Abstract
The role of Pleistocene forest refugia and rivers in the evolutionary diversification of tropical biota has been the subject of considerable debate. A range-wide analysis of gorilla mitochondrial and nuclear variation was used to test the potential role of both refugia and rivers in shaping genetic diversity in current populations. Results reveal strong patterns of regional differentiation that are consistent with refugial hypotheses for central Africa. Four major mitochondrial haplogroups are evident with the greatest divergence between eastern (A, B) and western (C, D) gorillas. Coalescent simulations reject a model of recent east–west separation during the last glacial maximum but are consistent with a divergence time within the Pleistocene. Microsatellite data also support a similar regional pattern of population genetic structure. Signatures of demographic expansion were detected in eastern lowland (B) and Gabon/Congo (D3) mitochondrial haplogroups and are consistent with a history of postglacial expansion from formerly isolated refugia. Although most mitochondrial haplogroups are regionally defined, limited admixture is evident between neighboring haplogroups. Mantel tests reveal a significant isolation-by-distance effect among western lowland gorilla populations. However, mitochondrial genetic distances also correlate with the distance required to circumnavigate intervening rivers, indicating a possible role for rivers in partitioning gorilla genetic diversity. Comparative data are needed to evaluate the importance of both mechanisms of vicariance in other African rainforest taxa.
Keywords: control region, mitochondrial, phylogeography, refugium
Mechanisms underlying evolutionary diversification in tropical forests have intrigued biologists for more than a century. Numerous hypotheses have been proposed (1–2), of which the Pleistocene forest refugium and riverine barrier hypotheses have provoked considerable interest and controversy (3–7). Whereas most studies have focused on the Amazon and Australian wet tropics (5, 8–10), data on central African rainforest taxa remain relatively sparse.
According to Pleistocene refuge theory, forest fragmentation during glacial maxima led to the isolation and subsequent diversification of forest-associated taxa (3). During periods of climate amelioration and population expansion, zones of secondary contact may have also formed between neighboring refugial populations. Although palynological and biogeographical data have been used to infer forest refugia (11–16), their precise location and role in Pleistocene diversification is controversial. Several molecular studies suggest that refugia may have played an important role in structuring montane birds (17–19), primates (20–22), and trees (23). However, other forest species such as chimpanzees (24–26) and elephants (27, 28) show relatively weak regional genetic structure, suggesting that wide-ranging and/or savannah-tolerant species may be poor indicators of range changes in tropical forest cover. One criticism of the Pleistocene refuge hypothesis argues that species divergence times often predate the Pleistocene, undermining the role of refugia in geographical speciation (29, 30). However, it could also be argued that the recent time frame of Pleistocene events requires a population-genetic rather than a species-level approach. Another criticism centers on the real difficulties of pinpointing the precise location of putative refugia (4) and discriminating between competing modes of diversification (5). Molecular data can, however, be used to infer signatures of past history and, in so doing, test several important predictions of Pleistocene hypotheses (3, 7, 31): (i) allopatric fragmentation is evident and coincident with the possible location of hypothesized refugia; (ii) signature(s) of demographic expansion within refugial populations are apparent; and (iii) haplotype exchange is coincident with boundaries of adjacent refugia.
In contrast, the riverine barrier hypothesis argues that tropical rivers limit species distributions (32, 33) and shape intraspecific patterns of diversification (34, 35). To date, support for this hypothesis is equivocal and depends heavily on the ecology and dispersal abilities of individual taxa. In central Africa, the Sanaga River is an important biogeographic boundary for several groups, including some primates and forest duikers (25, 36). For chimpanzee subspecies, however, this proposed barrier may be incomplete (37). Other studies have also reported that rivers influence genetic differentiation between mandrill (21) and bonobo (38) populations. Where sampling is of sufficient intensity, molecular data can be used to test predictions of the riverine barrier hypothesis, namely, that genetic variation is structured across different river banks, and that genetic differentiation decreases from the mouth to the headwaters of a given watercourse.
Gorillas are good candidates for testing geographically explicit hypotheses of vicariance because of their strong association with closed canopy forest and their restricted ability to traverse savannah–forest mosaic habitats (39). Gorillas may also be good models for testing the isolating effects of rivers because the taxonomic boundaries of many primates appear coincident with major river courses (33, 36). By drawing on data from a range-wide analysis of gorilla populations, we aim to uncover the relative importance of rainforest refugia and rivers in shaping gorilla population structure and, in so doing, provide insight into historical processes that underlie the evolution of tropical forest biota.
