Abstract
We present here a vector system to obtain homozygous marker-free transgenic plants without the need of extra handling and within the same time frame as compared to transformation methods in which the marker is not removed. By introducing a germline-specific auto-excision vector containing a cre recombinase gene under the control of a germline-specific promoter, transgenic plants become genetically programmed to lose the marker when its presence is no longer required (i.e. after the initial selection of primary transformants). Using promoters with different germline functionality, two modules of this genetic program were developed. In the first module, the promoter, placed upstream of the cre gene, confers CRE functionality in both the male and the female germline or in the common germline (e.g. floral meristem cells). In the second module, a promoter conferring single germline-specific CRE functionality was introduced upstream of the cre gene. Promoter sequences used in this work are derived from the APETALA1 and SOLO DANCERS genes from Arabidopsis (Arabidopsis thaliana) Columbia-0 conferring common germline and single germline functionality, respectively. Introduction of the genetic program did not reduce transformation efficiency. Marker-free homozygous progeny plants were efficiently obtained, regardless of which promoter was used. In addition, simplification of complex transgene loci was observed.
Selectable marker genes are used in nearly all transformation procedures. They are required for efficient generation of transgenic plants, but serve no purpose once plants have been obtained that are homozygous for the transgene. On the contrary, their continued presence can pose technological problems because it precludes retransformation with the same marker system and can raise safety and public concerns. Already a number of strategies for the removal of the selectable marker after selection exist. Delivery of the transgene and selectable marker via cotransformation and the subsequent segregation of both in the progeny was one of the earliest methods developed (Hare and Chua, 2002; Puchta, 2003; Miki and McHugh, 2004; Darbani et al., 2007). A second category of strategies is based on homologous recombination (Puchta, 2000; Zubko et al., 2000). Although interesting from a scientific point of view, it seems to be a system that, at least at this point, does not work efficiently enough. A third class of strategies is based on recombination reactions catalyzed by site-specific recombinases, which excise any given DNA sequence that is flanked on both sides by the target sequence of the recombinase if oriented in a direct repeat. The best-known recombination systems are CRE/lox from bacteriophage P1 (Hoess et al., 1982; Hoess and Abremski, 1985), FLP/frt from Saccharomyces cerevisiae (Cox, 1983; Senecoff et al., 1985), and R/RS from Zygosaccharomyces rouxii (Araki et al., 1985). This third class of strategies can be subdivided according to the placement of the recombinase gene, either on a vector different from the one containing the transgene and the selectable marker, or on the same vector between the recombination sites. The latter is often referred to as auto-excision. In the first subcategory, the recombinase can be delivered to a transgenic plant by retransformation (Odell et al., 1990; Dale and Ow, 1991; Lyznik et al., 1996), by sexual crosses (Bayley et al., 1992; Russell et al., 1992; Kilby et al., 1995; Kerbach et al., 2005), or by transiently expressing the recombinase (Gleave et al., 1999; Kopertekh et al., 2004a, 2004b; Kopertekh and Schliemann, 2005; Jia et al., 2006). Recently, the company Renessen received U.S. regulatory approval (http://www.aphis.usda.gov/brs/aphisdocs2/04_22901p_com.pdf) for the transgenic line LY038 from which the selectable marker, originally present between tandemly oriented lox sites, was removed through introduction of the cre gene by a sexual cross (Ow, 2007). In the auto-excision strategy, activation of the recombinase can be induced either chemically (Sugita et al., 2000; Zuo et al., 2001; Schaart et al., 2004; Sreekala et al., 2005; Zhang et al., 2006) or by heat shock (Kilby et al., 1995; Hoff et al., 2001; Zhang et al., 2003; Wang et al., 2005; Cuellar et al., 2006). In the available strategies, a lot of extra work has to be invested to obtain marker-free homozygous plants. An additional step to introduce or to activate the recombinase is required, such as retransformation, agroinfiltration, application of a chemical, performing heat shock, etc. Moreover, an extra regeneration step is often necessary, which is labor intensive, lengthens the time in which marker-free plants are obtained, and may introduce somaclonal variation. These limitations impede efficient removal of the selectable marker when its presence becomes superfluous.
In the approach presented here, marker-free transgenic plants are obtained via genetically programmed auto-excision without any extra handling and in the same time frame as compared to conventional transformation protocols in which the marker is not removed. This genetic programming is established by introducing a germline-specific auto-excision vector (GSA) in which a germline-specific promoter is used to control the CRE/lox recombination system. Germline is used as a collective term for those cells of which at least one descendent cell is a progenitor of a gamete and the gamete itself; DNA modification in the germline is thus passed on to the next generation through the gametes. Prior to their introduction in the auto-excision vector, the functionality of candidate promoters was evaluated via a test system. After this evaluation, two promoters with a different germline functionality profile were introduced in the auto-excision vector, resulting in two types of GSA vectors and hence two modules of this genetic program. In the first module, a promoter is used that is functional in the common germline—at this point, descendent cells can both lead to pollen or egg cells (e.g. floral meristem)—or in both germlines. In a second module, we used a single germline-specific promoter. Additionally, the GSA vector contains a counter-selectable marker between the target sites of the site-specific recombinase. The presence of this counter-selectable marker is not necessary to obtain marker-free transgenic plants, but further decreases the effort to obtain them.
Recently, Mlynarova et al. (2006) and Luo et al. (2007) showed that it was possible to remove transgenes (selectable markers and others) efficiently by using an auto-excision vector in which a promoter that was specifically functional during microsporogenesis, in pollen or in seed, was placed upstream of a site-specific recombinase gene. More efficient transmission of the recombined allele to the progeny was observed compared to previously described auto-excision strategies that rely on chemical or physical induction of the recombinase. The results presented here, together with the results obtained by Mlynarova et al. (2006) and Luo et al. (2007), clearly indicate that germline-specific auto-excision is an efficient, flexible, and versatile system to remove selectable markers from transgenic plants.
RESULTS
Testing the Functionality of the Promoters
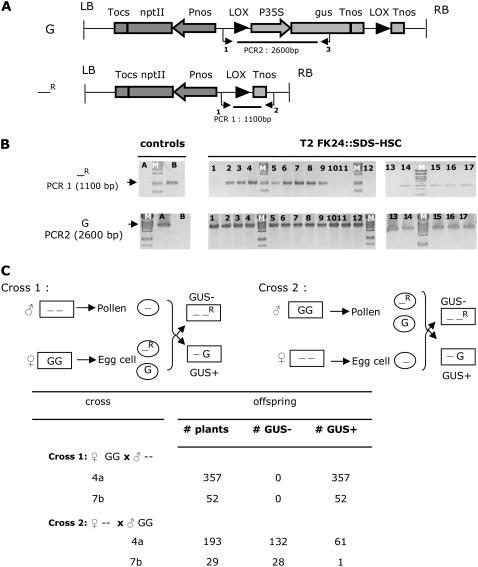
To assess the functionality of the promoters prior to their introduction in the auto-excision vector, we used a transgenic Arabidopsis (Arabidopsis thaliana) line, FK24 (De Buck et al., 2004), which is homozygous for a single insertion of the K2L610 T-DNA (De Buck et al., 1998; Fig. 1A) that contains a p35S-gus chimeric gene between two tandemly oriented lox sites, referred to as a gus allele. This line was supertransformed using the floral-dip method (Clough and Bent, 1998), with promoter-cre fusions after introducing the promoters in the HSC vector (Marjanac et al., 2007), a Gateway-compatible plant transformation vector in which promoters can easily be introduced upstream of the cre gene. A 1.9-kb promoter region of the APETALA1 (AP1; At1g69120) gene and a 2.2-kb promoter fragment of the SOLO DANCERS (SDS; At1g14750) gene of Arabidopsis were used to construct the AP1-HSC and SDS-HSC vectors, respectively. Primary FK24∷AP1-HSC and FK24∷SDS-HSC supertransformants (T1) were obtained after sowing seeds on selective medium for the HSC vector. To analyze the functionality of the promoter in the T1 germline, T2 plants were analyzed by PCR and GUS staining. A first PCR, with primers adjacent to the lox sites, could be used to screen for a recombined allele. A second PCR, with one primer inside and one outside the lox cassette, could be used to check the presence of the original allele (Fig. 1A).
