Abstract
The development of novel transformation vectors is essential to the improvement of plant transformation technologies. Here, we report the construction and testing of a new multifunctional dual binary vector system, pCLEAN, for Agrobacterium-mediated plant transformation. The pCLEAN vectors are based on the widely used pGreen/pSoup system and the pCLEAN-G/pCLEAN-S plasmids are fully compatible with the existing pGreen/pSoup vectors. A single Agrobacterium can harbor (1) pCLEAN-G and pSoup, (2) pGreen and pCLEAN-S, or (3) pCLEAN-G and pCLEAN-S vector combination. pCLEAN vectors have been designed to enable the delivery of multiple transgenes from distinct T-DNAs and/or vector backbone sequences while minimizing the insertion of superfluous DNA sequences into the plant nuclear genome as well as facilitating the production of marker-free plants. pCLEAN vectors contain a minimal T-DNA (102 nucleotides) consisting of direct border repeats surrounding a 52-nucleotide-long multiple cloning site, an optimized left-border sequence, a double left-border sequence, restriction sites outside the borders, and two independent T-DNAs. In addition, selectable and/or reporter genes have been inserted into the vector backbone sequence to allow either the counter-screening of backbone transfer or its exploitation for the production of marker-free plants. The efficiency of the different pCLEAN vectors has been assessed using transient and stable transformation assays in Nicotiana benthamiana and/or Oryza sativa.
Plant transformation technologies are fundamental to state-of-the-art plant molecular genetics and crop improvement through genetic engineering (Vain, 2006). Over the past 30 years, the development of novel transformation vectors has been seminal to many breakthroughs in plant transgenesis (for review, see Vain, 2007). In the 1980s, the engineering of Agrobacterium tumefaciens Ti plasmids—namely, the removal of oncogenes and the use of a chimerical gene(s)—led to the production of the first fertile transgenic plants (Zambryski et al., 1983) and the development of binary vectors for plant transformation (Hoekema et al., 1983; Bevan, 1984). Additional types of vectors were developed for direct transfer of DNA into the plant nuclear genome (Paszkowski et al., 1984) and later on into the plastome (Svab et al., 1990). Vector development is particularly important for Agrobacterium-based technologies as binary vectors must comply with, and best exploit, the natural mechanisms and interactions between Agrobacterium and plant cells (Gelvin, 2003). In recent years, the further understanding of T-DNA integration into the plant nuclear genome, combined with an increasing demand for precise and efficient transformation technologies, has created a new opportunity to develop plant transformation vectors with improved characteristics.
Since the 1980s, binary vectors for Agrobacterium-mediated transformation have been optimized to contain a wide range of selectable marker and reporter genes (Rogers et al., 1987; Becker et al., 1992; Jones et al., 1992; McCormac et al., 1997), as well as incorporate features facilitating their engineering (Karimi et al., 2002) or multiplication in both Escherichia coli and Agrobacterium species (for review, see Hellens et al., 2000a). Vectors have been engineered to improve stable or transient transformation efficiency with the addition of virulence (vir) genes (Hiei et al., 1994; Vain et al., 2004), suppressors of gene silencing (Hellens et al., 2005), or overdrive sequences near the border sequences (Podevin et al., 2006). Vectors aiming at the transformation of particular plant species, such as cereal crops (Hiei et al., 1994; Wang et al., 1998), have also been designed. Binary vectors with small T-DNAs (Düring, 1994; Barrell and Conner, 2006), tandem border repeats (Kuraya et al., 2004; Podevin et al., 2006), or containing only plant-derived DNA (Rommens et al., 2004) have also been engineered to minimize the insertion of unwanted DNA sequences into the plant genome. Finally, many vector series were produced for specialized uses, such as RNA silencing (Wesley et al., 2001), overexpression of heterologous genes (Gleave, 1992), functional genomics (for review, see Walden, 2002), cotransformation of multiple transgenes (Goderis et al., 2002; Tzfira et al., 2005), transfer of large DNA fragments (Simoens et al., 1986; Hamilton et al., 1996), or the production of marker-free plants (Dale and Ow, 1990; Sugita et al., 2000). More recently, binary vectors enabling and/or exploiting the delivery of multiple DNA fragments at linked or unlinked locations in the plant nuclear genome have been developed. Such binary vectors permit the delivery of multiple T-DNAs from a single binary (Komari et al., 1996; Xing et al., 2000) or two binary (Miller et al., 2002; Vain et al., 2003) vectors and have been successfully used to produce plants free of selectable marker genes in a range of species. Alternatively, single binary vectors containing transgenes in both the T-DNA and the backbone have been used for maize (Zea mays) transformation (Huang et al., 2004). In this later system, the recovery of transgenic plants is assured by cotransformation of the T-DNA and backbone components. Progeny plants free of selectable marker genes can also be recovered when cotransformation occurs at unlinked genomic locations. It would be beneficial if many of the principles successfully trialed and tested in these vector series could be combined into a single vector system to facilitate the further development of controlled and efficient transformation technologies.
Here, we describe the development and evaluation of a dual binary vector system named pCLEAN based on the pGreen/pSoup system. The pCLEAN-G/pCLEAN-S vectors can be mixed and matched with existing pGreen/pSoup plasmids in a single Agrobacterium. pCLEAN vectors enable the delivery of multiple transgenes from two distinct T-DNAs and/or backbone sequences while minimizing the insertion of superfluous DNA sequences into the plant nuclear genome. The efficiency of the different pCLEAN vectors has been assessed using transient and stable transformation assays in Nicotiana benthamiana and/or rice (Oryza sativa).
