 or 2 for T-DNA 3732; third identifier, N for T-DNA containing pnos-cre or B for pBigmac-cre. When more than one callus was selected, the last identifier indicates the callus number of which the plants originated. Individual lanes underneath a bracket show different kanamycin-resistant progeny plants from a single regenerated R1 shoot. Lane P contains digested plasmid DNA of exchange construct pSDM3327 mixed with digested C24 chromosomal DNA; M, DIG-labeled molecular weight marker. Arrows indicate the expected RMCE fragment 7.0 kb and the target fragment 1.3 kb (A); the expected RMCE fragment 10.8 kb and the target fragment 5.7 kb (B); the expected 10.8-kb RMCE fragment (D); and CE fragments of 1.2 kb and 8.0 kb, the target fragment 5.7 kb, and the 4.4-kb fragment specific for exchange T-DNA, not recombined at the lox5171 site (E).
A PstI digest was hybridized with the 35S probe that allows the simultaneous detection of both the loxP (1.2 kb) and lox5171 (8.0 kb) recombinant junctions, although the presence of both fragments does not confirm they are physically linked, as already shown in previous blots (Figs. 1D and 3E). In the target line, a 5.7-kb fragment was found as expected. Indeed, all lines showed evidence of correct recombination of both junctions. Interestingly, line 52N1, in which we were unable to detect a correct RMCE fragment, did contain both recombinant junctions, confirming earlier PCR analysis. Summarizing, these data clearly show that the five R1 plants identified on the basis of RMCE-PCR are indeed the result of correct RMCE events.
Analysis of Additional T-DNA Integration Events
We analyzed the presence of additional random insertions for the five lines that resulted from a correct RMCE event. Southern-blot analysis revealed that in three of five analyzed recombination events (51N3, 52N3, and 41B), the cre gene, which was cotransformed with the exchange cassette, was also integrated in the plant genome (Fig. 3C). The cre DNA sequence does not contain SalI or XhoI sites, allowing us to estimate the number of T-DNA insertions. For lines 51N3 (two insertions) and 52N3 (one insertion), the cre insertions were able to segregate away from the RMCE locus (Fig. 3C, lane 4 of line 51N3 and lane 13 of line 52N3). We cannot comment at this stage on segregation of the cre gene for line 41B because we only analyzed one offspring plant for this recombinant. Our data clearly show that a strategy with transiently provided Cre on a cotransforming T-DNA is effective and can result in easy identification of stable RMCE plants.
Random integration of the exchange T-DNA was examined by hybridization of the NsiI blot with a codA probe (Fig. 3F). Random integration of complete exchange T-DNAs of pSDM3327 or pSDM3732 will result in detection of an 8.2-kb fragment (Fig. 1B). In these experiments, we did not apply selection against expression of codA. Surprisingly, only one of the five lines showed a signal when hybridized with the codA probe. For line 52N3 in three of the four analyzed R2 plants, a codA hybridizing fragment coincidently of similar size as the 10.8-kb RMCE fragment (Fig. 3, B and D) was detected, indicating that exchange T-DNA had inserted also randomly. The fact that the fragment is not of the expected size can easily be explained by truncation of the T-DNA, resulting in the loss of an NsiI site at one of the distal ends of the T-DNA. SalI/XhoI and PstI digests allowed simultaneous visualization of RMCE and random inserts in this line 52N3 using a 35S probe. In agreement with the codA blot, in the same offspring plant of 52N3 (lane 13 in all blots), no random fragment is detected (Fig. 3, A and E). Figure 3C shows that the cre gene is also absent in this offspring plant, resulting in a clean single-copy RMCE event. For lines 51N2 and 41B, however, additional hybridizing fragments were detected using 35S and gus probes. 51N2 contains one additional insert that hybridizes with gus and 35S probes (SalI/XhoI, NsiI, and PstI digest). This T-DNA could clearly segregate from the RMCE locus because it was only detected in one of the three analyzed plants, again resulting in a clean single-copy RMCE recombinant. 41B is more difficult to interpret because the 35S probe hybridizes with the RMCE fragment, with two copies of the randomly integrated cre gene (Fig. 3C) as well as the randomly integrated exchange cassette. However, the data also show that one additional copy of the exchange cassette had integrated as deduced from the blot hybridized with a gus probe. The absence of codA and fragment sizes that differ from the expected sizes for random integration is consistent with integration of truncated T-DNAs of the exchange vector in 51N2, 52N3, and 41B.