Figure 1.
Determining CRE functionality of the SDS promoter. A, Schematic presentation of the K2L610 T-DNA (De Buck et al., 1998) containing the p35S-gus chimeric gene, referred to as the gus allele (G) and the recombined allele (—R). The FK24 plant line is homozygous for a single-copy insertion of the K2L610 T-DNA. Primers used for PCR analysis are indicated below the constructs. B, PCR results on DNA purified from leaf material of T2 FK24∷SDS-HSC plants (lanes 1–17). Lane A, FK24; lane B, P35S-HSC; lane M, λ-DNA cut with PstI. PCR1, Primer LoxuitKP3 (primer 1) and primer Loxdel2 (primer 2); PCR2, primer LoxuitKP3 (primer 1) and primer GusR (primer 3). C, Reciprocal crossing scheme between a BAR-expressing plant and a homozygous T2 FK24∷SDS-HSC plant to determine in which germline the SDS promoter is functional. Boxed “--”: Genotype of the BAR expressing plant; −: absence of the nonrecombined and recombined allele; boxed “GG”: genotype of the T2 FK24∷SDS-HSC plant containing SDS-HSC; G: gus allele as presented in A; circled “-”: genotype of the gametes formed in the BAR-expressing plant; circled “G”: genotype of the gametes formed in the T2 FK24∷SDS-HSC when no recombination occurred in the respective germline; circled “-R”: genotype of gametes formed in the T2 FK24∷SDS-HSC when recombination occurred in the respective germline; boxed “--R”: recombined allele as presented in A. GUS+ and GUS−, Plants scored as GUS positive and GUS negative after GUS staining. Tnos, Polyadenylation signal of the nopaline synthase gene; Pnos, promoter of the nopaline synthase gene; nptII, neomycin phosphotransferase gene; P35S, CaMV 35S promoter; Tocs, polyadenylation signal of the octopine synthase gene; gus, gus gene.
Of 44 analyzed FK24∷AP1-HSC T2 plants, derived from 15 independent transgenic lines, 23 did not contain an original gus allele anymore (i.e. they were azygous for the gus gene) and 14 contained both a recombined allele and an original allele (heterozygous). The other seven plants were homozygous for the gus allele (data not shown). These results are consistent with expression data obtained by RNA in situ hybridizations and GUS reporter analyses, which indicated that the AP1 promoter is uniformly active in young flower primordia (Mandel et al., 1992; Hempel et al., 1997). It is therefore expected that male and female gametes containing a recombined allele are formed, resulting in azygous T2 progeny plants.
RNA in situ hybridization data showed that the SDS gene was transcribed both in male and female meiocytes. Additionally, reverse transcription-PCR data showed the presence of SDS mRNA in young floral buds, but not in roots, leaves, floral stems, old floral buds, and open flowers, indicating that the SDS gene is expressed only in meiocytes (Azumi et al., 2002). After PCR analysis of 92 T2 FK24∷SDS-HSC offspring plants derived from 29 independent transgenic T1 lines, no plants azygous for the gus allele were obtained, whereas 69 plants contained a recombined allele and an original allele (Fig. 1B). GUS staining on leaves from T2 plants containing a recombined allele and an original allele were consistent with the heterozygous presence of the gus allele as the leaves stained blue. Analysis of T3 plants by PCR, GUS staining, and Southern blot showed that the recombined allele of the T2 generation was transmitted to the T3 generation as T3 azygous plants were found (Supplemental Figs. S1 and S2). These results led to the hypothesis that functionality of the SDS promoter was restricted to one germline. From some T2 plants, which were scored as heterozygous via PCR, no azygous T3 plants were obtained. Southern-blot analysis on DNA prepared from those T2 plants did not result in a band indicative for a recombined allele in contrast to PCR analysis (Supplemental Fig. S2). We therefore think that, in those cases, the PCR band indicative for the recombined allele was caused by background somatic excision in a small number of leaf cells.
To confirm the hypothesis that the functionality of the SDS promoter was restricted to one germline and to determine which germline this was, reciprocal crosses were performed between T2 FK24∷SDS-CRE plants homozygous for the gus allele and a BAR-expressing plant line (Fig. 1C). Offspring was selected on the selectable marker delivered via the pollen. In a first cross, pollen from the BAR-expressing plant was transferred to the FK24∷SDS-HSC plant. In a second cross, pollen from the T2 FK24∷SDS-HSC plant was transferred to the BAR-expressing plant. These experiments were done with T2 plants from two independent FK24∷SDS-HSC lines (lines 4a and 7b), of which already a large number of other T2 plants and subsequent T3 offspring had been analyzed by PCR and GUS staining, clearly indicating a functional pSDS-cre fusion (Supplemental Fig. S1). Subsequently, offspring of the reciprocal crosses were grown and analyzed via GUS staining. GUS-positive offspring indicated that no excision occurred, whereas GUS-negative offspring indicated excision did occur in the respective germline of the FK24∷SDS-HSC plant. Offspring from cross 1 resulted for both lines only in GUS-positive offspring. All 357 and 52 plants for the 4a and 7b lines, respectively, stained blue, indicating an efficiency <0.3% in the female germline of line 4a. This is in contrast to the second cross in which pollen from the FK24∷SDS-HSC plants were used where the large majority of offspring were GUS negative. For line 4a, 132 plants of a total of 193 (68%) were GUS negative; for line 7b, this was the case for 28 of 29 analyzed plants (Fig. 1C). These results clearly indicate efficient functionality in the male germline. No functionality in the female germline was observed.