RESULTS AND DISCUSSION
Overview of the pCLEAN Vector System
The pCLEAN vectors have been developed from the dual binary system pGreen/pSoup, which enables the coexistence of two binary vectors within a single Agrobacterium. The pCLEAN-G plasmids have a pGreen-type backbone structure (including a bacterial kanamycin resistance gene) and therefore can be used as a pGreen-like vector. The pCLEAN-S vectors contain a pSoup-type backbone configuration (including a bacterial tetracycline resistance gene) and consequently can be utilized as a pSoup-like vector. However, the backbone of pCLEAN vectors generally contains additional genes or altered sequences outside the border repeats of the T-DNA. The pCLEAN-G and pCLEAN-S vectors are stable both in E. coli and A. tumefaciens and can be maintained individually in E. coli. In Agrobacterium the pCLEAN-G plasmids cannot replicate on their own and require the presence of a pCLEAN-S plasmid (or any other pSoup-based vector) to provide the replication function in trans as described previously for the pGreen/pSoup dual binary system (Hellens et al., 2000b). pCLEAN, pGreen, and pSoup vectors can be mixed and matched in a single Agrobacterium as follows: (1) pCLEAN-G and pSoup, (2) pGreen and pCLEAN-S, or (3) pCLEAN-G and pCLEAN-S vectors.
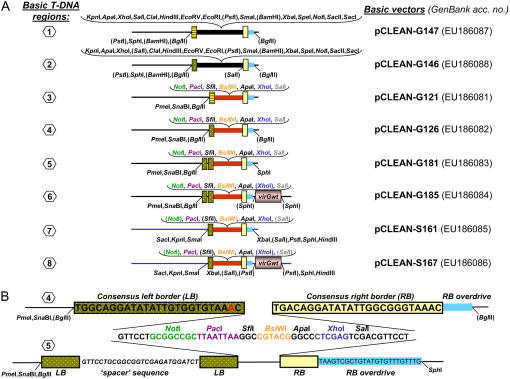
Eight different types of T-DNA regions have been constructed for the pCLEAN system and are detailed in Figure 1. These regions are referred to as numbers 1 to 8 depending upon the type of inner T-DNA, the sequence of the left border (LB), the presence of an additional vir outside the right border (RB), as well as the presence of unique restriction enzyme recognition sites within and outside the T-DNA. T-DNA regions numbers 1 to 6 and numbers 7 and 8 are corresponding to pCLEAN-G and pCLEAN-S vectors, respectively. The complete sequences of all basic pCLEAN vectors can be obtained from GenBank (Figs. 1 and 2).
Figure 1.
Schematic illustration of T-DNA regions constructed for the pCLEAN vector series. A, Six types of T-DNA regions (nos. 1–6) were developed for pCLEAN-G vectors and two types of T-DNA regions (nos. 7 and 8) for pCLEAN-S vectors. T-DNA regions numbers 1 and 2 contain the original large inner T-DNA of 728 nt from pGreenII (black box), and regions numbers 3 to 8 contain the minimal pCLEAN inner T-DNA of 52 nt (red box). Unique restriction endonuclease recognition sites within and surrounding the T-DNA regions are displayed. Brackets indicate restriction sites that are not single cutters. T-DNA regions numbers 1 and 3 contain the pGreenII original suboptimal 24-nt-long LB (yellow striped box), regions numbers 2 and 4 to 8 a 25-nt-long nopaline-type consensus LB (yellow textured box), and regions numbers 5 and 6 harbor a double LB. The RB is represented as a light yellow-colored box and its adjacent overdrive sequence (24 nt long) is shown as a light blue box. T-DNA regions numbers 6 and 8 contain a wild-type virG gene (pink-colored box) outside the RB/overdrive. For each type of T-DNA region, the corresponding basic pCLEAN vector plasmid is specified. B, Sequence details of the pCLEAN minimal T-DNA regions numbers 4 and 5. T-DNA region number 5 includes a double LB separated by a 26-nt-long spacer sequence. Additional sequence data for pCLEAN plasmids and their T-DNA regions are available at http://www.jic.ac.uk/staff/philippe-vain/vectors.htm and GenBank, respectively.
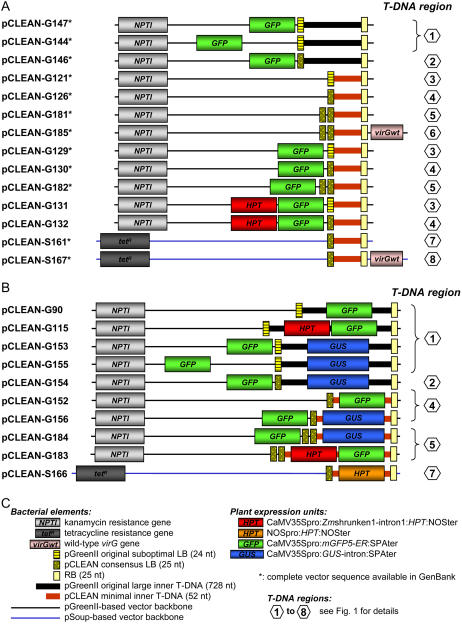
Figure 2.
Schematic illustration of the dual binary pCLEAN vector series A, Basic pCLEAN vectors (i.e. 12 pCLEAN-G and two pCLEAN-S vectors containing the T-DNA regions nos. 1–6 and nos. 7 and 8, respectively; Fig. 1). B, pCLEAN vectors containing selectable marker and/or reporter genes within their T-DNA. The T-DNA region into which the transgene(s) was inserted is indicated. C, Components constituting the different pCLEAN vectors.
The pCLEAN-G vectors are derived from a modified pGreen vector named pGreenII0000 that exhibits improved plasmid stability when multiplied in bacteria. The pCLEAN-S vectors are based on the pSoup vector (Hellens et al., 2000b). The smallest generic pCLEAN plasmids ready for accepting DNA fragments to be transferred into the plant nuclear genome are pCLEAN-G121 (2,638 nucleotides [nt]), pCLEAN-G126 (2,645 nt), and pCLEAN-G181 (2,718 nt) containing T-DNA regions numbers 3, 4, and 5, respectively, as well as pCLEAN-S161 (9,406 nt) harboring T-DNA region number 7 (Fig. 1A). The 52-nt-long multiple cloning site (MCS) found in T-DNA regions numbers 3 to 8 consists generally of the unique restriction sites NotI, PacI, SfiI, BsiWI, ApaI, XhoI, and SalI (Fig. 1). Unique restriction sites have also been engineered directly outside the LB sequence (e.g. PmeI, SnaBI, and/or BglII in T-DNA regions nos. 2–6; Fig. 1A) as well as outside the RB repeat (e.g. SphI in T-DNA region no. 5; Fig. 1A) to enable the insertion of additional transgenes into the vector backbone sequence.