Summarizing, line 51N5 was the result of a clean RMCE event, with no additional insertions of cre or exchange vector DNA. Line 51N3 contained additional cre insertions that could be segregated away easily. Similarly, recombinant 51N2 contained an additional random T-DNA insert that was lost in part of the offspring. Although we detected random insertions of cre and exchange vector in plants of lines 41B and 52N3, clean RMCE offspring could easily be obtained for such lines as shown for 52N3.
DISCUSSION
Transgene integration in plants, independent of the transformation method used, is beyond our control and takes place at random, unknown positions in the genome. Because directed integration by homologous recombination is extremely inefficient, site-specific recombination strategies have been developed to target transgenes to predetermined genomic locations. Targeted integration is a useful tool to eliminate transgene instability and variation in gene expression caused by transgene integration position and integration patterns and provides a means to obtain stable transgenic plants. The development of efficient methods for site-specific integration, using different protocols for transformation, is important for both fundamental research and crop improvement. Several groups have shown that, indeed, targeting to specific predetermined genomic locations, both in mammalian cells as well as in plants, resulted in reproducible gene expression levels (Fukushige and Sauer, 1992; Feng et al., 1999; Srivastava et al., 2004; Chawla et al., 2006). Chawla et al. (2006) showed that consistent gene expression levels were obtained in single-copy rice lines during several generations and that an allelic gene dosage effect nearly doubled gene expression levels. An additional advantage of directed integration is the elimination of unwanted mutations due to random insertion of transgenes in essential genes or essential genomic regions. Here, we provide evidence for successful targeting using a Cre-based RMCE approach. This strategy uses Agrobacterium-mediated transformation and is based on selection for kanamycin resistance as a result of precise exchange of a T-DNA cassette with a genomic target cassette that was placed in the genome via a first round of transformation. Introduction of target sites at particular genomic locations, for instance, via homologous recombination, is difficult. However, randomly introduced good target sites can be preselected based on expression levels and analysis of the integration site, allowing efficient future targeting to such a site.
The natural gene delivery system of Agrobacterium is a preferred method for the transformation of a wide range of plants. As a consequence of the highly efficient random T-DNA integration process, a tight selection system is required for identification of site-specific, single-copy integration events. Here, we used a promoter trap strategy that targets a promoterless nptII marker that will become expressed after specific integration downstream of a promoter sequence present at the target site. Stop codons in three reading frames were introduced upstream of the lox-nptII coding region to further diminish selection of unwanted random integration events as a result of fusion to endogenous DNA promoter sequences. In several small-scale RMCE experiments (each up to four plates with small, excised root explants) in which cre T-DNA was provided by a second Agrobacterium strain, we identified a total of 50 kanamycin-resistant calli. Using PCR analysis, which detected the fully exchanged cassette, we showed that 44% of these kanamycin-resistant calli carried putative RMCE events, indicating the stringency of the selection procedure. The recombinant nature of offspring obtained from five of these events was confirmed by Southern analysis.
Different site-specific recombination systems, namely, R/RS and Cre/lox, have been used in plants to obtain site-directed integration. Integration at a single genomic recombination site in the genome requires circular double-stranded DNA also harboring a single compatible recombination site. Plasmid DNA, or any double-stranded DNA that can be circularized upon entry in the host cell by a recombinase-mediated reaction to prevent integration of vector sequences (Srivastava and Ow, 2002), is a good substrate to be delivered via direct DNA transfer methods. A protoplast-based transformation protocol for tobacco has been used successfully for site-specific integration (Albert et al., 1995); however, protoplast-dependent transformation protocols are not widely applicable. Particle bombardment is a preferred method for transformation of cereals (Christou, 1995) and site-specific integration in rice using biolistics has been successful using a combination of mutant lox sites and disruption of cre expression (Srivastava and Ow, 2002; Srivastava et al., 2004). The single-stranded linear T-DNA transferred by Agrobacterium, however, is in principle a poor substrate for a site-specific integration reaction. In addition, Agrobacterium T-DNA has evolved to integrate highly efficiently into the genome at random positions. It is therefore not surprising that direct DNA transfer methods result in much higher ratios of site-specific to random integration (Albert et al., 1995) than A. tumefaciens-based methods. The efficiency of RMCE in this study, compared to random integration of control T-DNA, was on the order of 0.3%. However, we detected total numbers of integrants at the same order of magnitude as that described after bombardment of rice (Srivastava and Ow, 2002; Srivastava et al., 2004). Nanto et al. (2005) and Nanto and Ebinuma (2007) applied an RMCE strategy in plants and introduced an exchange T-DNA cassette using the R/RS recombinase system in tobacco. These authors used a completely different strategy than reported here, which was based rather on the accumulation of exchange events after a prolonged time of selection for rooted shoots (up to 6 months). Whereas here we used a promoter trap strategy to preferentially select kanamycin-resistant calli containing recombination events that occurred at the target site only, Nanto and colleagues used an hpt expression cassette on the exchange T-DNA, also allowing the selection of hygromycin-resistant calli due to random integration events. Due to the high efficiency of Agrobacterium-mediated random T-DNA integration, RMCE in this case is therefore most likely due to exchange with prior randomly integrated copies. The rationale for identification of single-copy RMCE events was the specific placement of three identical RS sites on the exchange T-DNA, containing the RS-flanked hpt exchange cassette and the RS-flanked R and ipt genes, combined with the use of the ipt gene as a negative selectable marker. RMCE events were detected among rooting shoots with a normal phenotype, which indicated the R-mediated deletion of the ipt and linked R gene, as well as nontruncated random events. Comparison of the efficiency of different existing protocols for site-specific integration is difficult due to the use of different strategies, DNA transfer methods, and selection procedures. The high efficiency of the Agrobacterium transfer method and the ease of the procedure, combined with a stringent selection system, make our Agrobacterium-RMCE-based site-specific integration method a very simple and practical procedure.
