Abstract
We investigated whether complex T-DNA loci, often resulting in low transgene expression, can be resolved efficiently into single copies by CRE/loxP-mediated recombination. An SB-loxP T-DNA, containing two invertedly oriented loxP sequences located inside and immediately adjacent to the T-DNA border ends, was constructed. Regardless of the orientation and number of SB-loxP-derived T-DNAs integrated at one locus, recombination between the outermost loxP sequences in direct orientation should resolve multiple copies into a single T-DNA copy. Seven transformants with a complex SB-loxP locus were crossed with a CRE-expressing plant. In three hybrids, the complex T-DNA locus was reduced efficiently to a single-copy locus. Upon segregation of the CRE recombinase gene, only the simplified T-DNA locus was found in the progeny, demonstrating DNA had been excised efficiently in the progenitor cells of the gametes. In the two transformants with an inverted T-DNA repeat, the T-DNA resolution was accompanied by at least a 10-fold enhanced transgene expression. Therefore, the resolution of complex loci to a single-copy T-DNA insert by the CRE/loxP recombination system can become a valuable method for the production of elite transgenic Arabidopsis thaliana plants that are less prone to gene silencing.
To obtain transgenic plants, the Agrobacterium-mediated transformation system is commonly applied (Gelvin, 2003; Veluthambi et al., 2003; Tzfira and Citovsky, 2006). The transgenic loci are considered to be less complex than those resulting from direct gene transfer. Nevertheless, integration of multiple T-DNA copies into direct and inverted repeats is fairly common (De Neve et al., 1997; De Buck et al., 1999; Tzfira et al., 2004). Over the past 15 years, the correlation between inverted T-DNA repeat structures and transgene silencing has been demonstrated repeatedly (Hobbs et al., 1993; Jorgensen et al., 1996; Muskens et al., 2000; De Buck et al., 2001). Although single-copy transgenes can induce silencing as well (Elmayan and Vaucheret, 1996; Day et al., 2000; Meyer, 2001), they generally have, at least in Arabidopsis (Arabidopsis thaliana), a uniform and stable transgene expression (De Buck et al., 2004; Schubert et al., 2004). Thus, transgenic plants with one single copy of the introduced transgene DNA are preferred to those with multiple copies. Therefore, several approaches, such as agrolistics, treatment with niacinamide, use of transposable elements as transgene transporters, or site-specific recombination, have been proposed to enrich for single-copy transgenic lines (Hansen and Chilton, 1996; De Block et al., 1997; Hansen et al., 1997; Koprek et al., 2001; Ow, 2002). However, conventionally, single-copy transformants are screened among a large pool of transformants (De Buck and Depicker, 2004).
Site-specific recombination systems have many applications in plants (Ow, 2002). First, they have been successfully employed for the removal of selectable markers from transgenic plants (Hoa et al., 2002; Zhang et al., 2003; Srivastava and Ow, 2004; Sreekala et al., 2005; Wang et al., 2005; Jia et al., 2006; Moore and Srivastava, 2006), and, second, they can be used to precisely integrate single-copy transgenes into the plant genome (Albert et al., 1995; Vergunst et al., 1998; Vergunst and Hooykaas, 1998; Day et al., 2000; Srivastava and Ow, 2002; Lyznik et al., 2003; Chawla et al., 2006). This latter approach requires two rounds of transformation: in the first, a target site, such as the loxP site, is randomly introduced into the plant genome, and, in the second, a loxP-containing DNA construct is integrated into this genomic target site. CRE-mediated gene integration has been obtained only at a low frequency after transformation with Agrobacterium, while it is seemingly more efficient when DNA is introduced by particle bombardment (Srivastava et al., 2004; Chawla et al., 2006). Third, site-specific recombination has been utilized to reduce the tandem arrays of loxP-flanked gene copies occurring after direct gene transfer to a single unit (Srivastava et al., 1999; Srivastava and Ow, 2001). Finally, the CRE/loxP system can be used to allow controlled and inducible transgene expression (Joubès et al., 2004).
The question remains whether a site-specific resolution of complex T-DNA loci is a possible strategy to efficiently obtain elite single-copy T-DNA transgene events. Therefore, we designed a special T-DNA construct and applied the recombination-mediated resolution on complex T-DNA loci. Additionally, we determined whether the resolution to single T-DNA copies had an influence on transgene expression levels.
RESULTS
Experimental Setup
To determine whether the CRE/loxP recombination system is capable of resolving complex T-DNA loci into a single T-DNA insert, an SB-loxP T-DNA was constructed (Fig. 1A). This SB-loxP T-DNA harbors a multicloning site flanked by two invertedly oriented loxP recombination sites inside and immediately adjacent to the left and right T-DNA border ends. For proof of concept, we introduced the NPTII (neomycin phosphotransferase II gene) selectable marker and the 35S-GUS reporter in between the loxP sites, generating the Ksb T-DNA (Fig. 1B; De Buck et al., 2000a). Regardless of the number and orientation of the SB-loxP-derived T-DNA copies inserted at one locus, recombination between the outermost loxP sequences in direct orientation should resolve multiple copies into a single T-DNA copy (Fig. 1C). Seven transformants (PA23, PA22, PA25, PA29, PA1, PA5, and PA16) with multiple Ksb T-DNA inserts on a single locus were identified based on DNA gel-blot and segregation analyses (Table I). The transgenic Arabidopsis line PA23 contained two Ksb T-DNAs in tandem orientation, with in between the complete vector backbone DNA of the plasmid Ksb, presumably due to read through at the left border (LB; Table I; Fig. 2, A and B). In line PA22, four Ksb T-DNA copies were inserted, of which two were oriented in an inverted repeat about the right border (IRRB) and two in tandem repeat with the complete vector backbone in between (Table I; Fig. 3A). Transformant PA25 contained two T-DNA copies in an IRRB (Table I; Fig. 3B), and transformant PA29 harbored four to six Ksb T-DNA copies, of which at least two formed an IRRB (Table I; Fig. 3C). In transformants PA1, PA5, and PA16, four or more T-DNA copies were integrated (Table I).
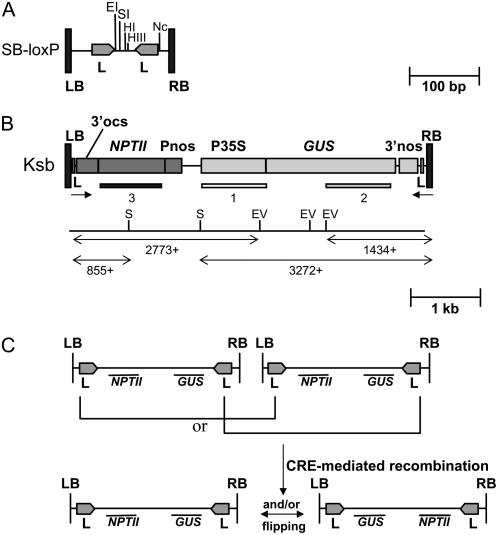
Figure 1.
Schematic outlines of the SB-loxP T-DNA, Ksb T-DNA, and the resolution principle. A, Representation of the SB-loxP T-DNA. The two loxP (L) sequences, each indicated by an arrow that gives its orientation, are located next to the 25-bp LB and RB repeats, delineated by vertical black boxes. In between these loxP sites, a multicloning site is present. B, Representation of the Ksb T-DNA. After the plant DNA had been digested with the EcoRV (EV) or SphI (S) enzymes, whose restriction sites are indicated, T-DNA/plant junctions of at least 1,434 or 2,773 bp (EV) and 3,272 or 855 bp (S) were observed after hybridization with the GUS or NPTII probe, respectively. The probes corresponded to the numbered boxes below the Ksb T-DNA. C, Outline of the resolution principle with tandemly repeated T-DNAs. Regardless of the number of T-DNAs integrated at one locus, recombination between the outermost loxP sequences in direct orientation resolved the integrated T-DNAs into a single copy. When the CRE recombinase was still active in the plant, inversion of the sequence between the loxP sequences was possible. Abbreviations: CRE, recombinase of Cre/loxP recombination system; EI, EcoRI; EV, EcoRV; HI, HpaI; HIII, HindIII; L, loxP sequence recognition site of the Cre/loxP recombination system; 3′nos, 3′ end of the nopaline synthase gene; 3′ocs, 3′ end of the octopine synthase gene; P35S, cauliflower mosaic virus promoter; GUS, GUS-encoding gene; Nc, NcoI; Pnos, promoter of the nopaline synthase gene; S, SphI; SI, SalI.