Overall, the pCLEAN vector series provides the following additional features compared to the original pGreen/pSoup dual binary vector system. (1) Most of the pCLEAN vectors contain a minimal T-DNA sequence (102 nt including a 52-nt MCS) in contrast to the original large pGreen-based T-DNA (777 nt including a 728-nt MCS). (2) The sequence homology between pCLEAN-G and pSoup vectors has been reduced. (3) The pCLEAN vector system facilitates the delivery of multiple T-DNAs from a single Agrobacterium as both pCLEAN-G and pCLEAN-S vectors harbor an independent T-DNA. (4) The LB sequence of the majority of pCLEAN plasmids has been modified to accommodate a consensus nopaline-type LB repeat. (5) pCLEAN vectors are also available that contain a double consensus LB. (6) To enable visual counter-screening of backbone transfer, several pCLEAN vectors include a visual marker gene outside the LB. (7) To make use of vector backbone transfer for the production of marker-free plants, reporter and selectable marker genes have been introduced directly outside the LB. (8) To improve transformation efficiency, some pCLEAN vectors harbor an additional virulence gene in a super dual binary vector configuration in addition to the 24-nt-long overdrive sequence already present outside the RB. The binary vectors described in this article are available from the authors and further information is available at http://www.jic.ac.uk/staff/philippe-vain/vectors.htm.
Engineering of LB Sequence and Reducing Vector Backbone Transfer with pCLEAN Vectors
T-DNAs from previous pGreen-, pGreenII-, and pSoup-based vectors contain a 24-nt-long suboptimal LB sequence missing an adenosine residue in their LB repeat (Hellens et al., 2000b; Afolabi et al., 2004) when compared to the consensus 25-nt-long terminal sequences of octopine- and nopaline-type T-DNA regions (Slightom et al., 1985) and, in particular, when compared to border repeats of the nopaline vector pTiT37. The LB sequence in the T-DNA regions numbers 2 and 4 to 8 (i.e. in vectors pCLEAN-G126, -G130, -G132, -G146, -S161, and -S167, as well as their derivatives pCLEAN-G152, -G154, -G156, -S166; Fig. 2) has been modified to match this consensus sequence (Fig. 1B). The suboptimal LB sequence was also used in several pCLEAN vectors (T-DNA regions nos. 1 and 3: pCLEAN-G90, -G115, -G121, -G129, -G131, -G144, -G147, and -G155; Fig. 2) for comparison purposes. In addition, some pCLEAN vectors (i.e. pCLEAN-G181 to -G185; Fig. 2) have been constructed to contain two nopaline-type consensus LB sequences separated by a 26-nt-long spacer sequence of nonbacterial origin (T-DNA regions nos. 5 and 6; Fig. 1B). This feature was added to the pCLEAN vectors as Kuraya et al. (2004) and Podevin et al. (2006) previously showed that multiple LB sequences improve the correct recognition of the LB sequence by Agrobacterium and reduce the read-through of LB sequences in rice and Arabidopsis, respectively. The consensus RB (25 nt) and the overdrive sequence (24 nt) directly outside the RB (Peralta et al., 1986; Fig. 1B) have not been altered compared to the original pGreen/pSoup T-DNAs (T-DNA region no. 1; Fig. 1B).
To assess the effect of altering the LB sequence from a suboptimal sequence (TGGCAGGATATATTGTGGTGTAA.C) to a consensus nopaline-type LB sequence (TGGCAGGATATATTGTGGTGTAAAC) on vector back- bone transfer, transient expression assays were carried out in N. benthamiana. This was achieved by monitoring GFP expression levels in leaves infiltrated with Agrobacterium strains containing the basic pSoup binary vector and the different pCLEAN-G vectors harboring the cauliflower mosaic virus (CaMV) 35S:mGFP5-ER:SPA expression unit outside their T-DNAs with a suboptimal or consensus LB sequence (Supplemental Fig. S1). In these experiments, Agrobacterium strains containing binary vectors with the GFP gene inside the minimal or large T-DNA (i.e. pCLEAN-G90, -G152, and/or -G115) were used as positive controls. Background fluorescence was measured by observing leaves infiltrated with either the infiltration buffer alone or an Agrobacterium strain that contained a construct with a GUS expression unit inside the T-DNA (pGVT5; Supplemental Fig. S1). The comparison of GFP fluorescence detected using constructs with a suboptimal LB T-DNA versus the one with a consensus LB repeat, i.e. (1) pCLEAN-G129 versus -G130, (2) pCLEAN-G131 versus -G132, (3) pCLEAN-G147 versus -G146, and (4) pCLEAN-G153 versus -G154 (Supplemental Fig. S1), showed that the modification of the sequence of the LB repeat did not significantly modify (Kruskal-Wallis test, P < 0.05) the frequency of vector backbone transfer and that the introduction of a GFP expression unit outside the T-DNA enabled the facile and nondestructive detection of vector backbone transfer. The level of backbone transfer was further reduced by 30% to 60% when a transgene was inserted into a large or minimal empty T-DNA (Fig. 3; Supplemental Fig. S1).
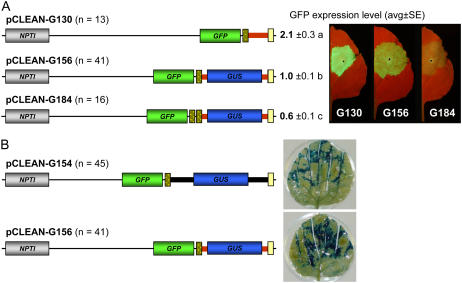
Figure 3.
Evaluation of pCLEAN vectors using transient expression assays in N. benthamiana. A, Effect of the number of consensus LB repeats and the presence of a transgene in the T-DNA on the level of backbone transfer into plant cells. Fluorescence levels (average ± se) indicate delivery and expression of the GFP gene located in the backbone sequence. Values followed by different letters are significantly different (Kruskal-Wallis test, P < 0.05). B, Expression of a reporter gene (GUS) in large and minimal T-DNAs. “n” represents the total number of leaves analyzed. For further details of the vector components, refer to Figure 2C.