Germline-Specific Auto-Excision of the Selectable Marker
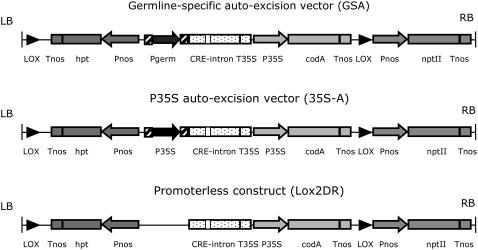
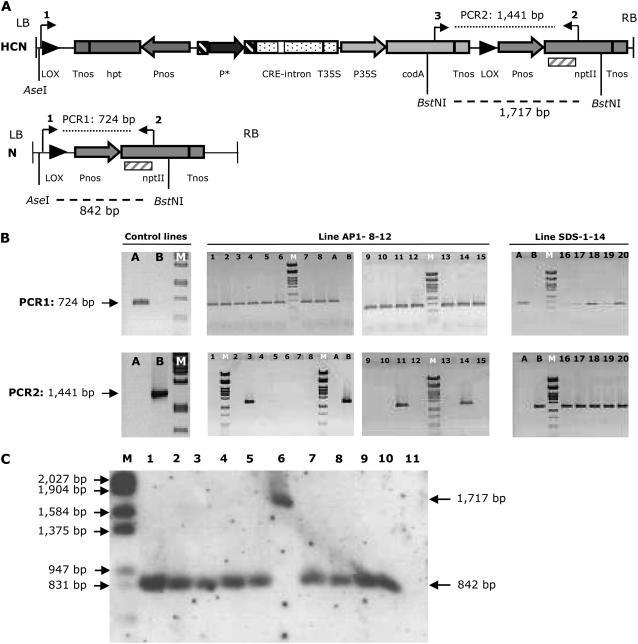
The GSA vector, which is schematically presented in Figure 2, contains three transcriptional units between tandemly oriented lox sites. A first unit comprises the cre gene containing an intron (cre-i; Joubès et al., 2004) under control of a germline-specific promoter. A second transcriptional unit contains the hygromycin phosphotransferase (hpt) gene under the control of a nopaline synthase promoter (Pnos) as a positive selection marker for plant transformation. The third unit present between the lox sites is the cytosine deaminase (codA) gene (Stougaard, 1993; Kobayashi et al., 1995) placed under the control of a cauliflower mosaic virus (CaMV) 35S promoter as a counter-selectable marker. As a gene of interest (GOI), we used the neomycin phosphotransferase (nptII) gene placed outside the lox cassette. By introducing this construct, the plant becomes genetically programmed to produce marker-free gametes and hence marker-free offspring without the need for extra handling to activate the recombinase. The original nonrecombined allele or T-DNA is referred to as “HCN” (contains the hpt, codA, and nptII genes), whereas the recombined allele is referred to as “N” (because it has lost the hpt, codA, and cre-i genes and only contains the nptII gene). As control constructs, on the one hand, we used an auto-excision construct containing the constitutive CaMV 35S promoter (35S-A) and, on the other hand, a construct without a promoter (Lox2DR) upstream of the cre recombinase gene (Fig. 2).
Figure 2.
Schematic representation of the GSA vector and control constructs: the P35S auto-excision vector (35S-A) and the promoterless construct (Lox2DR). LB and RB, Left border and right border of the T-DNA; Tnos, polyadenylation signal of the nopaline synthase gene; hpt, hygromycin phosphotransferase gene; Pnos, promoter of the nopaline synthase gene; T35S, CaMV 35S polyadenylation signal; codA, cytosine deaminase gene; nptII, neomycin phosphotransferase gene; CRE-intron, intron containing the cre recombinase gene; Pgerm, germline-specific promoter; P35S, CaMV 35S promoter. The germline promoters and P35S are introduced via an LR reaction (Gateway); this results in attB1 and attB2 sites upstream and downstream of the promoter sequence, respectively (hatched boxes).
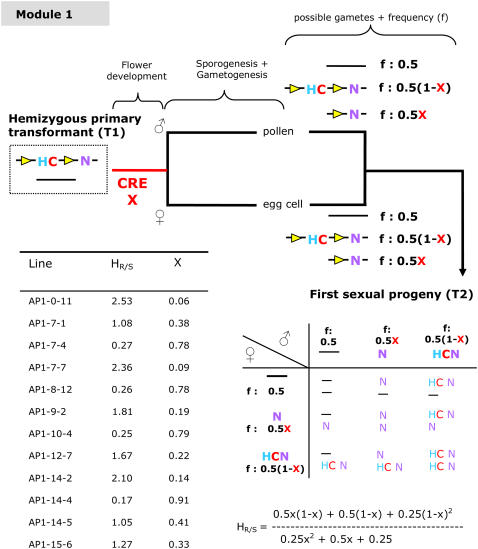
To obtain marker-free transgenic plants via module 1 (Fig. 3), we used the 1.9-kb AP1 promoter fragment described above as a germline-specific promoter in the GSA vector. As a consequence, both male and female marker-free gametes, containing the GOI, were formed, leading to marker-free transgenic plants in the first sexual progeny. We could not, however, discriminate between homozygous and hemizygous plants for the GOI via qualitative PCR. This can only be achieved by analyzing the progeny of those plants or by using quantitative PCR.
Figure 3.
Determining the CRE efficiency in module 1 (common germline or double germline CRE functionality). Yellow triangle, lox site; C, codA cassette, counter-selection on 5FC; H, hpt cassette, resistance to hygromycin; N, nptII cassette, resistance to kanamycin; between the lox sites, a 1.9-kb promoter fragment of the Arabidopsis AP1 gene is present upstream of the cre gene containing an intron. HR/S, Ratio resistant-to-sensitive T2 plants on hygromycin (n = ±200 seeds). X, Efficiency with which the CRE/lox recombination reaction occurs in the common germline (indicated as a red bar) leading to a marker-free gamete. A complete overview of the segregation data are given in Supplemental Table S3. For each line, X was calculated from the observed HR/S ratio.
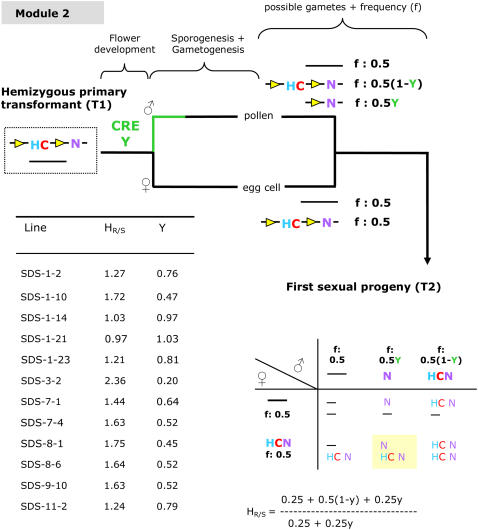
For obtaining marker-free transgenic plants via module 2 (Fig. 4), we used the same 2.2-kb promoter fragment of the SDS gene described above as a germline-specific promoter in the GSA vector. This resulted in transmission of the hygromycin resistance marker through one of the two germlines (in casu the female germline), which confers, counterintuitively, two advantages. First, it allows the identification of T2 seed stocks, which contain one active transgene locus, in which efficient excision of the selectable marker occurred. Second, it allows to easily obtain plants that are homozygous for the GOI.
Figure 4.
Determining the CRE efficiency in module 2 (single germline CRE functionality). Yellow triangle, lox site; C, codA cassette, counter-selection on 5FC; H, hpt cassette, resistance to hygromycin; N, nptII cassette, resistance to kanamycin; between the lox sites is a 2.2-kb promoter fragment of the Arabidopsis SDS gene present upstream of the cre gene containing an intron. HR/S, Ratio of resistant-to-sensitive first sexual progeny plants on hygromycin (n = ±200 seeds). Y, Efficiency with which the CRE/lox recombination occurs in a single germline (indicated in green; e.g. the male germline) leading to a marker-free gamete. Transmission of the marker to the T2 progeny through one germline results in a HR/S close to 1:1 if the transgene is present in one locus and the excision in the other germline occurred efficiently. Homozygous plants, containing a recombined (N) and a nonrecombined (NHC) allele, can easily be discriminated by PCR (highlighted). A complete overview of the segregation data are given in Supplemental Table S5. For each line, Y was calculated from the observed HR/S ratio (see also “Materials and Methods”).