Table I.
Overview of the transgenic plant lines used
In all transformants, the T-DNAs were inserted into one locus. ND, Not determined.
| Transformant | T-DNA | No. of T-DNAsa | Configurationsb |
|---|---|---|---|
| PA23 | Ksb | 2 | Read through |
| PA22 | Ksb | 4 | IRRB, read through |
| PA25 | Ksb | 2 | IRRB |
| PA29 | Ksb | 4–6 | At least one IRRB |
| PA1 | Ksb | 4–6 | At least one IRRB, read through |
| PA5 | Ksb | 4–6 | IRRB, read through |
| PA16 | Ksb | 3–4 | Read through |
| CRE13 | Cre | ND | ND |
| HSC6 | Cre | ND | ND |
T-DNA copy number determined on the basis of the number of T-DNA/plant DNA junction fragments on the DNA gel blots.
Read through, vector backbone sequence inserted between two T-DNAs in tandem orientation, presumably because of read through at the LB.
Figure 2.
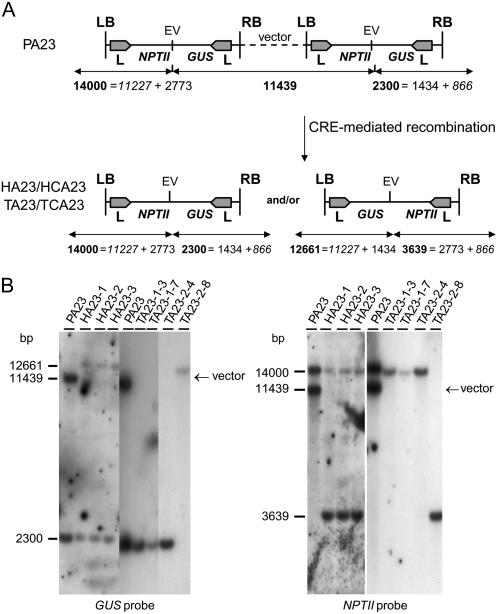
DNA gel-blot analysis on EcoRV-digested DNA of PA23, HA23, and TA23. A, Schematic representation of the complex locus PA23 and the resolved/inverted T-DNA in HA23/HCA23 and TA23/TCA23 seedlings. The PA23 transformant carried two T-DNAs in direct orientation with vector backbone sequences in between, resulting in two fragments after hybridization with the GUS (11,439 bp and 2,300 bp) and NPTII probes (11,439 bp and 14,000 bp). For the two T-DNA/plant junctions, the T-DNA fragment was known (2,773 bp and 1,434 bp for the LB and RB regions, respectively; Fig. 1), and the plant fragment, in italics, could be calculated (11,227 bp and 866 bp for the LB and RB plant regions, respectively). After resolution of the complex T-DNA locus, two different T-DNA configurations could be found: the original T-DNA configuration with the same T-DNA/plant junction fragments as in the parental line and the inverted T-DNA configuration. In this latter configuration, the length of the fragments could be predicted again because both T-DNA and plant fragments of the junctions were known: for the NPTII and GUS probes, a fragment of 3,639 bp (2,773 bp of the NPTII T-DNA fragment and 866 bp for the RB/plant junction) and a fragment of 12,661 bp (11,227-bp LB/plant junction + 1,434-bp T-DNA fragment), respectively. B, DNA gel-blot analysis on DNA of parental (P), hybrid (H), and progeny (T) plants of A23 with the GUS (left) and NPTII probes (right). The hybrids were obtained after crossing the parental plants with the CRE13 line. On top of each lane, the name is given of the transformant from which DNA was prepared. Abbreviations: EV, EcoRV; L, loxP sequence recognition site of the Cre/loxP recombination system; vector, read through at the LB results in a tandem repeat of two Ksb T-DNAs with the entire vector backbone sequence in between.
Figure 3.
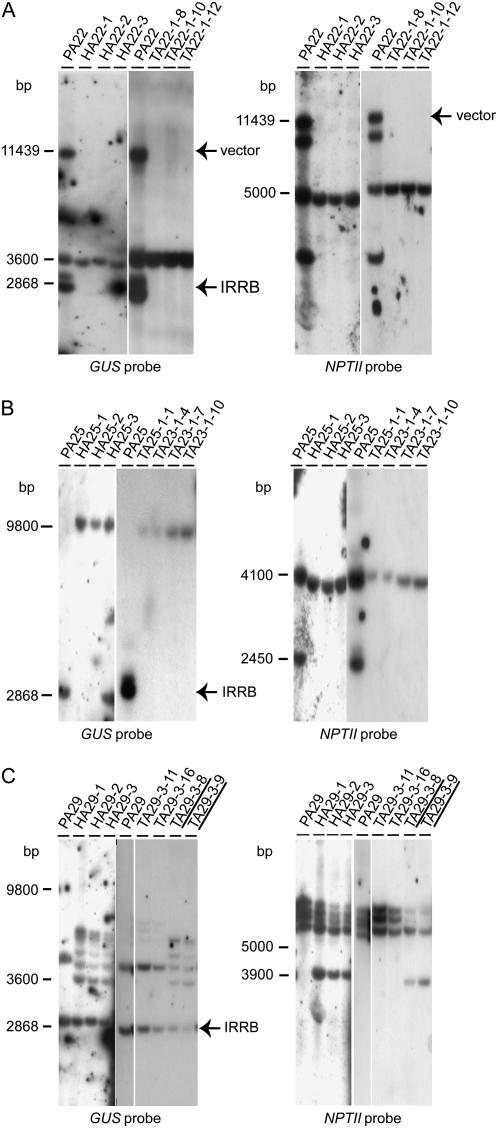
DNA gel-blot analysis on EcoRV-digested DNA. DNA gel-blot analysis on DNA of parental (P), hybrid (H), and F2 progeny (T) plants of A22 (A), A25 (B), and A29 (C), detected with the GUS (left) and NPTII probes (right), is shown. The hybrids were obtained after crossing the parental plants with the CRE13 line. On top of each lane, the name of the transformant is given from which DNA was prepared. When the Cre T-DNA is present in the TA transformant, the name of that transformant is underlined. Abbreviations are as in Figure 2.
To combine the target complex loci with the CRE recombinase, the PA transformants and the homozygous CRE-expressing lines CRE13 and HSC6 were crossed (“Materials and Methods”; Table II), resulting in hybrids HA and HCA, respectively. These HA and HCA hybrids are hemizygous for both the Ksb and the CRE loci (“Materials and Methods”). Results obtained after crossing with both CRE-expressing lines were analogous and, therefore, are not discussed separately. Additionally, neither the F1 hybrids nor the F2 progeny plants displayed any detectable effect of CRE expression on the phenotype.
Table II.
Nomenclature of the obtained hybrid and progeny plants
| Transgenic Plant | Nomenclature |
|---|---|
| PA × CRE13 | HA hybrids (Col-4 × C24 ecotype) |
| PA × HSC6 | HCA hybrids (Col-4 × Col-0 ecotype) |
| HA self-fertilized progeny | TA seedlings |
| HCA self-fertilized progeny | TCA seedlings |
Both hybrids and F2 progeny plants were analyzed by DNA gel-blot analysis and PCR to verify the T-DNA integration pattern, the possible resolution of the complex T-DNA locus, and the correctness of the loxP sequences. Additionally, real-time PCR gave an idea about the CRE expression in the progeny plants. GUS activity measurement indicated whether resolution was accompanied with an increased transgene expression level. Because the results for lines PA29, PA1, PA5, and PA16 were very similar, only those of PA29 will be discussed in more detail.
The CRE/loxP-Mediated Recombination System Resolved Complex SB-loxP-Derived T-DNA Loci into Single-Copy T-DNA Loci
The T-DNA integration patterns in hybrid and F2 progeny plants were compared with those in the original parental transformants (Figs. 2 and 3) after digestion with the methylation-insensitive enzymes EcoRV and SphI and hybridization with both the GUS and NPTII probes (“Materials and Methods”). In the hybrids of PA23, PA22, or PA25 and a CRE-expressing line and in their F2 progeny, resolution of the complex T-DNA loci to one T-DNA copy could be demonstrated (Figs. 2, A and B, and 3, A and B).