Results
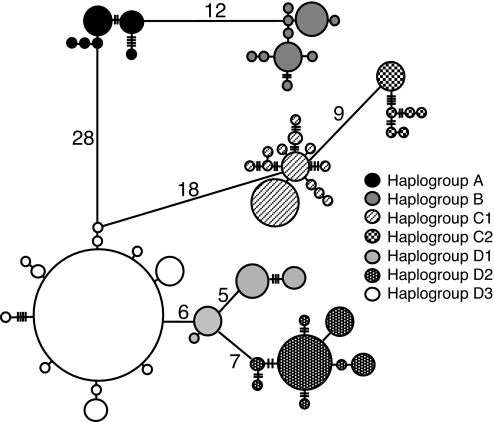
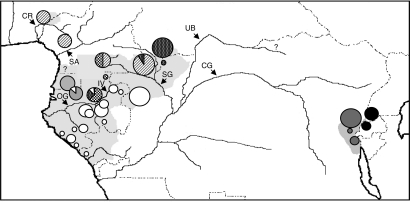
The minimum spanning network (Fig. 1) illustrates the deep divergence between eastern and western gorillas and the high levels of substructuring present in western gorillas relative to other gorilla subspecies. As previously observed (22), the divergence between eastern and western gorilla populations is far greater than that observed between mountain (A) and eastern lowland (B) gorillas. Phylogenetic analyses provide substantial statistical support for two regionally defined haplogroups (C, D) within western gorillas [supporting information (SI) Figs. 4 and 5]. The first (C) extends from the Cross River area in Nigeria to southeastern Cameroon and comprises two subgroups (C1, C2). Whereas C1 is distributed across the entire region, C2 is limited to Dja in central Cameroon, Minkébé in Northern Gabon, and the west bank of the Ivindo/Ayala River in central Gabon (Fig. 2). The second major haplogroup (D) extends from coastal Gabon eastwards to Congo and the southern tip of the Central African Republic (CAR). Within haplogroup D, three geographically defined subgroups are evident: (i) D1 in the montane regions of Equatorial Guinea and the adjacent Monts de Cristal in northwestern Gabon; (ii) D2 in the Dzanga-Sangha region of CAR; and (iii) D3 across much of Gabon and east to Lossi, Congo (Fig. 2). Although there is strong support for D2 and D3, subgroup D1 is only weakly differentiated.
Fig. 1.
Minimum spanning tree of mitochondrial HV1 haplotypes identified in all three major gorilla subspecies. The mutational steps separating haplotypes are indicated by using cross bars or by number. Major mitochondrial haplogroups (A–D) and their respective subdivisions (C1–2, D1–3) are based on haplogroups recovered in the phylogeny (see SI Figs. 4 and 5).
Fig. 2.
Geographic distribution of major mitochondrial haplogroups across central Africa. Important rivers are also indicated with an arrow. CR, Cross River; SA, Sanaga River; OG, Ogooué River; IV, Ivindo River; SG, Sangha River; UB, Ubangui River; CG, Congo River. The locations of two published museum samples (22, 57) are indicated by “?.”
Spatial analysis of molecular variation indicates that the among-group variance asymptotes at four groups (corresponding to the major haplogroups A–D). With seven groups, the among-group variance component reaches a near-maximal value of 85.44%. These seven groupings correspond well with the major branches recovered in phylogenetic analysis with the exception of the separation of haplogroups C1/2 and Ivindo (site 19). This latter population is made up of a mixture of highly divergent haplogroups (C2, D2, D3) and thus warrants separation from other population groupings. The only other areas where limited exchange between adjacent haplogroups is evident are in Lobéké, southeastern Cameroon (site 4), and the Monts de Cristal in northwestern Gabon (site 6).