Wild-type Arabidopsis Columbia-0 (Col-0) plants were transformed with the floral-dip method (Clough and Bent, 1998). Primary transformants (T1) were obtained after selection on hygromycin and transferred to the greenhouse. Transformation efficiency after selection on kanamycin and hygromycin was compared to verify whether premature excision of the hpt gene had occurred. For our promoterless construct, Lox2DR, the ratio of transformation efficiency on hygromycin (Heffic) to kanamycin (Keffic) was 1.2. For the control construct containing the 35S promoter, 35S-A, the ratio Heffic:Keffic was 0.14. For the AP1 and SDS constructs, the ratio Heffic:Keffic was 1.15 and 0.97, respectively. In all instances, around 10,000 seeds were sown on selective medium and transformation efficiencies between 0.5% and 1% were obtained after selection on kanamycin. These results indicate that the use of the AP1 and SDS promoter did not negatively influence the transformation efficiency in contrast to the 35S promoter, which resulted in a nearly 10-fold reduction of the transformation efficiency.
For first sexual progeny seeds (T2), collected from six primary transformants containing the Lox2DR construct, the ratio of resistant-to-sensitive plants was 3:1 on both kanamycin- and hygromycin-containing medium, indicating that no excision of the active transgene locus occurred in these lines (Supplemental Table S1). T2 seeds, collected from four 35S-A primary transformants selected on kanamycin and containing one active transgene locus, showed an excision efficiency of 100%, 99%, 97%, and 65%, whereas two other 35S-A lines showed no excision (Supplemental Table S2).
Module 1: Double Germline or Common Germline (AP1 Promoter) Excision
T2 seeds were sown on medium containing kanamycin to determine which transformants contained one active transgene locus and on medium with hygromycin, giving information about the efficiency of the CRE/lox recombination reaction (Fig. 3; Supplemental Table S3). Only the results from lines containing one active transgene locus are listed; 12 of 22 AP1 lines appeared to contain the T-DNA in one active transgene locus. The efficiency of the CRE/lox recombination reaction varied among independent lines with a maximum of 91%. Four lines having efficiencies >75% were retained for further characterization (AP1-7-4; AP1-8-12; AP1-10-4; AP1-14-4). Between 12 and 15 kanamycin-resistant plants for each of the four AP1 lines were transferred to the greenhouse and analyzed by PCR (Fig. 5). From the 57 plants analyzed by PCR, 41 contained only a recombined allele (N). We were unable to discriminate between homozygous and hemizygous plants concerning the GOI; therefore, we analyzed the progeny of those plants. In this experiment, we could use the nptII gene to identify homozygous plants. Therefore, 100 T3 progeny seeds from 38 marker-free lines were sown on kanamycin. For seven of the 38 lines, no kanamycin-sensitive progeny were found, indicating the presence of the GOI in a homozygous way. An overview of the complete segregation data can be found in Supplemental Table S4. When the presence of the GOI cannot be phenotypically evaluated, homozygous marker-free transgenic plants can, for example, be obtained by determining which T2 seed stocks have the lowest number of plants still containing the selectable marker. Those T2 seed stocks can then be screened by PCR for a 3:1 segregation ratio of plants containing the GOI to plants that do not. PCR analysis of T3 offspring plants will identify homozygous marker-free T3 seed stocks.
Figure 5.
Analysis of T2, T3, and T4 plants by PCR and Southern blot. A, Schematic representation of the original HCN allele and an N allele. Primers used for PCR analysis and restriction sites and probes for Southern analysis are indicated, respectively, above and below the constructs. B, PCR results on DNA purified from T2 leaf material. PCR1, Primers C-3300-F (primer 1) + nptII-SR (primer 2); PCR2, primers codA-1 (primer 3) + nptII-SR. Lane A, 35S-3-1; B, Lox2DR-2-6; M, λ/PstI marker. Lanes 1 to 15, DNA purified from kanamycin-resistant progeny of AP1-8-12 (K1–K15; module 1). Lanes 16 to 20, DNA purified from hygromycin-resistant progeny of SDS-1-14 (H1–H5; module 2; sometimes faint background excision was observed). The length of the lox cassette is 6,522 bp + the length of the promoter. For module 1, T3 seeds were collected from plants that were PCR1 positive and PCR2 negative. For module 2, T3 seeds were collected from a number of plants that were PCR1 and PCR2 positive (e.g. lanes 18 and 20). C, Southern-blot analysis of DNA purified from T3 plants of AP1 lines (lanes 8–11) and T4 plants of SDS lines of which the T3 plants were selected on 5FC (lanes 1–5). DNA was digested with AseI and BstNI, and the nptII-2 probe (hatched box) was used. M, Marker (λ-DNA cut with EcoRI-HindIII and fluorescein labeled); lane 1, SDS-1-2-H3-FC1; lane 2, SDS-1-14-H5-FC2; lane 3, SDS-1-21-H5-FC1; lane 4, SDS-1-23-H10-FC1; lane 5, SDS-11-2-H6-FC2; lane 6, Lox2DR-2-6; lane 7, 35S-3-1; lane 8, AP1-8-12-K1; lane 9, AP1-10-4-K1; lane 10, AP1-14-4-K1; lane 11, Col-0 wild type. P*, Germline-specific promoter (AP1 or SDS) or CaMV 35S promoter. Lox2DR is a promoterless construct.
Around 100 T3 seeds of the aforementioned 38 lines were sown on medium containing hygromycin. All of them were sensitive, consistent with the absence of the marker (Supplemental Table S4). DNA was purified from 20 kanamycin-resistant T3 plants and screened for the presence of the hpt marker gene by PCR. Additionally, DNA was isolated from leaf material of T3 progeny plants, originating from three transformation events (AP1-8-12, AP1-10-4, and AP1-14-4), for Southern-blot analysis. DNA purified from plants containing the promoterless construct Lox2DR and the 35S-A construct were used as positive and negative controls for the presence of the marker (Fig. 5). The results clearly indicate that, for the AP1 lines, the marker was no longer present because only a fragment characteristic for the recombined allele (N) was observed.
Module 2: Single Germline Excision (SDS Promoter)
In module 1, the marker cannot be used to identify lines containing a single transgene locus or to discriminate between homozygous and hemizygous transgenic plants. This can be resolved by using a promoter conferring single germline functionality.