In hybrids HA23 and HCA23, the band of 11,439 bp, corresponding to the GUS-vector-NPTII junction between both T-DNAs, disappeared, whereas, as expected, the left and right T-DNA/plant junction fragments remained (Fig. 2, A and B). Interestingly, an additional T-DNA/plant junction fragment was generated for both probes, one of 12,661 bp for the GUS probe and one of 3,639 bp for the NPTII probe (Fig. 2, A and B). The extra right and left T-DNA/plant junctions resulted from the constitutive CRE recombinase expression, causing continuous inversion of the DNA sequence between the two loxP sequences in inverted orientation. Because the lengths of the T-DNA and plant fragments in the parental T-DNA/plant junction were known, the length of the two extra T-DNA/plant junctions resulting from the inversion could be predicted (Fig. 2A) and corresponded to the extra bands found in the DNA gel-blot analysis (Fig. 2B). In the TA23/TCA23 F2 progeny plants in which the CRE recombinase gene segregated out, only one of the two T-DNA/plant junctions was present (Fig. 2, A and B). Indeed, in transformants TA23-1-3, TA23-1-7, and TA23-2-4, the T-DNA insert had its original orientation, whereas in transformant TA23-2-8 the inverted T-DNA insert was stabilized with the GUS and NPTII expression cassettes located next to the LB and right-border (RB) repeats, respectively (Fig. 2B). All these results support the conclusion that the new T-DNA/plant fragment in the hybrid at both the LB and the RB originated from an inversion and not from an additional T-DNA insertion.
Transgenic lines PA22 and PA25 contained an IRRB of two T-DNAs (Fig. 3, A and B). After hybridization with the GUS probe, the presence of the diagnostic 2,868-bp EcoRV fragment, consisting of twice the distance from the RB region until the EcoRV site, could be visualized (Figs. 1B and 3, A and B). PA22 contained two additional T-DNAs, of which one was linked in tandem with another T-DNA, interspersed with vector DNA, as predicted by the 11,439-bp EcoRV fragment visualized after hybridization with both the GUS and the NPTII probes. For the HA22/HCA22 and HA25/HCA25 hybrids, the resolution from multiple T-DNA copies to one T-DNA copy could be demonstrated based on, for instance, the reduction of four and two LB-T-DNA/plant junction fragments to one LB-T-DNA/plant junction fragment in HA22 and HA25 hybrids, respectively (Fig. 3, A and B). Continuous inversion of the DNA sequence in between the two remaining inversely oriented loxP sites did not occur.
In the hybrid plants HA29/HCA29, the LB and RB T-DNA/plant junction fragments were the same as those observed in the parental plant, albeit some with reduced intensity (Fig. 3C). In addition, a number of new fragments were visualized that resulted from CRE recombinase activity (Fig. 3C). Once the CRE recombinase gene segregated, the DNA integration pattern in the F2 progeny plants was the same as that of the parental plants or simplified, whereas, when the CRE recombinase was still present in the F2 progeny plants, the pattern was the same as in the F1 hybrid (Fig. 3C, TA29-3-8 and TA29-3-9). This observation indicates that deletion and/or inversion reactions might have taken place in some cells, but that most somatic cells of the analyzed leaf tissue and also the gametes had no resolved T-DNA locus.
The CRE/loxP Recombination System Can Generate New Isoforms from Complex SB-loxP-Derived T-DNA Loci
DNA gel-blot analysis clearly indicated that CRE activity resulted in recombination within a complex SB-loxP-derived T-DNA locus because the number of hybridizing fragments decreased both in the hybrid and progeny plants. This recombination might yield allelic T-DNA isoforms: inversion could result in a T-DNA harboring LB-GUS and RB-NPTII junctions, whereas resolution of an IRRB could lead to a T-DNA harboring two LB repeats/regions. To prove the resolution and the formation of allelic T-DNA isoforms, four PCRs were amplified with primers specific for regions encoding the LB and NPTII, the RB and GUS, the RB and NPTII, and the LB and GUS, and these PCR fragments were sequenced (Figs. 4, 5, and 6; “Materials and Methods”).
Figure 4.
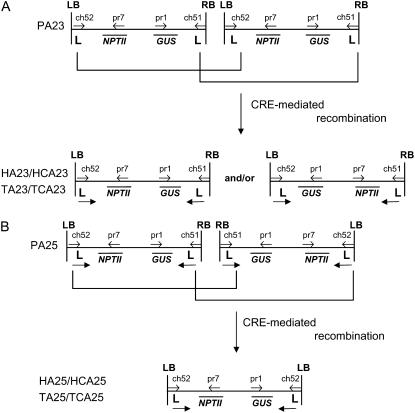
Schematic representation of the CRE/loxP-induced resolution of complex T-DNA loci. A, Resolution in HA23/HCA23 and TA23/TCA23 plants. Parental plant PA23 harbored two T-DNAs in tandem orientation. Because of the CRE/loxP-induced recombination between the two outermost loxP sequences, reduction to one T-DNA copy was obtained. Because in both hybrids and progeny plants the CRE recombinase could still be expressed, inversion of the T-DNA cassette was possible, resulting in the newly formed LB-GUS and RB-NPTII junctions. B, Resolution in the HA25/HCA25 and TA25/TCA25 plants. The parental plants PA25 contained two T-DNAs oriented as an inverted repeat over the RB. Upon resolution, deletion between the outermost loxP sequences was performed, and a newly formed “T-DNA” harboring two LB regions was obtained. The primers ch51, ch52, pr1, and pr7 are shown as arrows. Abbreviation: L, loxP sequence recognition site of the CRE recombinase.
Figure 5.
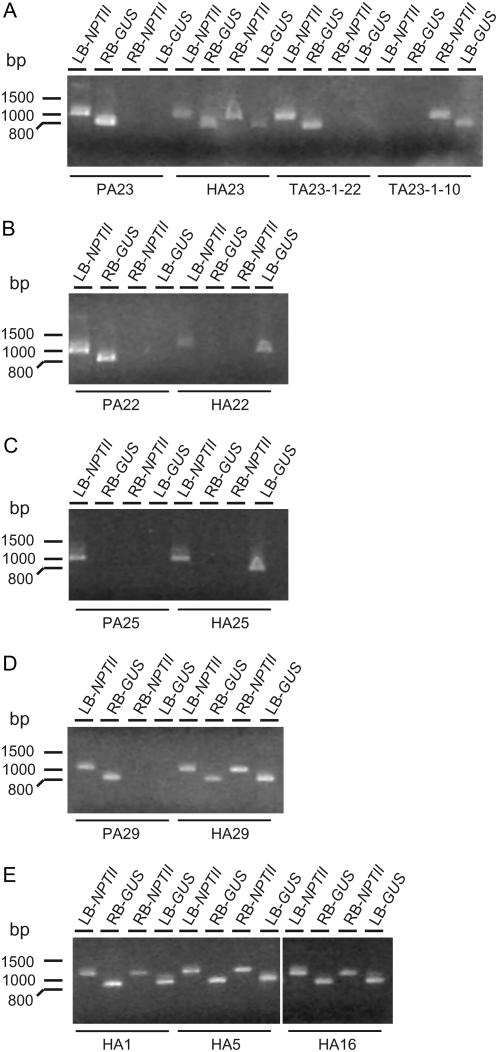
PCR analysis to identify the newly formed T-DNA configurations. PCR analysis on DNA of parental, hybrid, and progeny plants of A23 (A), A22 (B), A25 (C), A29 (D), and HA1, HA5, and HA16 (E) is shown. Ten microliters of PCR product was loaded on a 1% agarose gel. On top of each lane, the name of the transformant and the corresponding PCR reaction are given. LB-NPTII, Primer combination ch52 + pr7 amplified an LB-NPTII junction of 1,142 bp; RB-GUS, primer combination ch51 + pr1 amplified an RB-GUS junction of 905 bp; RB-NPTII, primer combination ch51 + pr7 amplified a RB-NPTII junction of 1,133 bp; LB-GUS, primer combination ch52 + pr1 amplified an LB-GUS junction of 914 bp.