Despite the fact that T-DNA may not be a perfect substrate for site-specific integration, it was shown that A. tumefaciens T-DNA could be successfully targeted to a single genomic lox site (Vergunst and Hooykaas, 1998; Vergunst et al., 1998b) or via RMCE (Nanto et al., 2005; Nanto and Ebinuma, 2007; this article). The single-stranded nature of T-DNA may not be a problem because it was demonstrated that single-stranded T-DNA can become double stranded extrachromosomally and genes located on T-DNA become expressed and recombine efficiently among each other (Offringa et al., 1990; Narasimhulu et al., 1996; Tzvira et al., 2003; Marillonnet et al., 2004). In addition, efficient selection of T-DNA targeted to a single genomic lox site was obtained by placing an additional lox site on the T-DNA integration vector to allow Cre-mediated circularization of the T-DNA prior to site-specific integration (Vergunst et al., 1998b). This circularization could have occurred prior to or after random integration. RMCE, in principle, does not require circularization of the exchange cassette. We anticipated that in an RMCE strategy circle formation prior to exchange might also be advantageous. Therefore, we tested whether placing an additional loxP site adjacent to the opposite T-DNA border sequence to allow circularization prior to or after random integration of the exchange T-DNA would improve RMCE frequency. Sequence analysis showed that correct circularization at the loxP sites in the exchange DNA after cotransformation with cre T-DNA to wild-type root explants had occurred. However, comparison of the efficiencies of RMCE after transformation with an exchange T-DNA with or without an additional loxP site for circularization did not show any difference (Table I, experiments 5 and 6). Circle formation in this strategy, therefore, did not improve RMCE frequency.
Site-specific integration strategies, using any transformation method, require that recombinase activity is controlled in some way, which can be combined with the use of specific mutant lox sites to prevent reversible recombination events. A single lox-targeting strategy involving displacement of cre from its promoter turned out to be insufficient to completely stabilize T-DNA targeting events (Vergunst et al., 1998b). To increase stability of integrants, here, Cre was provided from cotransferred T-DNA, combined with an RMCE strategy using incompatible loxP and lox5171 sites (Lee and Saito, 1998) that flanked both the target cassette and the exchange cassette in inverted orientation. Siegel et al. (2001) showed, however, that incompatibility between loxP and lox5171 sites was not complete and we indeed observed some instability between the loxP and lox5171 sites of the exchange cassette in the presence of Cre in A. tumefaciens and E. coli. Therefore, we physically separated the targeting construct and the cre expression construct by using two Agrobacterium strains to deliver T-DNAs. Highly efficient delivery of two different T-DNAs into the same cell has been described. In Nicotiana benthamiana, cotransfer efficiencies of up to 90% were detected in infiltrated leaves using nonselectable markers (Marillonnet et al., 2004). De Buck and colleagues showed, using a different approach in Arabidopsis, that in 50% of selected transformants a second T-DNA was transiently expressed (De Buck et al., 2000). Here, genuine cassette exchange was mediated by transiently expressed cre T-DNA. From detailed Southern-blot analysis of five RMCE events, we concluded that two of these lines indeed contained the correctly recombined exchange cassette without additional cre sequences inserted in the genome. For three other RMCE events, the cre gene was detected in the genome, but in at least two of these lines, cre segregated independently from the RMCE locus in the offspring. This allowed recovery of RMCE plants free of cre sequences.