Transmission of the hygromycin resistance marker through one of the two germlines allows identification of T2 seed stocks that contain one active transgene locus and in which efficient excision of the selectable marker occurred. T2 progeny from plants with one transgene locus and with high excision efficiency in one germline will segregate on hygromycin-containing medium in a ratio of resistant-to-sensitive plants (HR/S) that is close to 1:1. The higher the excision efficiency, the more the HR/S ratio approaches 1 (Fig. 4). A HR/S that is close to 3:1 indicates the presence of two active transgene loci of which the marker has efficiently been excised in one germline, or the presence of one active transgene locus of which the marker is not removed. Moreover, once the T2 seed stocks with a HR/S close to 1 are identified, T2 plants homozygous for the GOI can easily be obtained: single locus plants containing a recombined (N) and a nonrecombined allele (HCN) are de facto homozygous for the GOI (Fig. 4). Plants containing both an N and an HCN allele can conveniently be identified by PCR (see below). Although not required to obtain homozygous marker-free transgenic T3 plants, the presence of the codA gene between the lox sites made it easier to identify marker-free T3 offspring plants homozygous for the GOI. Its presence enabled counter-selection of T3 progeny plants containing the marker. In the absence of a counter-selectable marker, marker-free offspring need to be identified by PCR; at least 50% of the T3 offspring of a T2 plant containing an N and an HCN allele will still contain the selectable marker because it is transmitted via the female germline.
T2 progeny seeds were sown on kanamycin to determine the number of transgene loci from 30 SDS lines; 12 lines contained one active transgene locus. Segregation analysis of T2 seeds germinated on hygromycin revealed that the efficiency of the CRE/lox recombination reaction varied among the different lines. Nine of 12 lines showed an efficiency that was higher than 50% and a HR/S ratio close to 1:1 (Fig. 4; Supplemental Table S5).
Five SDS lines, showing the highest recombination efficiencies, were retained for further characterization: SDS-1-2, SDS-1-14, SDS-1-21, SDS-1-23, and SDS-11-2, with recombination efficiencies of 76%, 97%, 100%, 81%, and 79%, respectively. Of each retained line, around 12 hygromycin-resistant T2 plants were transferred to the greenhouse and analyzed by PCR (Fig. 5). As expected, two categories of progeny plants were observed: 28 plants contained only an HCN allele, whereas as many as 27 contained both an N and an HCN allele and were thus homozygous for the GOI (nptII). Because weak excision bands could be caused by somatic background excision (cfr. supra), T3 seeds were only collected from T2 plants showing a strong band for PCR indicative for the recombined allele. T3 seeds were sown on three types of medium (a complete overview of the segregation data can be found in Supplemental Table S6). First of all, around 100 seeds from 16 different seed stocks, distributed over the different lines, were sown on kanamycin. No sensitive plants were observed, indicating the homozygous status of the GOI. T3 seeds were additionally sown on medium containing either hygromycin or 5-fluorocytosine (5FC). If no excision occurred in the male germline of T2 plants, 25% of the seeds were expected to be hygromycin sensitive, 5FC resistant, and marker-free, whereas 50% of the seeds were expected to be hygromycin sensitive, 5FC resistant, and marker-free when a recombination efficiency of 100% in the male germline of the T2 plants was reached. From the progeny of the five lines that we retained for detailed analysis, >25% of the seeds were sensitive to hygromycin and resistant to 5FC, indicating that excision in the male germline of the T2 plants occurred. PCR analysis on DNA purified from 60 plants, surviving selection on 5FC and distributed evenly over the five different transformation events, confirmed the marker-free status because only the PCR specific for the N allele scored positive. Additionally, T4 seed was collected from 30 plants—six for each of the five different transformation events—surviving selection on 5FC and sown on hygromycin; no resistant plants were observed, consistent with the marker-free status (Supplemental Table S7). Southern-blot analysis on DNA from pooled T3 progeny plants surviving 5FC selection and from pooled leaf material collected from progeny of individual 5FC-resistant T3 plants confirmed molecularly the absence of the selectable marker (Fig. 5).
Simplification of Complex T-DNA Loci
An additional feature of recombination-based systems is the possibility that complex transgene loci can be simplified (Srivastava et al., 1999; Srivastava and Ow, 2001; De Buck et al., 2007). To investigate whether or not this was the case in this experiment, we compared the structure of the transgene locus of eight primary transformants and their marker-free progeny plants via Southern-blot analysis (Supplemental Fig. S3).
The SDS-1-2 primary transformant contained at least three copies of the T-DNA, leading to a direct repeat (DR) and an inverted repeat (IR) over the right border (RB; e.g. left border [LB]→RB LB→RB RB←LB). The complex locus of the primary transformant SDS-1-21 contained at least one DR and probably at least two IRs, one over the RB and one over the LB (Supplemental Fig. S3). The other lines of modules 1 and 2 appeared to contain a single copy of the T-DNA.
Southern-blot analysis on DNA from T4 plants, which was also used to verify the removal of the selectable marker (Fig. 5), was used to obtain information of the locus structure after the CRE/lox recombination reaction occurred. The results showed the locus was simplified in both the SDS-1-2 and SDS-1-21 line. In the progeny of the SDS-1-2 line, one copy of the T-DNA was removed by a recombination reaction between the two outer tandemly oriented lox sites present on the two T-DNAs forming the DR; two copies of T-DNA forming an IR over the RB were still present (Supplemental Fig. S3). The complex locus of the SDS-1-21 line was simplified to a single copy. This can be explained if the outer T-DNAs of the complex locus, as present in the primary transformant, are in a DR orientation (Supplemental Fig. S3).
DISCUSSION
There are some disadvantages to site-specific recombinase-based systems that depend on an additional step to introduce or to activate the recombinase. When auto-excision constructs are used, the recombinase can be activated by a chemical compound or by a heat shock in the shoots and seeds or during a subculture step and an extra regeneration step. The latter possibility lengthens the time to obtain marker-free transgenic plants and can introduce (additional) somaclonal variation, but is sometimes necessary. The efficiencies with which the recombinase is functional in the germline cells after chemical induction vary. In Arabidopsis (Zuo et al., 2001), an efficiency of 29% to 66% was reported, whereas for tomato (Solanum lycopersicum; Zhang et al., 2006) this was 15%. In rice (Oryza sativa), the recombinase was not induced in germline cells when seeds were placed on an inductive medium, whereas the efficiency of the recombination reaction was 30% when the recombinase was activated during a subculture step, although in a number of transformants incomplete recombination of multicopy loci was reported (Sreekala et al., 2005). It has been reported that heat shock promoters conferred leaky expression of the recombinase (Hoff et al., 2001; Zhang et al., 2003; Wang et al., 2005). Only Zhang et al. (2003) analyzed the influence of this leaky expression on the transformation efficiency; a nearly 20-fold reduction was reported (Zhang et al., 2003).
To circumvent problems with chemically or physically inducible promoters, GSA vectors can be used to efficiently remove selectable markers from the genome. By introducing a GSA vector, transgenic plants become genetically programmed to lose the selectable marker when its presence is no longer required (i.e. after the initial selection of primary transformants). The nature of the genetic program is defined by the functionality of the germline-specific promoter. Here, we presented the results of two modules of this genetic program. In the first module, the use of the AP1 promoter, conferring common germline-specific functionality, leads to marker-free transgenic plants in the T2 generation. In the second module, the use of the SDS promoter, which is functional in one germline, leads to marker-free transgenic plants in the T3 generation. The use of the latter module had the advantage that lines with one active transgene locus and with efficient excision of the marker could easily be identified. Moreover, homozygous marker-free plants could be obtained more easily than in module 1. Germline excision was observed in 10 of 12 lines (83%) in module 1 and in 12 of 12 lines (100%) in module 2. Within the independent lines, varying efficiencies from relatively low to high were observed. As shown here, an element that further decreased the work necessary to obtain marker-free transgenic plants by using module 2, although not essential for it, was the presence of a counter-selectable marker between the lox sites of the GSA vector.