Figure 6.
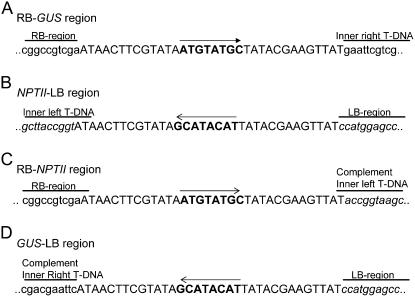
Sequence analysis of the RB-GUS region (A), NPTII-LB region (B), RB-NPTII region (C), and the GUS-LB region (D). The loxP sequence is indicated in capital letters, and the 8-bp spacer region of the loxP sequence in bold and its orientation by an arrow. The presence of the inverted T-DNA orientation was also clearly demonstrated: the RB-NPTII fragment was composed of the original RB-loxP region and the complement sequence of the inner LB T-DNA region, while the LB-GUS fragment harbored the complement inner RB region and the original loxP-LB region.
From parental line PA23, as expected, only the LB-NPTII and RB-GUS T-DNA junctions could be amplified, and in the hybrid plants two additional new PCR products, the LB-GUS and RB-NPTII junctions, could be demonstrated (Figs. 4A, 5A, and 6, A and B). That the hybrids were chimeric for the inverted and original T-DNA orientation was confirmed by the presence of the four different PCR products (Fig. 5A). The presence of the T-DNA isoform was also corroborated by sequence data. The RB-NPTII fragment consisted of the original RB-loxP region and the complement sequence of the inner LB T-DNA region (Fig. 6C), and the LB-GUS fragment harbored the LB-loxP region and the original complement inner RB region (Fig. 6D). As described above, in TA23/TCA23 F2 progeny plants, the CRE recombinase could segregate away from the target T-DNA locus, and in these plants only one of the two possible T-DNA isoforms was present (Figs. 4A, 5A, and 6, A and B). Transformant TA23-1-22 had the original T-DNA configuration (Fig. 5A), and only PCR fragments for LB-NPTII and RB-GUS junctions were amplified. The F2 progeny plant TA23-1-10 had the inverted T-DNA construct, and, correspondingly, only the newly formed RB-NPTII and LB-GUS junctions could be detected (Fig. 5A).
In parental line PA22, with four Ksb T-DNAs in one locus of which at least two were oriented in an IRRB, the expected LB-NPTII and RB-GUS fragments were amplified (Fig. 5B). In the parental line PA25, containing two T-DNAs in an IRRB, the LB-NPTII junction could be amplified, but not the RB-GUS junction, most probably because of intrastrand annealing of this region close to the center of the palindrome during the primer annealing step of the reaction (Figs. 4B and 5C). However, for both HA22/HCA22 and HA25/HCA25 hybrids, the recombination was clearly demonstrated by the amplification of the LB-NPTII and LB-GUS junctions, but not of the RB-GUS or of the RB-NPTII fragments, indicating resolution to a single T-DNA copy flanked by two LB regions and suggesting that the two outermost T-DNAs in parental lines PA22 and PA25 were oriented in an IRRB (Figs. 4B and 5, B and C). Although the DNA sequence in between the two remaining inversely oriented loxP sites were seemingly not continuously inverted in the hybrids, the loxP sequences at both LB regions of HA22/HCA22 and HA25/HCA25 did not contain any deletion or mutation (Fig. 6).
Finally, PCR and sequence analysis of HA29/HCA29, HA1, HA5, and HA16 revealed that also in these hybrids not only the original LB-NPTII and RB-GUS T-DNA junctions but also the LB-GUS and RB-NPTII junctions could be amplified (Fig. 5, D and E). In addition, sequencing of these PCR fragments revealed that the loxP sequences next to at least one LB and one RB region were not deleted or mutated and had the correct sequence (Fig. 6). In conclusion, newly formed T-DNA isoforms could clearly be demonstrated by PCR after CRE-mediated resolution of complex SB-loxP-derived T-DNA loci.
Simplification of Invertedly Repeated T-DNAs into a Single T-DNA Copy by the CRE/loxP Recombination System Can Result in Enhanced Transgene Expression
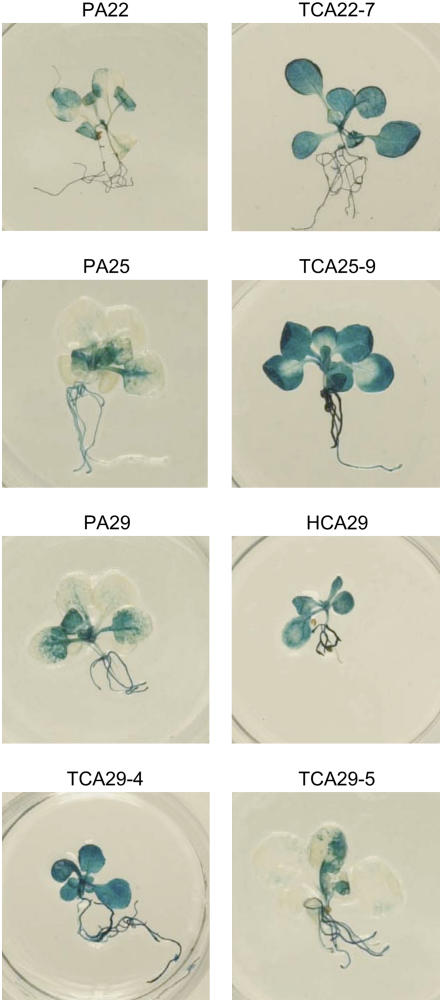
To determine whether the different T-DNA isoforms exhibited the same or different transgene expression levels and whether simplification of complex T-DNA loci resulted in an increased transgene expression, the GUS activity in the parental plants was compared with that in hybrid and progeny plants (Table III). The clearest effect of the reduced T-DNA copy number on transgene expression was seen in the hybrids and F2 progeny plants of PA22 and PA25. In parental lines PA22 and PA25, the GUS activity was low (<40 units of GUS/mg total soluble protein [TSP] and <10 units of GUS/mg TSP, respectively), both in 4- and 10-week-old seedlings (Table III), most probably because of the presence of at least one IRRB that efficiently triggers posttranscriptional gene silencing (De Buck et al., 2001). The low GUS expression in both parental lines was also reflected in a patchy pattern after histochemical staining (Fig. 7). Upon reduction of the complex locus into one T-DNA copy, GUS activity dramatically increased (3-fold to more than 10-fold) in both HA22/HCA22 and HA25/HCA25 hybrids, and the high levels were maintained in TA22/TCA22 and TA25/TCA25 F2 progeny plants (Table III). This stable and high transgene expression was also confirmed by a uniformly blue GUS staining of 2-week-old seedlings (Fig. 7). Additionally, the presence of the CRE-expressing T-DNA had no significant impact on the GUS activity levels (data not shown). For the different isogenic progeny plants, some variation in expression could be observed, possibly caused by the zygosity of the resolved locus, physiological effects, and experimental conditions (De Buck et al., 2004).
Table III.
GUS activity in parental, hybrid, and F2 progeny plants
| Sample | GUS Activity (Units of GUS/mg TSP)a
|
|
|---|---|---|
| 4-Week-Old Seedlings | 10-Week-Old Seedlings | |
| PA23 | 1,794 ± 506 (5) | 2,975 ± 1,226 (5) |
| HCA23 | 131 ± 39 (5) | 161 ± 83 (5) |
| TCA23-6 | 252 ± 170 (10) | 184 ± 62 (8) |
| TCA23-7 | 169 ± 98 (10) | 390 ± 288 (9) |
| PA22 | 31 ± 13 (5) | 2 ± 3 (5) |
| HCA22 | 174 ± 82 (5) | 451 (1) |
| TCA22-3 | 172 ± 124 (10) | 580 ± 404 (9) |
| TCA22-7 | 304 ± 173 (10) | 862 ± 530 (10) |
| PA25 | 6 ± 3 (4) | 0 ± 0 (4) |
| HCA25 | 178 ± 137 (5) | 514 ± 221 (4) |
| TCA25-1 | 286 ± 203 (10) | 1,008 ± 582 (7) |
| TCA25-9 | 497 ± 331 (10)b | 703 ± 454 (8)b |
| PA29 | 0 ± 0 (5) | 2 ± 1 (5) |
| HCA29 | 213 ± 227 (5) | 325 ± 276 (5) |
| TCA29-3 | 0 ± 0 (5) | 0 ± 0 (4) |
| TCA29-4 | 486 ± 991 (10) | 947 ± 1,722 (10) |
| TCA29-5 | 18 ± 6 (9) | 95 ± 211 (7) |
Means ± sd. The number of seedlings analyzed is indicated between parentheses.