The effect of adding a second consensus LB sequence on backbone transfer was also assessed in N. benthamiana as described above. Transient assays showed that adding a second LB further reduced levels of vector backbone transfer, leading to the lowest level of backbone-derived GFP fluorescence obtained with pCLEAN vectors (pCLEAN-G184; Fig. 3).
Additional transient agroinfiltration assays were performed in N. benthamiana to investigate whether the position of the GFP gene within the vector backbone has an effect on backbone transfer. Agrobacterium strains carrying either a construct with the GFP gene directly adjacent to LB or in the middle of the vector backbone (i.e. about 750 nt from the suboptimal LB) were analyzed for their vector backbone-derived GFP fluorescence levels. Experiments undertaken either with empty T-DNAs (pCLEAN-G147 versus -G144) or with a GUS gene inserted into the T-DNAs (pCLEAN-G153 versus -G155) showed that the position of the GFP gene within the vector backbone sequence has no significant effect (Kruskal-Wallis test, P < 0.05) on the detection of integrated plasmid backbone as very similar levels of GFP fluorescence were monitored from the constructs differing in their GFP-backbone configuration (Supplemental Fig. S1). In these assays, it was again observed that, as mentioned above, T-DNAs containing a transgene are less prone to backbone transfer than empty T-DNAs.
Overall, backbone transfer can be reduced with pCLEAN vectors by using a double LB strategy. Inserting a vital reporter gene such as GFP into the plasmid backbone sequence is also a very efficient way to rapidly counter-screen transformation events containing backbone sequences. The alteration of the LB sequence itself or the position of the reporter gene within the vector backbone did not lead to significant improvements.
Reducing Superfluous T-DNA Sequences, Delivery of Multiple T-DNAs, and Use of Additional vir Genes with the pCLEAN Series
The pCLEAN vectors have been designed to minimize the introduction of superfluous DNA sequences into the plant nuclear genome with a particular emphasis on avoiding the insertion of unnecessary inner T-DNA sequences and facilitating the elimination of selectable marker genes. Most of the pCLEAN vectors (i.e. pCLEAN-G121, -G126, -G129 to -G132, -G181, -G182, -G185, -S161, -S167, and their derivatives; Fig. 2) contain a minimal T-DNA sequence of 102 nt in length comprising an inner T-DNA sequence of seven unique restriction sites (52 nt) flanked by two 25-nt-long direct border repeats (T-DNA regions nos. 3–8; Fig. 1A). In contrast, the original pGreen/pSoup inner T-DNA sequence (Hellens et al., 2000b; Afolabi et al., 2004) is 728 nt in length (T-DNA regions nos. 1 and 2 [Fig. 1A] in plasmids pCLEAN-G144, -G146, and -G147 and their derivatives; Fig. 2). Superfluous sequences such as the β-galactosidase (lacZ) gene as well as other sequences derived from pBluescript SKII+/pBin19 in the pGreen/pSoup plasmids have been removed. The exclusion of nonessential sequences from transformation vectors and, in particular, from their T-DNAs is desirable to improve the biosafety and public acceptance of genetically modified crops/products used as food, feed, biomaterials, or source of bioenergy. In the past, several series of transformation vectors aimed at reducing the size of the T-DNA were developed (e.g. vector pSR8-30; Düring, 1994). Recently, pMOA vectors with minimal T-DNAs (i.e. a 141-nt inner T-DNA region including a 90-nt-long stretch with 14 unique restriction sites) have been used for potato (Solanum tuberosum) transformation (Barrell and Conner, 2006). To investigate whether the minimal T-DNA of the pCLEAN vectors is enabling the efficient delivery of a transgene(s), the expression of the GFP and GUS genes were monitored via transient transformation experiments. Agroinfiltration of N. benthamiana leaves demonstrated that the minimal T-DNA containing the GUS gene allowed high levels of GUS activity (Fig. 3, pCLEAN-G156), which is comparable to transient GUS expression derived from the GUS gene located in the large pGreen-T-DNA (Fig. 3, pCLEAN-G154). Similarly, pCLEAN-G184 (GUS gene in minimal T-DNA with double LB) showed equally high GUS expression levels in transient assays in N. benthamiana (data not shown). Likewise, the minimal T-DNA also enabled the expression of GFP to high levels in transient agroinfiltration assays in N. benthamiana. There was no significant difference between the average transient GFP expression levels originating from the large or minimal T-DNA (i.e. GFP expression from pCLEAN-G90 versus -G152; Kruskal-Wallis test, P < 0.05; Supplemental Fig. S1). Finally, the use of a minimal T-DNA (T-DNA regions nos. 3–8) resulted in a significant reduction in the homology between pCLEAN-G and pCLEAN-S/pSoup vectors. Formerly, a 400-nt-long sequence from pBluescript was present in both the pGreen/pSoup inner T-DNA and the pSoup backbone regions (Hellens et al., 2000b; Afolabi et al., 2004). This improvement minimizes putative homologous recombination events between the two types of binary vectors.
The pCLEAN vector system is also designed to facilitate the delivery of multiple T-DNAs from a single Agrobacterium as pCLEAN-G and pCLEAN-S each harbor a T-DNA. This permits “clean-gene” approaches to generate transgenic plants free of selectable marker genes. In this strategy, unwanted selectable marker genes contained within one of the pCLEAN-S vectors (e.g. HPT gene in pCLEAN-S166; Fig. 2B) can be eliminated by segregation in the progeny plants from the transgene(s) of interest carried in one of the pCLEAN-G binary vectors, because single or different T-DNAs frequently integrate at two or more independent loci (Cluster et al., 1996; Vain et al., 2003; Afolabi et al., 2004). This “one-strain and two-binary vector” approach is an attractive strategy to produce plants free of selectable marker genes as it circumvents limitations linked to the construction of vectors containing multiple T-DNAs and/or mixing of Agrobacterium strains using a “two-strain and two-binary” strategy.
In earlier studies (Vain et al., 2004), the addition of the wild-type virG gene (virGwt) in the vector backbone sequence nearly doubled the overall performance of the pGreen/pSoup vector system when considering transformation frequency, absence of backbone sequence integration, and expression of unselected transgenes. Therefore, some pCLEAN vectors were constructed (i.e. pCLEAN-G185 and pCLEAN-S167; Fig. 2A) that contain the wild-type virG gene outside the RB/overdrive sequence in the vector backbone sequence with the aim to improve transformation efficiency.