Results obtained by Mlynarova et al. (2006) and Luo et al. (2007), who reported efficient transgene removal by using auto-excision vectors with microspore, pollen, or seed-specific promoters, together with the results presented here, clearly show the advantages of these new types of auto-excision vectors. First of all, it makes any additional handling to activate the recombinase unnecessary, saving a lot of time and effort and avoiding the previously mentioned disadvantages. Second, activating the recombinase in the germline leads to more efficient transmission of the marker-free transgene allele to the progeny. As a consequence, elimination of the marker can perfectly coincide with obtaining transgenic plants as such, allowing faster retransformation, field trials, and/or commercialization of transgenic plants.
A next step is testing the applicability of these systems in other plant species and crops. Several elements will contribute to the overall efficiency of the system. First of all, introduction of the GSA vector may not lead to a substantial decrease in transformation efficiency. Therefore, a promoter with no or very low somatic background excision will have to be used. Second, a promoter conferring very efficient functionality has to be used. This can either be a promoter functional in both germlines (module 1) or in one germline (module 2). Using a visual marker, such as gfp instead of a counter-selectable marker, would avoid the need of germination of T3 seeds in module 2. T3 seeds, collected from T2 plants containing both a recombined and a nonrecombined allele, which do not contain gfp (marker-free seeds), could easily be distinguished from T3 seeds still containing the gfp marker (e.g. by fluorescence microscopy [Stuitje et al., 2003]). This would allow obtaining homozygous and marker-free transgenic seeds containing one active transgene locus one generation earlier. If, in a certain plant species or crop, no promoter would be readily available conferring high efficiency, the fusion between lox and frt sites could lead to higher recombination efficiencies (Luo et al., 2007). So, clearly, the GSA vector system offers great potential for obtaining marker-free transgenic plants efficiently, without the need for extra handling, and this in the same time frame as compared to transformation protocols where the selectable marker is not removed and in all plant species and crops that reproduce sexually.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 plants and FK24 (De Buck et al., 2004) were grown on Murashige and Skoog medium (Duchefa) and placed in a growth chamber (temperature 23°C ± 2°C; 16-h/8-h photoperiod at 27–30 μmol m−2 s−1) prior to their transfer to the greenhouse.
Cloning of the GSA Vector and Control Constructs (35S-A and Lox2DR)
The vector is constructed using classical cloning techniques. Restriction enzymes and T4 DNA ligase from Fermentas were used following the manufacturer's protocol. If necessary, vector backbone was dephosphorylated by antarctic phosphatase (New England Biolabs) prior to ligation.
Cloning of the promoter of the Arabidopsis genes AP1 and SDS in pDONR201 (Invitrogen): A 1.9-kb (upstream of the ATG) promoter fragment of the AP1 gene (locus no. At1g69120) and a 2.2-kb (upstream of the ATG) promoter fragment of the SDS gene (locus no. At1g14750) were amplified from genomic DNA of Arabidopsis Col-0 using primers containing the attB1 and attB2 sites. The PCR product was introduced in pDONR201 via a BP reaction (Invitrogen) following the manufacturer's protocol, resulting in an entry clone.
Primer Sequences
Gateway primers used to clone the promoters of AP1 and SDS (underlined is the sequence-specific part) were as follows: AP1 forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCTTGGGATGTTGTCTTCAAGG-3′; AP1 reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCAAACAAAACAAAGACCCCC-3′; SDS forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTAGGAAGCGTATTGCTCGACTC-3′; and SDS reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTTTCTCCGTACGAAAGCTTG-3′.
Cloning of the GSA vector, 3SS-A, and Lox2DR: Two lox sites in tandem orientation were introduced in the pCambia3300 vector in two consecutive steps via an adaptor as an AseI-EcoRI fragment and a HindIII-PvuI fragment, respectively.
The positive selectable marker cassette, Pnos-hpt-Tnos, was inserted between the lox sites, after the cassette was amplified via PCR, as a BamHI-EcoRI fragment. The cre-i-T35S cassette was mobilized to the vector, after being subcloned in pUC19, as a PstI fragment. The cre-i was the same as used by Joubès et al. (2004). The Pnos-nptII-Tnos cassette was introduced outside the lox sites, after the cassette was amplified via PCR, as a BglII fragment. The CaMV 35S promoter-codA-T35S cassette, originating from the pNE3 plasmid (Stougaard, 1993), was introduced between the lox sites as a HindIII fragment. At this stage, the vector referred to as Lox2DR (Fig. 2) was obtained. Finally, the Gateway cassette was introduced, resulting in a destination vector. The AP1 and SDS promoters were introduced as germline-specific promoters, resulting in a GSA vector, and the introduction of the CaMV 35S promoter resulted in 35S-A (Fig. 2).
PWO polymerase (Hoffmann-La Roche) was used for PCR amplification reactions. All clones were verified by sequencing.
Plant Transformation
Wild-type Arabidopsis Col-0 and FK24 (De Buck et al., 2004) plants were transformed using the floral-dip method (Clough and Bent, 1998) with Agrobacterium strain C58C1RifR (pMP90) (Koncz and Schell, 1986).
Selection and Segregation Analysis
Selection was performed on Murashige and Skoog medium supplemented with 15 mg/L hygromycin (Duchefa), 75 mg/L kanamycin (Sigma), or 500 mg/L 5FC (Sigma; Kobayashi et al., 1995). For selection of primary transformants, 150 mg/L timentin (GlaxoSmithKline) was added to the selection medium to avoid Agrobacterium growth. To determine the number of active transgene loci, around 100 T2 seeds were sown on kanamycin. The ratio of resistant-to-sensitive plants on hygromycin (HR/S) was used to calculate the efficiency of CRE-mediated marker removal (see Figs. 3 and 4). Therefore, around 200 T2 seeds were sown on hygromycin. In module 2, a maximal HR/S ratio of 1 is expected, corresponding to 100% excision in one germline. Due to random variation, an HR/S >1 and a calculated efficiency of >100% might be obtained. This was the case for one line (SDS-1-21; module 2), where an efficiency of 103% was found (93 plants were resistant and 96 were sensitive). Because a value >100% has no physical meaning, we decided to report an efficiency of 100% in the main text. The χ2 test was used as a statistical tool.
Molecular Analysis
PCR Analysis
Genomic DNA was prepared from leaf material by using the GenElute Plant Genomic DNA miniprep kit (Sigma). Around 50 ng of genomic plant DNA was used as template for the PCR reactions. TaKaRa Ex Taq polymerase was used.
Primers
Primers used were as follows: LoxuitKP3, 5′-CCACACATTATACGAGCCGGAAGCAT-3′; Loxdel2, 5′-TGATCCATCTTGAGACCACAGGCCCAC-5′; GusR, 5′-GAGCGTCGCAGAACATTACA-3′; C-3300-F, 5′-GCGGACGTTTTTAATGTACTGAATTAACG-3′; nptII-SR, 5′-CCGCATTGCATCAGCCATGATGG-3′; and codA-1, 5′-GTCGCCAACCCGCTGGTCAATATTC-3′.