One seedling of TCA25-9 was not taken into account to calculate the mean ± sd because the GUS activity level was above 8,000 units/mg and thus very different from the other values.
Figure 7.
Histochemical GUS staining of 2-week-old seedlings. Transformants with low GUS activity (PA22, PA25, PA29, and TCA29-5) had a patchy pattern, while transformants with a stable and high GUS expression (TCA22-7, TCA25-9, HCA29, and TCA29-4) were uniformly stained. Nontransgenic Col-0 plants had no blue staining (De Buck et al., 2004).
Because the GUS activity was high in the parental PA23 plants, reduction of a tandem repeat to a single T-DNA copy did not result in a further increase but rather in a decrease (Table III). The reason might be the copy dosage effect because the homozygous PA23 plants had four GUS transgenes whereas the hybrid HA23/HCA23 plants had only one. In all PA23, HA23/HCA23, and TA23/TCA23 plants, GUS staining was uniformly blue, indicating the absence of transgene silencing in these lines (Fig. 7).
For transformant PA29, the situation was more complex. The GUS activity was very low to zero, both in 4- and 10-week-old seedlings (Table III), and the histochemical GUS staining pattern was patchy, implying transgene silencing (Fig. 7). In the hybrid plants, GUS activity levels were variable; in some hybrids GUS activity increased more than 100-fold whereas in others it remained low (Table III). In the TCA29 progeny plants, three categories could be discerned (Table III). All TCA29-3 progeny plants had no GUS activity, notwithstanding the presence of several T-DNA copies as in the parental transformant (Table III). TCA29-4 progeny plants displayed variable GUS expression levels, varying from low (8 units of GUS/mg TSP) to very high (>1,000 units of GUS/mg TSP), irrespective of the presence of the CRE recombinase. Finally, in TCA29-5 progeny plants, GUS activities were higher than those measured in the parental line, but still too low (<20–100 units GUS/mg TSP) to be classified as high and stable (Table III). Again, GUS activities were not influenced by CRE recombinase activity. We postulate that in these HA29/HCA29 hybrids and TA29/TCA29 F2 progeny plants, the CRE/loxP recombination system reorganized the transgene locus in a variable number of somatic leaf cells, resulting in high transgene expression variability.
CRE mRNA Levels Were Very Comparable in the Different F2 Progeny Plants
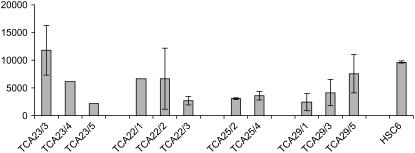
To investigate whether the observed lack of resolution or inversion in some transformants could be caused by differential CRE expression levels, RNA extracts from pools of seven to 10 2-week-old F2 seedlings were analyzed with a real-time quantitative PCR analysis (“Materials and Methods”; Fig. 8). CRE mRNA levels were very comparable in the progeny seedlings harboring the HSC T-DNA and derived from the different parental lines (Fig. 8). Indeed, the CRE mRNA levels in the progeny plants of A23, with a resolved locus, were not drastically different from the mRNA levels in the TCA29 seedlings in which no or very partial resolution was detected. The observed variation in CRE mRNA levels is probably related to the different Cre T-DNA zygosity in different F2 progeny plants.
Figure 8.
mRNA expression analysis of P35S-CRE in progeny seedlings and the CRE-expressing line HSC6. mRNA accumulation levels in seven to 10 pooled 2-week-old seedlings were determined by real-time PCR with ACTIN2 as constitutive control. The fold difference of expression in the analyzed lines compared to untransformed control is shown. Error bars are sd of two biological repeats.
DISCUSSION
We wanted to know whether the site-specific CRE recombinase in combination with a loxP-containing T-DNA vector could be an efficient method to obtain Arabidopsis transformants harboring a single T-DNA copy, implying enrichment for transformants with high and stable transgene expression. Indeed, the frequency with which single-copy transformants were obtained after floral-dip transformation varied between 4.8% and 22% (De Buck et al., 2004). Additionally, the majority of transformants contained these multiple T-DNAs at a single locus, implying that genetic segregation could not yield single-copy T-DNA segregants with a high frequency. Therefore, an SB-loxP T-DNA was designed in which two inversely oriented loxP sequences were located both immediately adjacent and inside of the LB and RB T-DNA ends. Seven randomly selected transformants, obtained after floral-dip transformation and containing two or more T-DNA copies at one genetic locus, were crossed with a CRE-expressing line. Three different complex loci were resolved to a single T-DNA copy in the hybrids and their progeny plants, demonstrating that the CRE-mediated resolution of three complex T-DNA loci in the F1 hybrid plants was transmitted to 100% of the progeny plants. Based on DNA gel-blot analysis, there was no indication that the released DNA fragment remained present extrachromosomally or was integrated at another position into the genome. In contrast, Srivastava and Ow (2003) reported that from one hybrid plant, four out of 20 F2 progeny plants not only showed incomplete resolution of the complex locus but also the presence of a circular loxP fragment. The frequency of observed site-specific mediated resolution of complex T-DNA loci can be compared with the frequency of resolution of transgene arrays obtained after direct gene transfer (Srivastava et al., 1999). Of the F2 progenies, 20% to 40% of three out of four wheat (Triticum aestivum) lines had maintained multiple copies of the transgenes after crossing the parental wheat line with a CRE-expressing line, suggesting that the germline cells were chimeric (Srivastava et al., 1999).
In four of the seven parental lines, CRE-mediated resolution of the complex T-DNA locus could not take place and only partial rearrangements were observed. The lack of resolution is likely not inhibited by the distance between the most outside-located loxP sites in tandem orientation because distance limitations seem to become relevant only when exceeding roughly 150 kb (Coppoolse et al., 2005). Several, mutually not exclusive, reasons might explain the lack of resolution in some lines. First, PCR and sequence analysis revealed that only LB and RB were linked to complete and nonmutated loxP sequences. Nevertheless, because the parental plants PA29, PA1, PA5, and PA16 contain multiple T-DNA copies, we cannot rule out that the most extreme loxP sequences were deleted, which makes resolution to a single copy impossible. Indeed, truncation of the left and/or right T-DNA ends is a common phenomenon (Brunaud et al., 2002; Windels et al., 2003). Second, although the CRE mRNA levels measured in the current study might possibly be sufficient to resolve the PA23, PA22, and PA25 loci, they might be too low to induce a complete resolution in the four other lines, as a previous analysis clearly indicated that a 2-fold difference in CRE can have a significant impact on the CRE-mediated excision frequency (Marjanac et al., 2007). Third, complex loci, consisting of repeated T-DNA units, might trigger heterochromatinization of the locus, making the loxP sequences inaccessible for the CRE recombinase and recombination and resolution very inefficient. Indeed, the chromosomal position of the loxP target has a strong impact and can result in a difference of deletion efficiency of at least 50-fold (Baubonis and Sauer, 1993). In addition, fusion of the loxP and FRT sequences and expression of the FLP and CRE alone increased deletion efficiency in tobacco (Nicotiana tabacum), suggesting that fused loxP-FRT sequences might enhance the alignment of the recognition sequences, DNA binding or cleavage, or the formation of a Holliday junction or DNA-recombinase complex (Luo et al., 2007). Therefore, we believe that packed chromatin in combination with insufficient CRE expression might be the major reasons for the lack of resolution of some complex loci.