In our strategy, we included the possibility of selecting against random integration of the exchange T-DNA by placing the codA gene adjacent to the exchange cassette. Selection for absence of codA was, however, not applied because limited numbers of kanamycin-resistant calli were already obtained. An advantage of omitting selection for loss of codA was that an offspring plant with a clean single-copy RMCE event could now be selected after segregation of codA sequences in the offspring, which otherwise would have been lost. Unexpectedly, two lines (51N2 and 41B) that did not contain codA sequences contained randomly integrated DNA shown by detection with gus and 35S probes. It might be that two copies of the exchange T-DNA integrated as an inverted repeat linked at their right borders. Excisional recombination by Cre would then lead to loss of both codA sequences. It is difficult to establish whether RMCE occurred directly with extrachromosomal T-DNA that had become double stranded or with randomly integrated copies of the exchange T-DNA. Any footprints, such as codA sequences, that may be left after prior random integration may equally have been the result of integration after extrachromosomal recombination of T-DNA.
In summary, among the five analyzed putative RMCE events, one clean RMCE event without additional cre or exchange T-DNA insertions (51N5) was obtained. In three other RMCE events, the randomly integrated T-DNA sequences easily segregated, resulting in clean RMCE events. The simplicity of our RMCE strategy, the stringency of selection, and the ease of detection of RMCE by PCR provide good potential to select target lines with high expression loci and to target transgenes to predetermined genomic positions to improve the repeatability of transgene expression levels.
MATERIALS AND METHODS
Bacterial Strains
Agrobacterium tumefaciens strain LBA1100 (C58C1 with a disarmed octopine-type pTiB6 plasmid; Beijersbergen et al., 1992) was used for RMCE and plant transformation experiments. Escherichia coli DH5α was used for cloning.
Recombinant DNA Techniques
Standard cloning techniques were carried out according to Sambrook et al. (1989). Restriction enzymes were purchased from New England Biolabs. Bacteria were grown in Luria culture medium (10 g L−1 bacto-tryptone, 5 g L−1 yeast extract, 8 g L−1 NaCl), with appropriate antibiotics. Antibiotics (Duchefa Biochemie BV) were used at concentrations of 100 mg L−1 kanamycin, 20 mg L−1 rifampicin, 250 mg L−1 spectinomycin, and 40 mg L−1 gentamycin for A. tumefaciens. For selection in E. coli, 100 mg L−1 carbenicillin, 10 mg L−1 gentamycin, or 25 mg L−1 kanamycin were used.
Plasmid Constructs
Cre expression vector pSDM3088 (pBigMac-cre) was described previously (Vergunst and Hooykaas, 1998) and contains the cre gene driven by the strong BigMac promoter placed between the T-DNA border repeats. A second cre expression vector, pSDM3021 (pnos-cre), was made as follows. An XhoI-EcoRI fragment containing a mannopine synthetase (mas) termination signal and a polylinker containing a multiple cloning site (MCS; 5′-GTCGACAAGCTTGGGAAGATCTAGTACTTTTGGGGTACCCCGCTCTAGAGCGAATTC [SalI and EcoRI sites underlined]) were inserted in the XhoI/SalI-digested pUC21. The nos promoter region of the 35S-lox-cre target plasmid (pSDM3110) described earlier (Vergunst et al., 1998b) was then inserted in the BglII/HindIII sites of the MCS, resulting in pSDM3018. The nos-MCS-mas cassette was cloned between the T-DNA borders of binary vector pSDM14 (Offringa, 1992) as a SalI-XhoI fragment, resulting in pSDM3019. Finally, the cre coding region of pUC19CRE (Mozo and Hooykaas, 1992) was cloned in the EcoRI/XbaI sites of the MCS of pSDM3019 after the SphI site that created an out-of-frame ATG just upstream of the ATG start codon in pUC19CRE had been removed (Klenow fragment of T4 DNA polymerase) via a subcloning step in pIC19H. This resulted in pSDM3021, harboring a pnos-cre-tmas cassette.