Southern-Blot Analysis
DNA from plant material (1 g) was purified using the Nucleon Phytopure DNA extraction kit (RPN8511; GE Healthcare). One to 2 μg of genomic DNA was cut with the respective enzymes and separated on a 1% Tris-acetate EDTA agarose gel. After depurination (0.25 m HCl), denaturation (0.5 m NaOH; 1.5 m NaCl; pH 14), and neutralization (1.5 m NaCl; 1 m Tris-HCl; pH 7–7.5), the DNA was transferred to a Hybond nylon filter (RPN203B; GE Healthcare) by upward capillarity using a high-salt blotting buffer (20× SSC). The depurination step was omitted in the Southern-blot hybridization, where the excision of the lox cassette was verified because the length of the expected fragments (1,717 and 842 bp) was smaller than 2,000 bp. The probe obtained after PCR and gel purification was labeled with fluorescein using the Gene Images random prime labeling kit (RPN3520; GE Healthcare). For obtaining the nptII probe (used in testing the functionality of the promoters), primers nptII probe F (5′-GTCGCTTGGTCGGTCATTTCGAAC-3′) and nptII probe R (5′-GAGAGGCTATTCGGCTATGACTGG-3′) were used. For obtaining the nptII-2 probe, primers S-nptII-F (5′-ATGATTGAACAAGATGGATTGCACGC-3′) and S-nptII-R2 (5′-TGATGCTCTTCGTCCAGATCATC-3′) were used. For obtaining the hpt probe, primers S-hpt-F (5′-GTCTGCTGCTCCATACAAGCCAACC-3′) and S-hpt-R (5′-GACGTCTGTCGAGAAGTTTCTGATC-3′) were used.
As molecular markers, we used λ-DNA cut with PstI and λ-DNA cut with EcoRI and HindIII. The latter was labeled with fluorescein (MIR 3023; Mirus).
Histochemical GUS Staining
For each analyzed plant, two leaves were stained in a separate reaction. Prior to incubation with GUS buffer, leaves were incubated in 90% acetone at 4°C for 15 to 30 min. After removing the acetone, leaves were washed three times with 0.1 m phosphate buffer. GUS buffer was added and vacuum was applied for 15 min. The leaves, together with the GUS buffer, were incubated for 16 h at 37°C. After incubation, GUS buffer was removed and leaves were washed with 0.1 m phosphate buffer. Chlorophyll was extracted by applying 90%, 80%, and 70% ethanol solutions in three consecutive steps (Jefferson et al., 1987).
GUS Buffer
The buffer was 1 mL of 0.1 m phosphate buffer (pH 7) with 10 mm EDTA + 5 μL 0.1 m K3[Fe(CN)6] + 5 μL 0.1 m K4[Fe(CN)6] + 1.5% dimethyl sulfoxide + 10 μL X-gluc of a 100 mm X-gluc solution (100 mm X-gluc solution: 100 mg 5-bromo-4-chloro-3-indolyl-β-d-GlcUA + 1.92 mL N,N-dimethylformamide).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Functionality of the pSDS-cre fusions in the lines used for the reciprocal crosses, 4a and 7b.
Supplemental Figure S2. Comparison between PCR data and Southern data indicating limited somatic background excision.
Supplemental Figure S3. Comparing the locus structure between primary transformants (T1), selected on hygromycin, and marker-free progeny plants via Southern-blot analysis.
Supplemental Table S1. Overview of the segregation analysis on kanamycin and hygromycin of T2 seeds, collected from primary transformants (T1) selected on hygromycin, of the promoterless Lox2DR control construct.
Supplemental Table S2. Overview of the segregation analysis on kanamycin and hygromycin of T2 seeds, collected from primary transformants (T1) selected on kanamycin (top) and hygromycin (bottom), respectively, of the 35S control lines.
Supplemental Table S3. Overview of the results of the segregation analysis on kanamycin and hygromycin of T2 seeds, collected from primary transformants (T1) selected on hygromycin, in module 1 (promoter fragment of the AP1 gene from Arabidopsis).
Supplemental Table S4. Overview of the results of the segregation analysis on kanamycin and hygromycin of T3 seeds, collected from T2 plants containing only a recombined allele (PCR1+ and PCR2−; Fig. 5), in module 1.
Supplemental Table S5. Overview of the results of the segregation analysis on kanamycin and hygromycin of T2 seeds, collected from primary transformants (T1) selected on hygromycin, in module 2 (promoter fragment of the SDS gene from Arabidopsis).
Supplemental Table S6. Overview of the segregation analysis on kanamycin, hygromycin, and 5FC of T3 seeds, collected from T2 plants containing a recombined and a nonrecombined allele (PCR1+ and PCR2+; Fig. 5), in module 2.
Supplemental Table S7. Overview of the segregation analysis on kanamycin and hygromycin of T4 seeds, collected from T3 plants resistant to 5FC.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Lieven Deveylder and Dr. Jens Stougaard for providing plasmids and Dr. Ann Depicker for advice and suggestions regarding analysis of the germline-specific promoters.