Because the final goal of the resolution of complex into single-copy T-DNA loci mediated by the CRE/loxP recombination was to obtain plants with high and stable transgene expression levels, the effect of the changed transgene locus structure upon transgene expression was also examined. Either the GUS expression could significantly change or the epigenetic status of the 35S-GUS transgenes in the original T-DNA locus could be maintained (De Neve et al., 1999; Fojtová et al., 2006), resulting in similar transgene expression levels of the parental locus and the resolved locus. The three F2 progenies that contained only one T-DNA copy displayed high and stable transgene expression levels. For the hybrids and F2 progenies of PA22 and PA25, a clear increase of GUS activity, even more than 10-fold, could be observed when compared to the parental plants with an inverted repeat of the GUS gene. Furthermore, the newly formed single-copy allelic loci had expression levels that were as high as those of independently obtained single-copy Ksb T-DNA transformants (De Buck et al., 2004). Similarly, in transgenic mice, a CRE/loxP resolution strategy resulted not only in a reduced copy number but also in a marked increase in transgene expression (Garrick et al., 1998). In parental PA23 plants, HA23/HCA23 hybrids, and F2 progeny plants, GUS activity levels correlated with the gene dosage (De Buck and Depicker, 2004). Indeed, homozygous PA23 plants contain four Ksb T-DNA copies that give rise to high and stable transgene expression. In the HA23/HCA23 hybrid plants with a resolved locus and one T-DNA copy in hemizygous condition, an approximately 4-fold dosage-dependent decrease in GUS activity was measured. For the complex PA29 loci, which could not be resolved to one T-DNA copy, the GUS activity levels remained below detection in the F2 progeny of one hybrid, while they varied in F2 progeny plants derived from another F1 plant from low (<20 units of GUS/mg TSP) to very high (>1,000 units of GUS/mg TSP). Part of the explanation for this variation might be that the presence of CRE in three-quarters of the F2 progeny plants mediates T-DNA locus rearrangements in the somatic cells. In this way, the inverted repeat of two GUS genes may be converted to a tandem repeat and this might result in enhanced GUS expression. The variation may relate to the time during development at which the inversion occurred, similarly to the variation of GUS expression observed in the F1 plant. However, still other unidentified changes are going on. Molecular analysis of some of the high expressing F2 progeny plants showed that, even in the absence of CRE, these plants had the same locus structure as the low expressing parental plants, suggesting that CRE can induce epigenetic changes in transgene expression.
In conclusion, upon Agrobacterium-mediated transformation with an SB-loxP-derived T-DNA, complex loci can efficiently be resolved into single-copy T-DNA inserts through site-specific CRE recombination. This CRE-mediated resolution of complex loci can differentially affect the transgene expression levels: (1) resolution of complex T-DNA loci with invertedly repeated transgenes and low expression levels into single-copy loci increases transgene expression 10- to 100-fold; (2) resolution of multiple-copy T-DNA loci with high transgene expression levels to a single-copy locus is correlated with dosage-dependent transgene expression levels; and (3) complex T-DNA loci that are not resolved have variable expression levels in different progeny plants with the same complex locus, indicating epigenetic modifications of the transgene expression levels. The developed T-DNA vector SB-loxP contains a multicloning site between two invertedly oriented loxP sequences, which makes the presented vector very suitable for cloning transgenes. Derived T-DNA loci can be resolved to single-copy elite events that express the gene of interest at a high and stable level.
MATERIALS AND METHODS
Constructs and Strains
An outline of the SB-loxP and Ksb T-DNAs is given in Figure 1. The plasmid pSB-loxP was derived from the plasmid pGSV5 (kind gift of Johan Botterman, Bayer Crop Science NV), in which all T-DNA sequences between the borders were deleted and substituted by a multicloning site. In a first step, compatible oligonucleotides containing two loxP sequences in inverted orientation with a multicloning site in between were inserted into the SalI/NcoI site of pGSV5 to yield the plasmid pSB-loxP. The sequences of the oligonucleotides were as follows: 5′ATAACTTCGTATAATGTATGCTATACGAAGTTAT3′ for the SalI-compatible/loxP fragment; 5′GAATTCGTCGACGTTAACAAGCTTACCGGT3′ for the multicloning site; and 5′ATAACTTCGTATAGCATACATTATACGAAGTTATC3′ for the loxP/NcoI fragment. In a second step, the P35S-gus-3′nos-Pnos-NPTII-3′ocs EcoRI/AgeI fragment of pXD610 (De Loose et al., 1995) was inserted into the EcoRI/AgeI-cut vector SB-loxP, yielding pKsb (De Buck et al., 2000a). For the construction of the plasmid pCre that harbors the Cre T-DNA, refer to De Buck et al. (2000b). The Cre T-DNA carries the phosphinothricin phosphotransferase gene (bar) driven by the promoter of the Rubisco small subunit gene of Arabidopsis (Arabidopsis thaliana [L.] Heynh.; PSSUARA) and a CRE recombinase (CRE; Dale and Ow, 1990, 1991) expression cassette under control of the 35S promoter of the Cauliflower mosaic virus (P35S; Odell et al., 1985).
To design the pHSC T-DNA, the CRE-coding sequence was amplified from the plasmid pMM23 (Dale and Ow, 1990, 1991) with primers including the SacII and NcoI restrictions sites. The sequences of the primers were as follow: 5′TCCCCGCGGGTTGACATGTCCAATTTACTGACC3′ and 5′CATGCCATGGGAATTCTTACTAATCGCCATCTTCC3′. The 1,049-bp-long CRE fragment was cloned into the SacII/NcoI sites of the plasmid pT35S that contained the 3′35S terminator sequence in the vector pGEM5Zf(+). Subsequently, the SacI/ApaI CRE-3′35S fragment was inserted into the SacI/ApaI restriction sites of the binary vector pPZP200 (Hajdukiewicz et al., 1994), already carrying the Pnos-hpt-3′nos expression cassette. This cloning resulted in the plasmid pHCreT35S. To create a GATEWAY-compatible vector, the rfa cassette (Invitrogen) was cloned upstream of the CRE recombinase by inserting an extra oligonucleotide, containing the PmlI and SwaI restriction sites, into the HindIII site of the plasmid pHCreT35S, in which the GATEWAY cassette of 1,700 nucleotides was cloned in the PmlI site of the oligonucleotide inserted, resulting in the destination vector pHGWC. The transcriptionally correct insert orientation was confirmed by restriction analysis. To obtain the P35S entry clone, the P35S sequence was amplified by PCR from pXD610 (De Loose et al., 1995). The attB1 and attB2 recombination sites were formed by two consecutive PCRs. First, template-specific primers were used containing 12 bases of the attB1 sites, followed by universal attB adapter primers to amplify the full attB1 and attB2 recombination sites (Invitrogen). The resulting PCR product, flanked by attB1 sites, was recombined in the vector pDONR201-KmR containing the attP1 and attP2 recombination sites with the BP clonase enzyme (Invitrogen). The T-DNA vector HSC was generated by an LR reaction in which the P35S entry clone was incubated with the destination vector pHGWC in the presence of the LR clonase enzyme (Invitrogen). Plasmids pKsb and pCre were transferred to the Agrobacterium tumefaciens strain C58C1RifR(pGV2260) by triparental conjugation and plasmid pHSC to the Agrobacterium strain C58C1RifR(pMP90) by electroporation.
Generation of Two CRE-Expressing Plants
The CRE13 line was obtained after transforming Arabidopsis (ecotype C24) root cells with the Cre T-DNA, as described by Valvekens et al. (1988). After selection on phosphinothricin (10 mg/L), 13 different primary transformants were allowed to self-fertilize. The locus number was determined and only plants containing one locus were retained for further analysis. The HSC6 line was obtained after floral-dip transformation (Clough and Bent, 1998) of Arabidopsis (ecotype Columbia [Col]-0) with the HSC T-DNA. Of the 22 HSC primary transformants, resistant to hygromycin and carrying the P35S-CRE-3′35S cassette, only plants that had one HSC locus were further analyzed. Transformant HSC6 was retained.