The structure of the T-DNA of vector pSDM3164 that was used to produce the target line is drawn in Figure 1A. The sequence of the DE35S-loxP-bar translational fusion is depicted in Figure 1E (top) and was constructed from the following sequences: As a source for the promoter-ATG-loxP fusion, we used a fragment of the p35S-ATG-lox-npt control vector (Vergunst and Hooykaas, 1998), which contains the CaMV 35S promoter sequence with a double enhancer and alfalfa mosaic virus (AMV) 5′-leader sequence (DE35S) transcriptionally fused to loxP. To be able to create an in-frame translational fusion with a bar-tml sequence present on a (partial) BglII fragment of excision vector pSDM3043 (Vergunst and Hooykaas, 1998), a BglII linker was added first that resulted in a frame shift and an additional ClaI site. This yielded the DE35S-loxP-bar-tml construct pSDM3145. Two oligos representing the lox5171 site (Lee and Saito, 1998) were annealed and subcloned as an XbaI-StuI fragment into pUC28 (pSDM3150), followed by ligation into vector pSDM3145, resulting in pSDM3162. Finally, the DE35S-loxP-bar-tml-lox5171 construct was cloned between the right- and left-border repeats of binary vector pSDM14 (Offringa, 1992), yielding pSDM3164.
The structure of the exchange T-DNA of vector pSDM3327 is depicted in Figure 1B, which was constructed from the following DNA fragments: A loxP site was cloned via multiple cloning steps to the nptII open reading frame devoid of the start codon (Fig. 1E, bottom). nptII with the octopine synthase transcription termination sequence originated from pSDM56 (De Groot, 1992). The nos transcription terminator region of pSDM3322 was introduced as an EcoR1-SacI fragment and further cloned to binary vector pSDM3014, which is a derivative of pSDM14 in which the NotI site was deleted by filling in the site using T4-DNA polymerase. To introduce three stop codons, a BamHI-KpnI linker was placed in the MCS preceding the nos terminator region yielding sequence 5′-GTCGACGGATCCTAGTCTAGACTAGGTACC in construct pSDM3325. The lox5171 site of pSDM3150 was cloned to the codA gene under control of the figwort mosaic virus 34S promoter with the CaMV 35S 3′-region, a derivative of pSDM5042 (Schlaman and Hooykaas, 1997). The codA expression cassette-lox5171 was cloned as an XhoI-NotI fragment into pSDM3325, yielding pSDM3159. pSDM3330 contains the gus gene under the control of the BigMac promoter and mannopine synthetase (mas) transcription terminator region of pGN7344 (Comai et al., 1990) and was cloned as a NotI fragment into the NotI site of pSDM3159, yielding pSDM3327.
The structure of the exchange T-DNA of vector pSDM3732 is depicted in Figure 1C. An additional loxP site was cloned as a SalI fragment of pMS103 (Snaith et al., 1995) into the XhoI site near the right border of pSDM3327 in direct orientation to the loxP site to allow Cre-mediated circularization of the exchange T-DNA prior to RMCE.
The control T-DNA vector pSDM3066, containing pDE35S-ATG-loxP-nptII, was described previously (Vergunst and Hooykaas, 1998). Target and exchange T-DNA constructs were verified by restriction enzyme analyses and sequencing.
Plant Material and Transformation
Arabidopsis (Arabidopsis thaliana) ecotype C24 and transgenic lines thereof were used. Greenhouse and tissue culture conditions and the Agrobacterium-mediated transformation protocol of Arabidopsis root explants were described by Vergunst et al. (1998a). Selection of target plant lines based on resistance for PPT (Duchefa Biochemie BV) followed the description of Vergunst et al. (1998a). Single-copy transformants were identified on the basis of genetic and Southern-blot analyses and further analyzed for the presence of the complete bar cassette by Southern analysis using a bar probe. One transgenic line was selected and the DNA sequence of the T-DNA insert junction was determined using TAIL-PCR (Liu et al., 1995). The ploidy level of the progeny plants was determined by flow cytometry (Plant Cytometry Services). Homozygous T3 seeds were germinated in liquid medium containing 5 mg/L PPT and roots were excised for use in cassette-exchange root-transformation experiments. To generate hemizygous roots, the homozygous line was back-crossed, using the transgenic line as pollen donor, to the wild type. The resulting seeds were germinated in liquid medium, supplemented with 5 mg/L PPT.
Homozygous or hemizygous root explants were incubated with a mixture of two Agrobacterium strains, LBA1100 (exchange T-DNA) and LBA1100 (Cre-T-DNA) in a 1:1 ratio at a final concentration of OD600 = 0.2, prior to cocultivation for 3 d on callus-inducing medium agar plates (Vergunst et al., 1998a). In control cocultivations in the absence of LBA1100 (Cre-T-DNA), a final concentration of OD600 = 0.1 of LBA1100 (exchange T-DNA) was used.