Molecular diversity is greatest in western gorilla haplogroups C1/2, D1, and the heavily admixed Ivindo population (SI Table 1). When gorilla populations were pooled into eastern or western groups, molecular diversity indices θ (40) and π (41) were almost 2-fold higher for western gorillas (θ = 0.047, π = 0.058) than for eastern gorillas (θ = 0.029, π = 0.038). Fu's F and Tajima's D statistic are significantly negative for the eastern lowland gorilla haplogroup B and western gorilla haplogroup D3. Only Tajima's D was significant for the geographically widespread haplogroup C. Mismatch distribution profiles provide a strong unimodal pattern for haplogroups A, B, and D3 (data not shown). In keeping with earlier recommendations (7) and the acknowledged upward bias in estimates of the growth parameter g (42), only haplogroups B and D3 demonstrated significant positive values of g that were at least three times their standard deviation. These estimates were robust to changes in initial parameter estimates and the number and length of the Markov chains.
Mantel tests for the association between molecular and Euclidian geographic distances indicate a highly significant isolation by distance effect within western lowland gorillas (Reynolds's genetic distance P ≤ 0.001; r2 = 0.496; ΦST P ≤ 0.001; r2 = 0.332). The association between Reynolds's distance and both cost matrices was also highly significant. This association was strongest when stream orders of 6 or greater were considered impenetrable (P ≤ 0.001; r2 = 0.574). In partial Mantel tests, the partial correlation between this cost matrix and Reynolds's genetic distance was still significant even when controlling for the effects of Euclidian distances between sites (P ≤ 0.001; r2 = 0.2337). MESQUITE simulations rejected the hypothesis of east–west divergence during the last glacial maximum but are consistent with a model of Pleistocene separation of 52,500–120,000 years or older. MDIV analyses support an older divergence time of ≈1.6 million years, although this date still falls well within the Pleistocene.
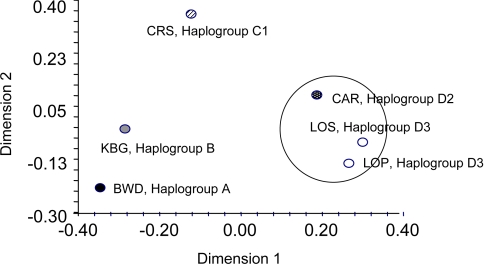
Cumulative probabilities of identity for the seven nuclear microsatellite loci used in the present study were <0.05 in all populations, even assuming full-sibling relationships. Multilocus genotypes with less than three loci or redundant genotypes were removed from the dataset, leaving a total of 91 genotype profiles with three or more typed loci (SI Table 2). Tests of Hardy–Weinberg equilibrium revealed no deviations after Bonferroni correction. All pair-wise FST comparisons showed significant population differentiation with the exception of the two populations within haplogroup D3 (Lopé and Lossi). Multidimensional scaling of microsatellite genetic distances between populations from the four major haplogroups (A–D) demonstrated strong regional differentiation at the nuclear level. In accordance with the mitochondrial data, three populations from haplogroup D (Lopé, Lossi, and Bai Hokou) formed the closest association (Fig. 3).
Fig. 3.
Multidimensional scaling plot of microsatellite distances between populations from all major gorilla haplogroups (A–D). In each case, the mitochondrial haplogroup affiliation is indicated.
Discussion
Our results suggest a role for both Pleistocene refugia and rivers in structuring gorilla genetic diversity. Evidence of a Pleistocene history of fragmentation and subsequent expansion is evident in eastern lowland gorillas, in keeping with earlier observations (20). However, patterns of demographic expansion were not observed in mountain gorillas, possibly because of the smaller sample sizes used here. Admixture is also evident along the borders between refugial populations, as predicted by refuge theory (3, 31). Whereas molecular genetic diversity is high within the western gorilla haplogroup C, there is very little evidence for signatures of historical fragmentation and population expansion. In contrast, regional allopatric differentiation within haplogroup D is well defined. Multiple lines of evidence also indicate strong signatures of population expansion in the geographically widespread haplogroup D3. In contrast, haplogroups D1 and D2 lack signatures of population expansion and appear to have a limited geographic distribution, although this may be a limitation of the present sampling strategy. Nuclear microsatellite data generally support patterns of regional genetic differentiation observed between major mitochondrial haplogroups.