The CRE13-transformed line contained a 35S-CRE gene in the C24 ecotype background (De Buck et al., 2000b), whereas the HSC6 transformant had a similar 35S-CRE gene in the Col-0 ecotype background. The CRE expression levels in CRE13 and HSC6 were very comparable as revealed by real-time PCR (Marjanac et al., 2007). To control for in vivo CRE activity in the two CRE-expressing lines, both CRE13 and HSC6 were crossed with a line containing a GUS gene between two loxP sites in the K T-DNA (De Buck et al., 1998). This K T-DNA harbors a GUS-expressing cassette surrounded by two loxP sequences in direct orientation (De Buck et al., 1998). Activity of the CRE recombinase removed this GUS cassette in the hybrids, indicating that the CRE recombinase was expressed both in CRE13 and HSC6 (data not shown).
Generation of Transgenic Arabidopsis Plants with a Complex Ksb T-DNA Integration Pattern
Transformants with Ksb T-DNA loci were obtained after a floral-dip transformation of Arabidopsis (ecotype Col-4-N944; Clough and Bent, 1998). Twenty-eight transformants were selected on K1 medium supplemented with kanamycin (50 mg/L) and screened for a simple locus. DNA gel-blot analyses with two different restriction enzymes (EcoRV and SphI) and the NPTII and GUS probes were performed (Fig. 1) to determine the number and configuration of the integrated T-DNAs (Figs. 2 and 3). The seven transformants with a complex locus that were retained for further analysis are presented in Table I. PA23 and PA25 were homozygous for the transgene locus, whereas PA22 and PA29 were heterogeneous seed stocks.
Introduction of the CRE Recombinase into the Transformants with a Complex Ksb T-DNA Locus
By crossing the parental transformants with the CRE-expressing line CRE13 or line HSC6, the CRE recombinase was introduced into the transformants with a complex Ksb T-DNA locus. For these crosses, the parental lines were used as male plants and the CRE-expressing plants as pollen acceptors. The hybrid seeds were selected on medium containing both kanamycin to select for the Ksb T-DNA locus, and phosphinothricin or hygromycin to select for the CRE13 and HSC6 T-DNA locus, respectively. Therefore, the hybrid seeds were hemizygous for both the Ksb and the CRE loci. The progeny plants of these hybrids were grown on medium containing only kanamycin, so that progeny plants with and without the CRE recombinase could be analyzed. The fact that the CRE13 and the parental plants had C24 and Col-4 ecotypes, respectively, was not an obstacle to obtain hybrids. Nevertheless, the hybrids had big leaves and stems appeared later than in the two parental plants and in the hybrids obtained after crossing the HSC6 (Col-0 ecotype) and PA (Col-4 ecotype). The hybrids HA and HCA (Table II) were allowed to self-pollinate and seeds were selected to grow plants in the next generation (designated TA and TCA; Table II).
Plant DNA Preparation, DNA Gel-Blot Analysis, and PCR Analysis to Estimate the T-DNA Copy Number and T-DNA Configurations in Parental, Hybrid, and Progeny Plants
DNA of Arabidopsis leaf material was prepared as described by De Neve et al. (1997) and the GUS (probe 1) and NPTII probes (probe 2) according to De Buck et al. (1999). Arabidopsis DNA (1 μg) was loaded in each lane of a 1% agarose gel. A nonradioactive method (GeneImages random prime-labeling module and GeneImages CDP star detection module; GE-Healthcare) was used to label and detect the DNA.
Presence of newly formed T-DNA isoforms was also monitored by PCR with border-specific primers (primer ch52 and primer ch51 for LB and RB regions, respectively) and primers specific for the GUS (pr1) or NPTII (pr7) expression cassettes. Approximately 100 ng of DNA was incubated with 250 ng of each primer in 1× Taq polymerase incubation buffer (Invitrogen) and 3 units of Taq polymerase in a final volume of 50 μL. Samples were heated to 94°C for 5 min before PCR, followed by 30 cycles of amplification. All primer combinations were denatured at 94°C for 1 min and the extension reaction was at 72°C for 1 min. For the primer combinations ch52 + pr7 and ch51 + pr1, annealing occurred at 54°C, for ch52 + pr1 at 52.6°C, and for ch51 + pr7 at 57.3°C, all reactions lasting 1 min. Sequences of the primers from 5′ to 3′ was as follows: primer ch51, CTCGGCCGTCGAATAACTTCG; primer ch52, TCAATTGTAAATGGCTCC; primer pr1, ATCACCTGCGTCAATGTAAT; and primer pr7, GTGCTCGACGTTGTCACTGAA.
Preparation of Protein Extracts and Determination of GUS Activity by Fluorimetric Measurement and Histochemical Staining
Protein extracts were prepared from 4- and 10-week-old fresh leaf material by grinding three leaves per seedling in 100 μL of buffer containing 50 mm phosphate buffer, pH 7, 10 mm 2-mercapthoethanol, 10 mm Na2-EDTA, and 0.1% Triton X-100. The ground plant tissue was centrifuged (15,000g) twice at 4°C for 10 min to remove insoluble material. GUS activity, expressed as units of GUS protein relative to the total amount of soluble extracted protein (units GUS protein mg−1), was determined as described by Bradford (1976) and Breyne et al. (1993).
Real-Time PCR Analysis
RNA was extracted from seven to 10 pooled 2-week-old Arabidopsis seedlings grown in K1 medium with TriZol reagent (MRC) according to the manufacturer's instructions. Per transgenic seed stock, two RNA samples were prepared. Poly d(T) cDNA was synthesized from 2 μg of total RNA with Superscript II reverse transcriptase (Invitrogen). Of the obtained cDNA, 1 μL was used for a PCR with ACTIN primers to check for contamination of residual DNA. The following primers were used: forward primer 5′ccacctgaaaggaagt3′ and reverse primer 5′aaaacaatgggactaaaacgcga3′. CRE expression was quantified on an iCycler real-time PCR detection system (Bio-Rad) with the qPCR core kit for SYBR Green I (Eurogentec). PCRs were carried out in triplicate. Relative expression levels were first normalized to ACTIN2 expression and then to the respective untransformed controls with the 2−ΔΔCT method (Livak and Schmittgen, 2001). Specific primer pairs were designed with Beacon Designer 4.0 (Premier Biosoft International): At3g18780/ACTIN2, 5′gttgactacgagcaggagatgg3′ and 5′acaaacgagggctggaacaag3′; and CRE, 5′tttcccgcagaacctgaagatg3′ and 5′atccgccgcataaccagtg3′.
Acknowledgments
We thank Anni Jacobs, Griet D en Herder, Annelies De Clercq, Marion Naudts, and Joke Devos for practical assistance; Dr. Mansour Karimi for help with GATEWAY cloning; Dr. Annick Bleys for critical reading of the manuscript and helpful comments; Karel Spruyt for photographic work and help by preparing the figures; and Martine De Cock for help in preparing the manuscript.
This work was supported by grants from the European Union BIOTECH program (QLRT–2000–00078), with additional cofinancing from the Flemish Community, the 6th framework program of the European Union “GENINTEG” (LSHG–CT2003–503303), and the “Bijzondere Onderzoeksfonds” of Ghent University (BOF 01111400).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ann Depicker (ann.depicker@psb.ugent.be).