Root explants, with a length of about 2 to 3 mm, were cultured on shoot-inducing medium with 50 mg/L kanamycin and 100 mg/L timentin (Vergunst et al., 1998a). Parts of the kanamycin-resistant calli were multiplied on callus-inducing medium and used for PCR analysis or for plant regeneration. The ploidy level of regenerated plants was determined using flow cytometry.
The random T-DNA integration frequency was estimated by cocultivation of target roots with LBA1100 (pSDM3066) constitutively expressing nptII in plant cells. The frequency of kanamycin-resistant green calli per root explant was determined 3 weeks after cocultivation. Kanamycin-resistant calli resulting from RMCE experiments were counted after 3 to 5 weeks.
DNA Isolation and Southern Analysis
Chromosomal DNA was isolated from 0.1 to 0.5 g of flower buds or leaves, using a Nucleon PhytoPure plant DNA extraction kit (GE) according to the manufacturer's protocol. Three micrograms of chromosomal DNA were digested with the appropriate restriction enzymes and separated on a 0.6% Tris borate/EDTA agarose gel. Twenty nanograms of digoxigenin (DIG)-labeled DNA molecular weight marker II, λ-DNA digested with HindIII (Roche Diagnostics), was used as a size marker. DNA blotting was performed on positively charged nylon membranes (Roche), and DIG dUTP was incorporated in the probes with the PCR DIG-labeling mix (Roche), according to the manufacturer's recommendations. Hybridization and immunological detection of the DIG signal (Neuhaus-Url and Neuhaus, 1993) was performed using the substrate CDP star (Roche).
PCR Analysis
Calli were cultured for at least 8 weeks on kanamycin-containing medium. Chromosomal DNA was isolated from callus or young leaves using a cetyl-trimethyl-ammonium bromide method according to Lassner et al. (1989). PCR reactions were performed in a T1 thermocycler (Whatman/Biometra) with primers as indicated (Fig. 1). A standard PCR protocol of 30 cycles was used: 1 min at 95°C, 1 min with annealing temperature (TA) of 55°C (or as indicated for specific reactions), and 1 min at 72°C. The reaction mixture (25 mL) contained 25 to 50 ng of template DNA, 0.4 μm of each primer, 200 μm dNTPs, and 0.5 units of RedTaq polymerase (Sigma) with the provided buffer.
PCR analysis to detect recombination site junctions used the following primers (5′-3′): LoxP-PCR, to detect the pDE35S-loxP-nptII junction site in the target locus, used primers p1 (GACGCACAATCCCACTATCCTTCGCAA) and n1 (TGATATTCGGCAAGCAGGCATC) at a TA of 64°C; and Lox5171-PCR, to amplify the gus-lox5171-right-border target plant junction, used primers g1 (CGCTGGACTGGCATGAACTTC) and r4 (CCATGAGTGATTAATAGAAGTCACACCTCGA) preceded by 10 touch-down cycles with a TA 60°C to 55°C, changing 0.5°C per cycle.
Primer t3 (GGTTCTTATAGGGTTTCGCTCATGTGT) was used in combination with n1 at a TA of 64°C to detect circle formation of T-DNA in plant cells.
To detect a 6.2-kb RMCE-specific fragment, the following protocol was developed: The PCR block was preheated to 98°C. A program of 35 cycles was preceded by a period of 4 min at 98°C. Cycles were composed of 45 s at 98°C, 30 s at 63°C, and 4 min at 72°C. The reaction mixture (50 mL) contained 25 to 100 ng of template DNA, 200 μm dNTPs, 7.9 mm MgCl2, 0.5 μm of primers p1 and r4, and 0.6 units Phusion DNA polymerase (Finnzymes) in the provided GC buffer.
To amplify the probes for Southern-blot analysis, several primer sets were used: primers p3 (GTGGGATTGTGCGTCATCCC) and p5 (GGATTGATGTGATAATTCCGATGGAGTC) were used to amplify the 35S probe; primers c3 (ACGCTGGTTAGCACCGCAGG) and c4 (CAGGCGCACCATTGCCCCTG) to amplify the cre probe; and primers CD3 (CGCTTCACCGTTGGGATACG) and p34S1 (CCGGTACAATAATGGGGAGG) for the codA probe; primers gusU (CAGCGAAGAGGCAGTCAACGGGGAA) and gusL (CATTGTTTGCCTCCCTGCTGCGGTT) for the gus probe.
Acknowledgments
We thank P. Hock and M.L. Brittijn for drawing the figures.
This work was supported by the Dutch Technology Foundation Stichting Toegepaste Wetenschappen, Applied Science Division of the Netherlands Organization for Scientific Research, and the Technology Program of the Ministry of Economic Affairs.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Paul J.J. Hooykaas (p.j.j.hooykaas@biology.leidenuniv.nl).