Making Use of Vector Backbone Transfer
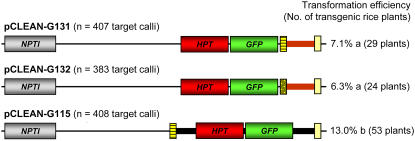
pCLEAN constructs have also been designed to exploit vector backbone transfer to produce marker-free transgenic plants. This strategy relies on the ability of Agrobacterium to frequently deliver a range of DNA fragments (i.e. T-DNA alone or T-DNA along with vector backbone sequence) at linked or unlinked locations in the plant nuclear genome. This enables selectable marker genes located outside the T-DNA (in the vector backbone) to be cotransformed with genes of interest located inside the T-DNA. When T-DNA and T-DNA plus backbone fragments integrate at different loci, progeny plants free of selectable marker gene (i.e. containing only the gene of interest) can be recovered. In this study, rice calli derived from immature embryos were transformed with Agrobacterium strains harboring the HPT and GFP marker genes either outside the T-DNA in the vector backbone sequence (i.e. vector pCLEAN-G131 or -G132 in combination with pSoup) or within the T-DNA (i.e. pCLEAN-G115 and pSoup). Transformation of rice calli with HPT and GFP positioned outside the T-DNA resulted in a transformation efficiency of about 50% of that derived from transformation with a construct containing the genes within its T-DNA (pCLEAN-G131/-G132 versus pCLEAN-G115; Fig. 4). As in the transient assays carried out in N. benthamiana and discussed above (Supplemental Fig. S1), the use of a consensus or suboptimal LB sequence did not significantly alter (χ2 test, P < 0.05) the delivery and/or integration of vector backbone sequences into the rice plant genome, as the transformation efficiency observed by using pCLEAN-G131 (suboptimal LB) and pCLEAN-G132 (consensus LB) was very similar (Fig. 4). It is also possible that not only vector backbone transfer (i.e. the failure of T-strand termination at the LB) resulted in marker gene insertion in our transgenic rice lines, but also that the T-strand was initiated at the LB as both LB and RB can initiate and terminate T-strands with a preference for initiation at the RB and termination at the LB (Huang et al., 2004). A similar strategy was previously successfully used in maize by Huang et al. (2004) and involved a single binary vector containing the selectable marker gene in the plasmid backbone and the gene of interest within the T-DNA to transform maize. This system was more efficient for the generation of marker-free plants than transformation with a single binary vector carrying multiple T-DNAs.
Figure 4.
Stable transformation of rice plants using either a T-DNA or the vector backbone to deliver transgenes. “n” indicates the total number of rice embryogenic calli used for Agrobacterium-mediated transformation. Transgenic plants are GFP+ and resistant to hygromycin. Values followed by different letters are significantly different (χ2 test, P < 0.05). For further details of the vector components, see Figure 2C.
CONCLUSION
The pCLEAN dual binary vector system enables the efficient delivery of a range of DNA fragments from single to multiple T-DNAs with, or without, vector backbone sequences into the nuclear genome of monocotyledonous and dicotyledonous species. It also minimizes the delivery of superfluous DNA sequences, such as selectable marker genes or unwanted inner T-DNA sequences. Basic pCLEAN vectors are amenable to the insertion of transgenes and can be easily further developed into RNA interference and/or Gateway-compatible systems. We believe that the basic set of pCLEAN vectors described here represents a versatile platform that can contribute to the further development of precise and efficient plant transformation technologies.
MATERIALS AND METHODS
Construction of pCLEAN-G and pCLEAN-S Vectors
Basic pCLEAN-G vectors with single LB and minimal T-DNA are pCLEAN-G121 and -G126 (Fig. 2A). The large inner T-DNA sequence of pGreenII0000 (GenBank accession nos. EF590266 for empty vector and EU048862 for large inner T-DNA) was removed using a HpaI-StuI digest and replaced by a HpaI-StuI-digested polylinker (i.e. two annealed complementary oligonucleotides, 5′-CTGGGACCTgcggccgcttaattaaGGCCGTACGGGCCCTCGAGTCGACGTTAACCTA and 5′-TAGGTTAACGTCGACTCGAGGGCCCGTACGGCCttaattaagcggccgcAGGTCCCAG), which inserted NotI (in lowercase and bold), PacI (in lowercase and underlined), BsiWI (in italics and bold), SwaI, SfiI, ApaI (underlined), XhoI (in bold), and SalI (in italics) restriction sites between the T-DNA border repeats (producing plasmid pRT117). Subsequently, pRT117 was used as a template to amplify PCR fragments with Fast Hifi Start DNA polymerase (Roche) and different primer pairs. The pCLEAN-G121 primers used were the forward primer 5′-CTGggatccGTTTAAACTACGTAAGATCTTGGCAGGATATATTGTGGTG introducing BamHI (in lowercase), PmeI (in bold), SnaBI (underlined), and BglII (in italics) sites outside the LB sequence, and the reverse primer 5′-ACGGGATCCCAAACAAACACATACAGCGAC inserting a BamHI site (in bold). The pCLEAN-G126 primers used were the forward primer 5′-ctgggatccGTTTAAACTACGTAAGATCTTGGCAGGATATATTGTGGTGTAAAC incorporating BamHI (in lowercase), PmeI (in bold), SnaBI (underlined), and BglII (in italics) sites and a consensus LB sequence, and the reverse primer 5′-ACGggatccAGATCTCAAACAAACACATACAGCGAC inserting BamHI (in lowercase) and BglII (in italics) sites. These PCR fragments were digested with BamHI and introduced into the BglII-digested pGreenII0000 vector backbone to give pCLEAN-G121 (GenBank accession no. EU186081) and pCLEAN-G126 (GenBank accession no. EU186082), respectively.