References
- Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7 649–659 [DOI] [PubMed] [Google Scholar]
- Baubonis W, Sauer B (1993) Genomic targeting with purified Cre recombinase. Nucleic Acids Res 21 2025–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Breyne P, De Loose M, Dedonder A, Van Montagu M, Depicker A (1993) Quantitative kinetic analysis of β-glucuronidase activities using a computer-directed microtiter plate reader. Plant Mol Biol Rep 11 21–31 [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 3 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla R, Ariza-Nieto M, Wilson AJ, Moore SK, Srivastava V (2006) Transgene expression produced by biolistic-mediated, site-specific gene integration is consistently inherited by the subsequent generations. Plant Biotechnol J 4 209–218 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coppoolse ER, de Vroomen MJ, van Gennip F, Hersmus BJM, van Haaren MJJ (2005) Size does matter: Cre-mediated somatic deletion efficiency depends on the distance between the target lox-sites. Plant Mol Biol 58 687–698 [DOI] [PubMed] [Google Scholar]
- Dale EC, Ow DW (1990) Intra- and intermolecular site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene 91 79–85 [DOI] [PubMed] [Google Scholar]
- Dale EC, Ow DW (1991) Gene transfer with subsequent removal of the selection gene from the host genome. Proc Natl Acad Sci USA 88 10558–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CD, Lee E, Kobayashi J, Holappa LD, Albert H, Ow DW (2000) Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev 14 2869–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M, Debrouwer D, Moens T (1997) The development of a nuclear male sterility system in wheat. Expression of the barnase gene under the control of tapetum specific promoters. Theor Appl Genet 95 125–131 [Google Scholar]
- De Buck S, De Wilde C, Van Montagu M, Depicker A (2000. a) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol Breed 6 459–468 [Google Scholar]
- De Buck S, De Wilde C, Van Montagu M, Depicker A (2000. b) Determination of the T-DNA transfer and the T-DNA integration frequencies upon cocultivation of Arabidopsis thaliana root explants. Mol Plant Microbe Interact 13 658–665 [DOI] [PubMed] [Google Scholar]
- De Buck S, Depicker A (2004) Gene expression and level of expression. In P Christou, H Klee, eds, Handbook of Plant Biotechnology, Vol 1. John Wiley & Sons, Chichester, UK, pp 331–345
- De Buck S, Jacobs A, Van Montagu M, Depicker A (1998) Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol Plant Microbe Interact 11 449–457 [DOI] [PubMed] [Google Scholar]
- De Buck S, Jacobs A, Van Montagu M, Depicker A (1999) The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J 20 295–304 [DOI] [PubMed] [Google Scholar]
- De Buck S, Van Montagu M, Depicker A (2001) Transgene silencing of invertedly repeated transgenes is released upon deletion of one of the transgenes involved. Plant Mol Biol 46 433–445 [DOI] [PubMed] [Google Scholar]
- De Buck S, Windels P, De Loose M, Depicker A (2004) Single-copy T-DNAs integrated at different positions in the Arabidopsis genome display uniform and comparable β-glucuronidase accumulation levels. Cell Mol Life Sci 61 2632–2645 [DOI] [PubMed] [Google Scholar]
- De Loose M, Danthinne X, Van Bockstaele E, Van Montagu M, Depicker A (1995) Different 5′ leader sequences modulate β-glucuronidase accumulation levels in transgenic Nicotiana tabacum plants. Euphytica 85 209–216 [Google Scholar]
- De Neve M, De Buck S, De Wilde C, Van Houdt H, Strobbe I, Jacobs A, Van Montagu M, Depicker A (1999) Gene silencing results in instability of antibody production in transgenic plants. Mol Gen Genet 260 582–592 [DOI] [PubMed] [Google Scholar]
- De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from ligation of separate T-DNAs. Plant J 11 15–29 [DOI] [PubMed] [Google Scholar]
- Elmayan T, Vaucheret H (1996) Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J 9 787–797 [Google Scholar]
- Fojtová M, Bleys A, Bedřichová J, Van Houdt H, Křižová K, Depicker A, Kovařík A (2006) The trans-silencing capacity of invertedly repeated transgenes depends on their epigenetic state in tobacco. Nucleic Acids Res 34 2280–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin DIK, Whitelaw E (1998) Repeat-induced gene silencing in mammals. Nat Genet 18 56–59 [DOI] [PubMed] [Google Scholar]
- Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67 16–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Hansen G, Chilton M-D (1996) “Agrolistic” transformation of plant cells: integration of T-strands generated in planta. Proc Natl Acad Sci USA 93 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen G, Shillito RD, Chilton M-D (1997) T-strand integration in maize protoplasts after codelivery of a T-DNA substrate and virulence genes. Proc Natl Acad Sci USA 94 11726–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa TTC, Bong BB, Huq E, Hodges TK (2002) Cre/lox site-specific recombination controls the excision of a transgene from the rice genome. Theor Appl Genet 104 518–525 [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Warkentin TD, DeLong CMO (1993) Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol 21 17–26 [DOI] [PubMed] [Google Scholar]
- Jia H, Pang Y, Chen X, Fang R (2006) Removal of the selectable marker gene from transgenic tobacco plants by expression of Cre recombinase from a Tobacco Mosaic Virus vector through agroinfection. Transgenic Res 15 375–384 [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA (1996) Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol Biol 31 957–973 [DOI] [PubMed] [Google Scholar]
- Joubès J, De Schutter K, Verkest A, Inzé D, De Veylder L (2004) Conditional, recombinase-mediated, expression of genes in plant cell cultures. Plant J 37 889–896 [DOI] [PubMed] [Google Scholar]
- Koprek T, Rangel S, McElroy D, Louwerse JD, Williams-Carrier RE, Lemaux PG (2001) Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barley. Plant Physiol 125 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CN Jr, McAvoy R, Jiang X, Wu Y, et al (2007) ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5 263–274 [DOI] [PubMed] [Google Scholar]
- Lyznik LA, Gordon-Kamm WJ, Tao Y (2003) Site-specific recombination for genetic engineering in plants. Plant Cell Rep 21 925–932 [DOI] [PubMed] [Google Scholar]
- Marjanac G, De Paepe A, Peck I, Jacobs A, De Buck S, Depicker A (2007) Evaluation of CRE-mediated gene excision in Arabidopsis thaliana. Transgenic Res (in press) [DOI] [PubMed]
- Meyer P (2001) Gene silencing in plants. In Nature Publishing Group, ed, Encyclopedia of Life Sciences. MacMillan Press, New York, pp 1–5
- Moore SK, Srivastava V (2006) Efficient deletion of transgenic DNA from complex integration locus of rice mediated by Cre/lox recombination system. Crop Sci 46 700–705 [Google Scholar]
- Muskens MWM, Vissers APA, Mol JNM, Kooter JM (2000) Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol Biol 43 243–260 [DOI] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua N-H (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812 [DOI] [PubMed] [Google Scholar]
- Ow DW (2002) Recombinase-directed plant transformation for the post-genomic era. Plant Mol Biol 48 183–200 [PubMed] [Google Scholar]
- Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing of Arabidopsis T-DNA transformants: the predominant role of gene-specific RNA sensing mechanism versus position effects. Plant Cell 16 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekala C, Wu L, Gu K, Wang D, Tian D, Yin Z (2005) Excision of a selectable marker in transgenic rice (Oryza sativa L.) using a chemically regulated Cre/loxP system. Plant Cell Rep 24 86–94 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Anderson OD, Ow DW (1999) Single-copy transgenic wheat generated through the resolution of complex integration patterns. Proc Natl Acad Sci USA 96 11117–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Ariza-Nieto M, Wilson AJ (2004) Cre-mediated site-specific gene integration for consistent transgene expression in rice. Plant Biotechnol J 2 169–179 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW (2001) Single-copy primary transformants of maize obtained through the co-introduction of a recombinase-expressing construct. Plant Mol Biol 46 561–566 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW (2003) Rare instances of Cre-mediated deletion product maintained in transgenic wheat. Plant Mol Biol 52 661–668 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Ow DW (2002) Biolistic mediated site-specific integration in rice. Mol Breed 8 345–349 [Google Scholar]
- Srivastava V, Ow DW (2004) Marker-free site-specific gene integration in plants. Trends Biotechnol 22 627–629 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V (2006) Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol 17 147–154 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Li J, Lacroix B, Citovsky V (2004) Agrobacterium T-DNA integration: molecules and models. Trends Genet 20 375–383 [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K, Gupta AK, Sharma A (2003) The current status of plant transformation technologies. Curr Sci 84 368–380 [Google Scholar]
- Vergunst AC, Hooykaas PJJ (1998) Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol Biol 38 393–406 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Jansen LET, Hooykaas PJJ (1998) Site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana mediated by Cre recombinase. Nucleic Acids Res 26 2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen B, Hu Y, Li J, Lin Z (2005) Inducible excision of selectable marker gene from transgenic plants by the Cre/lox site-specific recombination system. Transgenic Res 14 605–614 [DOI] [PubMed] [Google Scholar]
- Windels P, De Buck S, Van Bockstaele E, De Loose M, Depicker A (2003) T-DNA integration in Arabidopsis chromosomes. Presence and origin of filler DNA sequences. Plant Physiol 133 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Subbarao S, Addae P, Shen A, Armstrong C, Peschke V, Gilbertson L (2003) Cre/lox-mediated marker gene excision in transgenic maize (Zea mays L.) plants. Theor Appl Genet 107 1157–1168 [DOI] [PubMed] [Google Scholar]