References
- Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7 649–659 [DOI] [PubMed] [Google Scholar]
- Baer A, Bode J (2001) Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr Opin Biotechnol 12 473–480 [DOI] [PubMed] [Google Scholar]
- Bakkeren G, Koukolikov Z, Grimsley N, Hohn B (1989) Recovery of Agrobacterium tumefaciens T-DNA molecules from whole plants early after transfer. Cell 57 847–857 [DOI] [PubMed] [Google Scholar]
- Beijersbergen A, Den Dulk-Ras A, Schilperoort RA, Hooykaas PJJ (1992) Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256 1324–1327 [DOI] [PubMed] [Google Scholar]
- Bouhassira EE, Westerman K, Leboulch P (1997) Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood 90 3332–3344 [PubMed] [Google Scholar]
- Bundock P, Den Dulk-Ras A, Beijersbergen A, Hooykaas PJJ (1995) Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J 14 3206–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla C, Ariza-Nieto M, Wilson AJ, Moore SK, Srivastava V (2006) Transgene expression produced by biolistic-mediated site-specific gene integration is consistently inherited by the subsequent generations. Plant Biotechnol J 4 209–218 [DOI] [PubMed] [Google Scholar]
- Chilton MD, Que Q (2003) Targeted integration of Agrobacterium T-DNA into the tobacco genome at double-strand breaks: new insights in the mechanism of T-DNA integration. Plant Physiol 133 956–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou P (1995) Particle bombardment. Methods Cell Biol 50 375–382 [PubMed] [Google Scholar]
- Citovsky V, Guralnick B, Simon MN, Wall JS (1997) The molecular structure of Agrobacterium VirE2-single-stranded DNA complexes involved in nuclear import. J Mol Biol 271 718–727 [DOI] [PubMed] [Google Scholar]
- Comai L, Moran P, Maslyar D (1990) Novel and useful properties of a chimeric plant promoter combining CAMV-35S and Mas elements. Plant Mol Biol 15 373–381 [DOI] [PubMed] [Google Scholar]
- Corneille S, Lutz K, Svab Z, Maliga P (2001) Efficient elimination of selectable marker genes from the plastid genome by the Cre-lox site-specific recombination system. Plant J 27 171–178 [DOI] [PubMed] [Google Scholar]
- Day CD, Lee E, Kobayashi T, Holappa LD, Albert H, Ow DW (2000) Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev 14 2869–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, De Wilde C, Van Montagy M, Depicker A (2000) Determination of the T-DNA transfer and the T-DNA integration frequencies upon cocultivation of Arabidopsis thaliana root explants. Mol Plant Microbe Interact 13 658–665 [DOI] [PubMed] [Google Scholar]
- De Groot MJA (1992) Studies on homologous recombination in Nicotiana tabacum. PhD thesis. Leiden University, Leiden, The Netherlands
- Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE (1999) Site-specific chromosomal integration in mammalian cells: highly efficient Cre recombinase-mediated cassette exchange. J Mol Biol 292 779–785 [DOI] [PubMed] [Google Scholar]
- Francis KE, Spiker S (2005) Identification of Arabidopsis thaliana transformants without selection reveals a high occurrence of silenced T-DNA integration events. Plant J 41 464–477 [DOI] [PubMed] [Google Scholar]
- Fukushige S, Sauer B (1992) Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA 89 7905–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson L (2003) Cre-lox recombination: Cre-ative tools for plant biotechnology. Trends Biotechnol 21 550–555 [DOI] [PubMed] [Google Scholar]
- Gleave AP, Mitra DS, Mudge SR, Morris BAM (1999) Selectable marker-free transgenic plants without sexual crossing: transient expression of cre recombinase and use of a conditional lethal dominant gene. Plant Mol Biol 40 223–235 [DOI] [PubMed] [Google Scholar]
- Kim SI, Veena, Gelvin SB (2007) Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J 51 779–791 [DOI] [PubMed] [Google Scholar]
- Kohli A, Twyman RM, Abranches R, Wegel E, Stoger E, Christou P (2003) Transgene integration, organization and interaction in plants. Plant Mol Biol 52 247–258 [DOI] [PubMed] [Google Scholar]
- Kopertekh L, Juttner G, Schiemann J (2004) PVX-Cre-mediated marker gene elimination from transgenic plants. Plant Mol Biol 55 491–500 [DOI] [PubMed] [Google Scholar]
- Lassner MW, Peterson P, Yoder JI (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7 116–128 [Google Scholar]
- Lauth M, Spreafico F, Dethleffsen K, Meyer M (2002) Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res 30 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Saito I (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216 55–65 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y (2004) In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA 101 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozo T, Hooykaas PJJ (1992) Design of a novel system for the construction of vectors for Agrobacterium-mediated plant transformation. Mol Gen Genet 236 1–7 [DOI] [PubMed] [Google Scholar]