pCLEAN-G vectors containing transgene(s) within minimal/large T-DNA are pCLEAN-G90, -G115, and -G152 (Fig. 2B). pCLEAN-G90 was assembled by introducing the NotI-HindIII fragment (i.e. CaMV 35S promoter:mGFP5-ER:SPA terminator) of pGVT1 (Thole and Rawsthorne, 2003) into the NotI-ClaI-digested pGreenII0000 and by replacing the approximately 800-nt-long CaMV 35S promoter with the approximately 430-nt-long CaMV 35S promoter of pGVT5 (Thole and Rawsthorne, 2003). Subsequently, the CaMV 35S promoter:maize shrunken1-intron1:HPT:NOS terminator expression unit (Vain et al., 1996) was introduced into the SphI-digested pCLEAN-G90 to produce pCLEAN-G115. The blunted SalI-SpeI fragment of pCLEAN-G90 (i.e. CaMV 35S promoter:mGFP5-ER:SPA terminator) was introduced into the blunted NotI-digested pCLEAN-G126 to give pCLEAN-G152.
pCLEAN-G vectors containing selectable marker and/or reporter gene(s) in vector backbone outside single LB are pCLEAN-G129, -G130, -G131, -G132, -G144, -G146, and -G147 (Fig. 2A). pCLEAN-G129 (GenBank accession no. EU186090) and pCLEAN-G130 (GenBank accession no. EU186091) were constructed by inserting the blunted SalI-SpeI fragment of pCLEAN-G90 (i.e. CaMV 35S promoter:mGFP5-ER:SPA terminator) into the SnaBI-digested pCLEAN-G121 and pCLEAN-G126 plasmids, respectively. Inserting the blunted NotI-SalI fragment of pCLEAN-G115 (i.e. CaMV 35S promoter:maize shrunken1-intron1:HPT:NOS terminator) into the SnaBI-digested pCLEAN-G121 and pCLEAN-G126 vectors, respectively, resulted in the constructs pCLEAN-G131 and pCLEAN-G132. pCLEAN-G144 (GenBank accession no. EU186089) was assembled by introducing the blunted SalI-SpeI fragment of pCLEAN-G90 (i.e. CaMV 35S promoter:mGFP5-ER:SPA terminator) into the blunted DraIII-digested pGreenII0000. pCLEAN-G146 (GenBank accession no. EU186088) was cloned by inserting the HpaI-StuI fragment of pGreenII0000 (i.e. empty large inner T-DNA of 728 nt) into the blunted NotI-SalI-digested pCLEAN-G130. pCLEAN-G147 (GenBank accession no. EU186087) resulted from the insertion of the pGreenII0000-derived BglII fragment (i.e. empty large T-DNA including border repeats of 777 nt) into the blunted BglII-digested pCLEAN-G130.
pCLEAN-G vectors containing marker gene outside single LB along with reporter gene within minimal/large T-DNA are pCLEAN-G153, -G154, -G155, -G156, and -G184 (Fig. 2B). The blunted SpeI-HindIII fragment (i.e. CaMV 35S promoter:GUS-intron:SPA terminator) of pGVT5 was cloned into the SmaI-cut vectors pCLEAN-G147 (to give pCLEAN-G153), pCLEAN-G146 (to give pCLEAN-G154), and pCLEAN-G144 (to give pCLEAN-G155), as well as the blunted NotI-digested plasmids pCLEAN-G130 (to give pCLEAN-G156) and pCLEAN-G182 (to give pCLEAN-G184).
pCLEAN-S vectors with single LB and minimal T-DNA are pCLEAN-S161, -S166, and -S167 (Fig. 2, A and B). To clone pCLEAN-S161 (GenBank accession no. EU186085), the BglII fragment of pCLEAN-G126 containing the consensus LB, minimal T-DNA, RB repeat, and its overdrive sequence was introduced into the BamHI-digested pSoup vector (GenBank accession no. EU048870). The EcoRV fragment of pSLJ261 enclosing the NOS promoter:HPT:NOS terminator (Jones et al., 1992) was cloned into the blunted PacI-digested pCLEAN-S161 to give pCLEAN-S166. The PstI fragment of pCLEAN-S48 containing the wild-type virG (Vain et al., 2004) was cloned into the PstI-digested pCLEAN-S161 to result in pCLEAN-S167 (GenBank accession no. EU186086).
Basic pCLEAN-G vectors with double LB are pCLEAN-G181 and -G185 (Fig. 2A). To assemble pCLEAN-G181 (GenBank accession no. EU186083), several cloning steps were undertaken: (1) inserting from pRT172, a pCLEAN-S161 derivative, a blunted HindIII-SnaBI fragment containing the consensus LB, CaMV 35S terminator, RB, and its overdrive into the blunted BglII-SnaBI-digested pCLEAN-G126 (to give plasmid pRT174 while restoring SnaBI); (2) removing the 35S terminator-containing EcoRV-SmaI fragment from pRT174 (to give plasmid pRT175); and (3) deleting the additional SalI site outside the RB from pRT175 by XbaI-PstI digestion, blunting, and religation. To produce pCLEAN-G185 (GenBank accession no. EU186084), a PCR fragment encoding the virGwt was amplified from pCLEAN-S48 with the primers 5′-TAAGCATGCAGGCTCGCGGCGGACGCACGA (forward primer, SphI site un- derlined) and 5′-TATGCATGCGCAGTGTGATGGGCACATCG (reverse primer, SphI site underlined) and Fast Hifi Start DNA polymerase, digested with SphI, and introduced into the SphI-digested pCLEAN-G181.
pCLEAN-G vectors containing marker gene outside double LB and/or transgenes within minimal T-DNA are pCLEAN-G182, -G183, and -G184 (Fig. 2, A and B). pCLEAN-G182 (GenBank accession no. EU186092) was cloned by inserting the blunted SalI-SpeI fragment of pCLEAN-G90 (i.e. CaMV 35S promoter:mGFP5-ER:SPA terminator) into the SnaBI-digested pCLEAN-G181. To construct pCLEAN-G183, the NotI-SalI fragment of pCLEAN-G115 (i.e. CaMV 35S promoter:maize shrunken1-intron1:HPT:NOS terminator) was cloned into the NotI-SalI -digested pCLEAN-G181.
All cloning steps and PCR fragments were verified by sequencing using the BigDye Terminator Version 3.1 kit (Applera UK). The full-length sequences of vectors pCLEAN-G121, -G126, -G129, -G130, -G144, -G146, -G147, -G181, -G182, and -G185 and pCLEAN-S161 and -S167 are available in GenBank (see Fig. 1 for some examples), and further information about the pCLEAN vector system is available at http://www.jic.ac.uk/staff/philippe-vain/vectors.htm.