This work was supported in part by the Institute for the Promotion of Innovation by Science and Technology in Flanders (project no. GBOU 10067) and the Research Council of the Vrije Universiteit Brussel (project nos. OZR716 and OZR943).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Geert Angenon (geert.angenon@vub.ac.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Araki H, Jearnpipatkul A, Tatsumi H, Sakurai T, Ushio K, Muta T, Oshima Y (1985) Molecular and functional organization of yeast plasmid pSR1. J Mol Biol 182 191–203 [DOI] [PubMed] [Google Scholar]
- Azumi Y, Liu D, Zhao D, Li W, Wang G, Hu Y, Ma H (2002) Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J 21 3081–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley CC, Morgan M, Dale EC, Ow DW (1992) Exchange of gene activity in transgenic plants catalyzed by the Cre-lox site-specific recombination system. Plant Mol Biol 18 353–361 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated Arabidopsis thaliana transformation. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cox MM (1983) The FLP protein of the yeast 2 μm plasmid: expression of a eukaryotic genetic recombination system in Escherichia coli. Proc Natl Acad Sci USA 80 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar W, Gaudin A, Solorzano D, Casas A, Nopo L, Chudalayandi P, Medrano M, Kreuze J, Ghislain M (2006) Self-excision of the antibiotic resistance gene nptII using a heat inducible Cre-loxP system from transgenic potato. Plant Mol Biol 62 71–82 [DOI] [PubMed] [Google Scholar]
- Dale E, Ow D (1991) Gene-transfer with subsequent removal of the selection gene from the host genome. Proc Natl Acad Sci USA 88 10558–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbani B, Elimanifar A, Stewart CN, Camargo WN (2007) Methods to produce marker-free transgenic plants. Biotechnol J 2 83–90 [DOI] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A (1998) Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol Plant Microbe Interact 11 449–457 [DOI] [PubMed] [Google Scholar]
- De Buck S, Peck I, De Wilde C, Marjanac G, Nolf J, De Paepe A, Depicker A (2007) Generation of single-copy T-DNA transformants in Arabidopsis by the CRE/loxP recombination-mediated resolution system. Plant Physiol 145 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, Windels P, De Loose M, Depicker A (2004) Single-copy T-DNAs integrated at different positions in the Arabidopsis genome display uniform and comparable beta-glucuronidase accumulation levels. Cell Mol Life Sci 61 2632–2645 [DOI] [PubMed] [Google Scholar]
- Gleave A, Mitra D, Mudge S, Morris B (1999) Selectable marker-free transgenic plants without sexual crossing: transient expression of cre recombinase and use of a conditional lethal dominant gene. Plant Mol Biol 40 223–235 [DOI] [PubMed] [Google Scholar]
- Hare P, Chua N-M (2002) Excision of selectable marker genes from transgenic plants. Nat Biotechnol 20 575–580 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF (1997) Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124 3845–3853 [DOI] [PubMed] [Google Scholar]
- Hoess RH, Abremski K (1985) Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol 181 351–362 [DOI] [PubMed] [Google Scholar]
- Hoess RH, Ziese M, Sternberg N (1982) P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci USA 79 3398–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff T, Schnorr K-M, Mundy J (2001) A recombinase-mediated transcriptional induction system in transgenic plants. Plant Mol Biol 45 41–49 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Pang Y, Chen X, Fang R (2006) Removal of the selectable marker gene from transgenic tobacco plants by expression of cre recombinase from a tobacco mosaic virus vector through agroinfection. Transgenic Res 15 375–384 [DOI] [PubMed] [Google Scholar]
- Joubès J, De Schutter K, Verkest A, Inzé D, De Veylder L (2004) Conditional, recombinase mediated expression of genes in plant cell cultures. Plant J 37 889–896 [DOI] [PubMed] [Google Scholar]
- Kerbach S, Lorz H, Becker D (2005) Site-specific recombination in Zea mays. Theor Appl Genet 111 1608–1616 [DOI] [PubMed] [Google Scholar]
- Kilby NJ, Davies GJ, Michael RS, Murray JAH (1995) FLP recombinase in transgenic plants: constitutive activity in stably transformed tobacco and generation of marked cell clones in Arabidopsis. Plant J 8 637–652 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Hisajima S, Stougaard J, Ichikawa H (1995) A conditional negative selection for Arabidopsis expressing a bacterial cytosine deaminase gene. Jpn J Genet 70 409–422 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of the TL-DNA gene controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Kopertekh L, Jüttner G, Schliemann J (2004. a) Site-specific recombination induced in transgenic plants by PVX virus vector expressing bacteriophage P1 recombinase. Plant Sci 166 485–492 [Google Scholar]
- Kopertekh L, Jüttner G, Schliemann J (2004. b) PVX-Cre-mediated marker gene elimination from transgenic plants. Plant Mol Biol 55 491–500 [DOI] [PubMed] [Google Scholar]
- Kopertekh L, Schliemann J (2005) Agroinfiltration as a tool for transient expression of cre recombinase in vivo. Transgenic Res 14 793–798 [DOI] [PubMed] [Google Scholar]
- Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CN Jr, McAvoy R, Jiang X, Wu Y, et al (2007) “GM-Gene-deletor”: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5 263–274 [DOI] [PubMed] [Google Scholar]
- Lyznik LA, Rao KV, Hodges TK (1996) FLP-mediated recombination of frt sites in the maize genome. Nucleic Acids Res 24 3784–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360 273–277 [DOI] [PubMed] [Google Scholar]
- Marjanac G, De Paepe A, Peck I, Jacobs A, De Buck S, Depicker A (2007) Evaluation of CRE-mediated excision approaches in Arabidopsis thaliana. Transgenic Res (in press) [DOI] [PubMed]
- Miki B, McHugh S (2004) Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J Biotechnol 107 193–232 [DOI] [PubMed] [Google Scholar]
- Mlynarova L, Conner AJ, Nap JP (2006) Directed microspore-specific recombination of transgenic alleles to prevent pollen-mediated transmission. Plant Biotechnol J 4 445–452 [DOI] [PubMed] [Google Scholar]
- Puchta H (2000) Removing selectable marker genes: taking the shortcut. Trends Plant Sci 5 273–274 [DOI] [PubMed] [Google Scholar]
- Puchta H (2003) Marker-free transgenic plants. Plant Cell Tissue Organ Cult 74 123–134 [Google Scholar]
- Odell J, Caimi P, Sauer B, Russell S (1990) Site-directed recombination in the genome of transgenic tobacco. Mol Gen Genet 223 369–378 [DOI] [PubMed] [Google Scholar]
- Ow DW (2007) GM maize from site-specific recombination technology, what next? Curr Opin Biotechnol 18 115–120 [DOI] [PubMed] [Google Scholar]
- Russell S, Hoopes J, Odell J (1992) Directed excision of a transgene from the plant genome. Mol Gen Genet 234 49–59 [DOI] [PubMed] [Google Scholar]
- Schaart JG, Krens FA, Pelgrom KTB, Mendes O, Rouwendal GJA (2004) Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol J 2 233–240 [DOI] [PubMed] [Google Scholar]
- Senecoff JF, Bruckner RC, Cox MM (1985) The FLP recombinase of the yeast 2-μm plasmid: characterization of its recombination site. Proc Natl Acad Sci USA 82 7270–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekala C, Wu L, Gu Wang D, Tian D (2005) Excision of a selectable marker in transgenic rice (Oryza sativa L.) using a chemically regulated CRE/loxP system. Plant Cell Rep 24 86–94 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Anderson OD, Ow DW (1999) Single-copy transgenic wheat generated through the resolution of complex integration patterns. Proc Natl Acad Sci USA 96 11117–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Ow DW (2001) Single-copy primary transformants of maize obtained through the co-introduction of a recombinase-expressing construct. Plant Mol Biol 46 561–566 [DOI] [PubMed] [Google Scholar]
- Stougaard J (1993) Substrate-dependent negative selection in plants using a bacterial cytosine deaminase gene. Plant J 3 755–761 [Google Scholar]
- Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska EM, Nap J-P, Kneppers TJA (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1 301–309 [DOI] [PubMed] [Google Scholar]
- Sugita K, Kasahara T, Matsunaga E, Ebinuma H (2000) A transformation vector for the production of marker-free transgenic plants containing a single copy transgene at high frequency. Plant J 22 461–469 [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen B, Hu Y, Li J, Lin Z (2005) Inducible excision of selectable marker gene from transgenic plants by the Cre/lox site-specific recombination system. Transgenic Res 14 605–614 [DOI] [PubMed] [Google Scholar]
- Zhang W, Subbarao S, Addae P, Shen A, Armstrong C, Peschke V, Gilbertson L (2003) Cre/lox mediated marker gene excision in transgenic maize (Zea mays L.) plants. Theor Appl Genet 107 1157–1168 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li H, Ouyang B, Lu Y, Ye Z (2006) Chemical-induced autoexcision of selectable markers in elite tomato plants transformed with a gene conferring resistance to lepidopteran insects. Biotechnol Lett 28 1247–1253 [DOI] [PubMed] [Google Scholar]
- Zubko E, Scutt C, Meyer P (2000) Intrachromosomal recombination between attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat Biotechnol 18 442–445 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q-W, Møller SG, Chua N-H (2001) Chemical-regulated, site specific DNA excision in transgenic plants. Nat Biotechnol 19 157–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.