- Nanto K, Ebinuma H (2007) Marker-free site-specific integration in plants. Transgenic Res [DOI] [PubMed]
- Nanto K, Yamada-Watanabe K, Ebinuma H (2005) Agrobacterium-mediated RMCE approach for gene replacement. Plant Biotechnol J 3 203–214 [DOI] [PubMed] [Google Scholar]
- Narasimhulu SB, Deng XB, Sarria R, Gelvin SB (1996) Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus-Url G, Neuhaus G (1993) The use of the nonradioactive digoxigenin chemiluminescent technology for plant genomic southern blot hybridization: a comparison with radioactivity. Transgenic Res 2 115–120 [Google Scholar]
- Offringa R (1992) Gene targeting in plants using the Agrobacterium vector system. PhD thesis. Leiden University, Leiden, The Netherlands
- Offringa R, De Groot MJA, Haagsman HJ, Does MP, van den Elzen PJM, Hooykaas PJJ (1990) Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium-mediated transformation. EMBO J 9 3077–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow DW (2002) Recombinase-directed plant transformation for the post-genomic era. Plant Mol Biol 48 183–200 [PubMed] [Google Scholar]
- Pansegrau W, Schoumacher F, Hohn B, Lanka E (1993) Site-specific cleavage and joining of single-stranded DNA by virD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci USA 90 11538–11542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach C, Velten J (1991) Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol 17 49–60 [DOI] [PubMed] [Google Scholar]
- Rossi L, Hohn B, Tinland B (1996) Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 93 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schlake T, Bode J (1994) Use of mutated Flp recognition target (frt) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33 12746–12751 [DOI] [PubMed] [Google Scholar]
- Schlaman HRM, Hooykaas PJJ (1997) Effectiveness of the bacterial gene codA encoding cytosine deaminase as a negative selectable marker in Agrobacterium-mediated plant transformation. Plant J 11 1377–1385 [Google Scholar]
- Siegel RW, Jain R, Bradbury A (2001) Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett 505 467–473 [DOI] [PubMed] [Google Scholar]
- Snaith MR, Murray JAH, Boulter CA (1995) Multiple cloning sites carrying loxP and frt recognition sites for the Cre and Flp site-specific recombinases. Gene 166 173–174 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Anderson OD, Ow DW (1999) Single-copy transgenic wheat generated through the resolution of complex integration patterns. Proc Natl Acad Sci USA 96 11117–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Ariza-Nieto M, Wilson AJ (2004) Cre-mediated site-specific gene integration for consistent transgene expression in rice. Plant Biotechnol J 2 169–179 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW (2001) Single-copy primary transformants of maize obtained through the co-introduction of a recombinase-expressing construct. Plant Mol Biol 46 561–566 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW (2002) Biolistic mediated site-specific integration in rice. Mol Breed 8 345–350 [Google Scholar]
- Tinland B, Hohn B, Puchta H (1994) Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc Natl Acad Sci USA 91 8000–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzvira T, Frankman FR, Vaidya M, Citovsky C (2003) Site-specific integration of Agrobacterium T-DNA via double stranded intermediates. Plant Physiol 133 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, De Waal EC, Hooykaas PJJ (1998. a) Root transformation by Agrobacterium tumefaciens. In J Martinez-Zapater, J Salinas, eds, Arabidopsis Protocols. Humana Press, Totowa, NJ, pp 227–244 [DOI] [PubMed]
- Vergunst AC, Hooykaas PJJ (1998) Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol Biol 38 393–406 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Jansen LET, Hooykaas PJJ (1998. b) Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res 26 2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace HAC, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJH (2007) Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128 197–209 [DOI] [PubMed] [Google Scholar]
- Zhao X, Coats I, Fu P, Gordon-Kamm B, Lyznik LA (2003) T-DNA recombination and replication in maize cells. Plant J 33 149–159 [DOI] [PubMed] [Google Scholar]
- Ziemienowicz A, Merkle T, Schoumacher F, Hohn B, Rossi L (2001) Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Moller SG, Chua NH (2001) Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol 19 157–161 [DOI] [PubMed] [Google Scholar]
- Zupan JR, Citovsky V, Zambryski P (1996) Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc Natl Acad Sci USA 93 2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]