Maintenance of pCLEAN-G and pCLEAN-S Vectors in Escherichia coli and Agrobacterium tumefaciens
E. coli containing pCLEAN-G vectors were grown for about 12 to 16 h at 37°C in Luria broth (LB) supplemented with 50 mg/L kanamycin, and E. coli containing pCLEAN-S plasmids were grown for about 16 to 48 h at 37°C in LB media containing 7.5 mg/L tetracycline. In A. tumefaciens, colonies containing (1) pCLEAN-G and pSoup, (2) pGreen and pCLEAN-S, or (3) pCLEAN-G and pCLEAN-S vector combination grew after 48 to 72 h at 28°C in LB media supplemented with 50 mg/L kanamycin or a mixture of 50 mg/L kanamycin and 7.5 mg/L tetracycline.
Agrobacterium-Mediated Transformation of Rice
Embryogenic calli derived from mature seeds of rice (Oryza sativa L.) var. Nipponbare were used for transformation as described previously (Vain et al., 2003). Briefly, embryogenic calli were inoculated and cocultivated with Agrobacterium strain AGL1 containing different combinations of pCLEAN-G and pSoup vectors. Transgenic callus lines were selected on hygromycin (50 mg/L) during 6 weeks and plants were regenerated and analyzed using Standard Operating Procedures. Transformation efficiency obtained using pCLEAN-G131 or -G132 was compared to that of the control vector (pCLEAN-G155 containing transgenes within the T-DNA) using χ2 analysis.
Agrobacterium-Mediated Transient Assays in Nicotiana benthamiana
The different combinations of pCLEAN-G vectors together with the plasmid pSoup were introduced into A. tumefaciens strain AGL1 using a freeze-thaw method based on An et al. (1988). The different A. tumefaciens strains were grown overnight at 28°C in LB medium supplemented with 50 mg/L kanamycin and 150 μm acetosyringone (English et al., 1997). Cells were harvested by centrifugation (4,000 rpm, 4°C), resuspended in 10 mm MgCl2 and 150 μm acetosyringone, and diluted to an optical density (OD600 nm) of 1.0. The suspension of each Agrobacterium strain to be tested was mixed in a 1:1 ratio with a bacterial suspension carrying a vector containing the viral suppressor of gene silencing P38 (Thomas et al., 2003) in order to increase transient expression levels as described by Hellens et al. (2005). The mixed cultures were incubated at room temperature for 3 to 5 h before infiltration. On average, 16 leaves from 4- to 5-week-old wild-type N. benthamiana plants were agroinfiltrated with a 1-mL syringe. Control treatments included the infiltration of leaves with (1) 10 mm MgCl2 and 150 μm acetosyringone alone and/or an Agrobacterium strain harboring plasmid pGVT5 (Thole and Rawsthorne, 2003) containing NOS promoter:NPTII:NOS terminator and CaMV 35S promoter:GUS-intron:SPA terminator expression units as negative controls, and (2) an Agrobacterium strain containing a vector with the CaMV 35S promoter:mGFP5-ER:SPA terminator within its T-DNA (i.e. pCLEAN-G90 and/or pCLEAN-G152) as positive control (Supplemental Fig. S1). The infiltrated leaves were analyzed for GFP fluorescence and/or GUS activity 2.5 d postinfiltration. At least two independent experiments were conducted to assess each pCLEAN vector.
GFP and GUS Expression Analyses
The GFP fluorescence was monitored in cells and tissues using a MZ12 Leica dissecting microscope with a fluorescent module (Leica no. 10446093). The appropriate wavelength was adjusted using a filter block containing a 425/60-nm excitation filter, a 470-nm dichromatic beam splitter, and a G6457 emission barrier filter, over a high-voltage mercury lamp. The level of GFP expression in infiltrated leaf tissues was scored visually on a scale of 0 to 4 (i.e. 0 = no expression; 1 = low GFP expression; 2 = medium GFP expression; 3 = high GFP expression; and 4 = very high GFP expression). For each construct/strain tested, an average GFP expression level (±se) was calculated across all leaves observed. Statistical analyses, following the requirements of each test, were performed using Minitab 3.1 software package. GFP expression levels were compared using the Kruskal-Wallis test. The histochemical GUS assays were conducted according to Jefferson et al. (1987) using 5-bromo-4-chloro-3-indoyl glucuronide (Melford Laboratories Ltd.) as substrate.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU186081 (pCLEAN-G121), EU186082 (pCLEAN-G126), EU186083 (pCLEAN-G181), EU186084 (pCLEAN-G185), EU186085 (pCLEAN-S161), EU186086 (pCLEAN-S167), EU186087 (pCLEAN-G147), EU186088 (pCLEAN-G146), EU186089 (pCLEAN-G144), EU186090 (pCLEAN-G129), EU186091 (pCLEAN-G130), and EU186092 (pCLEAN-G182).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of the type of LB sequence on the level of backbone transfer and the position of the GFP gene in the backbone sequence for its detection.
Supplementary Material
Acknowledgments
We thank Aude Derevier and Shona Ross for constructing vectors pRT117 and pCLEAN-G115, respectively, as well as Lesley Fish for critical reading of the manuscript.
This work was supported by the Biotechnology and Biological Sciences Research Council and the Plant Sciences Research Programme (R8031) funded by the UK Department for International Development (DFID) and administered by the Centre for Arid Zone Studies for the benefit of developing countries. The views expressed are not necessarily those of DFID.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Philippe Vain (philippe.vain@bbsrc.ac.uk).
The online version of this article contains Web-only data.
References
- Afolabi AS, Worland B, Snape JW, Vain P (2004) A large-scale study of rice plants transformed with different T-DNAs provides new insights into locus composition and T-DNA linkage configurations. Theor Appl Genet 109 815–826 [DOI] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A3: 1–19
- Barrell PJ, Conner AJ (2006) Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. Biotechniques 41 708–710 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bevan M (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluster PD, O'Dell M, Metzlaff M, Flavell RB (1996) Details of T-DNA structural organisation from a transgenic petunia population exhibiting co-suppression. Plant Mol Biol 32 1197–1203 [DOI] [PubMed] [Google Scholar]
- Dale EC, Ow DE (1990) Intra- and intermolecular site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene 91 79–85 [DOI] [PubMed] [Google Scholar]
- Düring K (1994) A plant transformation vector with a minimal T-DNA. Transgenic Res 3 138–140 [DOI] [PubMed] [Google Scholar]
- English JJ, Davenport GF, Elmayan T, Vaucheret H, Baulcombe DC (1997) Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J 12 597–603 [Google Scholar]
- Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67 16–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goderis I, De Bolle MFC, Francois I, Wouters PFJ, Broekaert WF, Cammue BPA (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol 50 17–27 [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Lewis C, Tanksley SD (1996) Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci USA 93 9975–9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H (2000. a) A guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5 446–451 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000. b) pGreen: a versatile and flexible binary vector for Agrobacterium-mediated transformation. Plant Mol Biol 42 819–832 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir-region and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303 179–180 [Google Scholar]
- Huang S, Gilbertson LA, Adams TH, Malloy KP, Reisenbigler EK, Birr DH, Snyder MW, Zhang Q, Luethy MH (2004) Generation of marker-free transgenic maize by regular two-border Agrobacterium transformation vectors. Transgenic Res 13 451–461 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K (1992) Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res 1 285–297 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T (1996) Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J 10 165–174 [DOI] [PubMed] [Google Scholar]
- Kuraya Y, Ohta S, Fukuda M, Hiei Y, Murai N, Hamada K, Ueki J, Imaseki H, Komari T (2004) Suppression of transfer of non-T-DNA ‘vector backbone’ sequences by multiple left border repeats for transformation of higher plants mediated by Agrobacterium tumefaciens. Mol Breed 14 309–320 [Google Scholar]
- McCormac AC, Elliot MC, Chen DF (1997) pBECKS: a flexible series of binary vectors for Agrobacterium-mediated plant transformation. Mol Biotechnol 8 199–213 [DOI] [PubMed] [Google Scholar]
- Miller M, Tagliani L, Wang N, Berka B, Bidney D (2002) High efficiency transgene segregation in co-transformed maize plants using an Agrobacterium tumefaciens 2 T-DNA binary system. Transgenic Res 11 381–396 [DOI] [PubMed] [Google Scholar]
- Paszkowski J, Shillito RD, Saul M, Mandak V, Hohn T, Hohn B, Potrykus I (1984) Direct gene transfer to plants. EMBO J 3 2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta EG, Hellmiss R, Ream W (1986) Overdrive, a T-DNA transmission enhancer on the A. tumefaciens tumour-inducing plasmid. EMBO J 5 1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podevin N, De Buck S, De Wilde C, Depicker A (2006) Insights into recognition of the T-DNA border repeats as termination sites for T-strand synthesis by Agrobacterium tumefaciens. Transgenic Res 15 557–571 [DOI] [PubMed] [Google Scholar]
- Rogers SG, Klee HJ, Horsch RB, Fraley RT (1987) Improved vectors for plant transformation: expression cassette vectors and new selectable markers. Methods Enzymol 153 253–277 [Google Scholar]
- Rommens CM, Humara JM, Ye JS, Yan H, Richael C, Zhang L, Perry R, Swords K (2004) Crop improvement through modification of the plant's own genome. Plant Physiol 135 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Simoens C, Alliotte T, Mendel R, Müller A, Schiemann J, Van Lijsebettens M, Schell J, Van Montagu M, Inzé D (1986) A binary vector for trans-ferring genomic libraries to plants. Nucleic Acids Res 14 8073–8090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom JL, Jouanin L, Leach F, Drong RF, Tepfer D (1985) Isolation and identification of TL-DNA/plant junctions in Convolvulus arvensis transformed by Agrobacterium rhizogenes strain A4. EMBO J 4 3069–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita K, Kasahara T, Matsunaga E, Ebinuma H (2000) A transformation vector for the production of marker-free transgenic plants containing a single copy transgene at high frequency. Plant J 22 461–469 [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P (1990) Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA 87 8526–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V, Rawsthorne S (2003) Development of a strategy for transgenic studies and monitoring of transgene expression in two closely related Moricandia species possessing a C3 or C3-C4 intermediate photosynthetic phenotype. Physiol Plant 119 155–164 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Leh V, Lederer C, Maule AJ (2003) Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306 33–41 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503–516 [DOI] [PubMed] [Google Scholar]
- Vain P (2006) Global trends in plant transgenic science and technology (1973-2003). Trends Biotechnol 24 206–211 [DOI] [PubMed] [Google Scholar]
- Vain P (2007) Thirty years of plant transformation technology development. Plant Biotechnol J 5 221–229 [DOI] [PubMed] [Google Scholar]
- Vain P, Afolabi AS, Worland B, Snape JW (2003) Transgene behaviour in populations of rice plants transformed using a new dual binary vector system: pGreen/pSoup. Theor Appl Genet 107 210–217 [DOI] [PubMed] [Google Scholar]
- Vain P, Finer KR, Engler DE, Pratt RC, Finer JJ (1996) Intron-mediated gene expression enhancement in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep 15 489–494 [DOI] [PubMed] [Google Scholar]
- Vain P, Harvey A, Worland B, Ross S, Snape JW, Lonsdale D (2004) The effect of additional virulence genes on transformation efficiency, transgene integration and expression in rice plants using the pGreen/pSoup dual binary vector system. Transgenic Res 13 593–603 [DOI] [PubMed] [Google Scholar]
- Walden R (2002) T-DNA tagging in a genomics era. Crit Rev Plant Sci 21 143–165 [Google Scholar]
- Wang M, Li Z, Matthews PR, Upadhyaya NM, Waterhouse PM (1998) Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic 461 401–407 [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Xing A, Zhang Z, Sato S, Staswick P, Clement T (2000) The use of two T-DNA binary system to derive marker-free transgenic soybeans. In Vitro Cell Dev Plant 36 456–463 [Google Scholar]
- Zambryski P, Joos H, Genetello C, Leemans J, Van Montagu M, Schell J (1983) Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J 2 2143–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.