Although it is difficult to pinpoint the precise location of hypothesized refugia, Maley (15) provides a structural framework in which to infer the Pleistocene history of western lowland gorillas. The refugial origin of haplogroup D3 is most likely located in the Massif du Chaillu and Mont Doudou upland refugia of southern Gabon. Similarly, haplogroup D1 is tied very closely to a putative upland refugium in the Monts de Cristal in northwestern Gabon and adjacent Equatorial Guinea. Both upland areas are believed to have harbored species during glacial maxima and are centers of diversity for several endemic plant groups (13, 14, 43). Although haplogroup D2 is not associated with any upland region, its distribution may have nevertheless coincided with a fluvial refugium during the arid phases of the Pleistocene (22, 44). The widespread distribution of a relatively small number of haplotypes within haplogroup D3 suggests a history of past bottleneck(s) and rapid postglacial expansion. This observation is reinforced by strong signatures of population expansion.
Although it has been suggested that forest refugia may have been reservoirs of ancestral diversity rather than engines of diversification (45), we find little evidence to support the former hypothesis. Although it is difficult to date haplogroup divergence times with certainty, most previous estimates place the divergence of eastern and western gorillas within an early- to mid- Pleistocene time frame (e.g., refs. 46 and 47). In contrast, coalescent simulations in the present study cannot reject the hypothesis of a more recent minimum divergence time in keeping with other recent estimates based on genome-wide patterns of variation (49). It therefore seems plausible that western gorilla haplogroups arose recently during subsequent arid phases of the Pleistocene.
Although much attention has focused on forest refugia in the Neotropics, surprisingly little consideration has been given to this hypothesis in equatorial Africa (43). In Africa, the lower temperatures and greater aridity periodically experienced during the Plio-Pleistocene (50, 51) provide a historical precedent for ice age forest refugia (15). Marine sedimentary cores indicate that ≈2.8 million years ago, the climate began to change as ice sheets became large enough to influence climate at tropical latitudes (52). This shift toward a cooler, drier climate has been implicated in the evolution of African bovids (53) and hominids (54). Recent geomorphological data also provide support for lowland forest refugia during the arid phases of the Pleistocene (55).
Findings from this study also suggest that major river courses have played an important role in shaping boundaries of several regional haplogroups in gorillas, notably the partial genetic structuring evident across the Sangha River (C1 and D2), the Ogooué River (D1 and D3), and the Sanaga River (C2 but not C1). The Ivindo/Ayina River may have also influenced postglacial expansion by directing the southern extension of haplogroup C2 into northeastern Gabon. However, quantifying the degree to which a river constitutes a barrier to gene flow is difficult without systematically sampling from the headwaters to the estuary (e.g., refs. 8 and 35).
Although our data support a model of regional genetic differentiation consistent with the inferred location of Pleistocene refugia and distribution of major rivers, several important caveats apply. The paucity of palynological data makes it difficult to precisely pinpoint the location of hypothesized refugia. Rivers are also incomplete barriers to dispersal because of seasonal variation in water levels and historical shifts in drainage patterns. Several alternative diversification hypotheses have been proposed (2, 6), of which ecological gradients (EGH) (4) and riparian refugia (RRH) (16, 44, 56) have attracted considerable attention. Proponents of the EGH argue that areas of biological diversification may coincide with zones of ecological transition such as the savannah–forest ecotone (57) or regions of topographical complexity (58). Although EGH theory seems plausible for species that tolerate a broad range of habitats, gorillas are closed-canopy specialists whose distribution does not extend across the savannah–forest boundary. Furthermore, patterns of genetic differentiation and/or admixture do not appear coincident with any obvious zones of ecological transition, although montane areas may have been important centers of diversification. In contrast, the RRH posits that tropical lowland taxa may have persisted during the arid phases of the Pleistocene in riparian forest. The RRH is not new (16, 56) but remains largely untested in African ecosystems. According to this hypothesis, genetic variation should be partitioned by major watersheds and not by candidate refugia. Aside from the Dzanga-Sangha region in CAR, the present data do not support this hypothesis, because rivers appear to limit the distribution of genetic diversity rather than act as centers of evolutionary diversification. Geo-referenced samples of other rainforest mammals are needed in order to gain a more complete understanding of the most important mechanisms of diversification in central African rainforests.
Materials and Methods
This study features previous data (22) and uses a larger sample collected from 29 sites across the range of all three recognized subspecies: western lowland Gorilla gorilla gorilla, eastern lowland G. g. graueri, and mountain G. g. beringei gorillas (SI Table 3). Fecal and hair collections were made from sites across the core range of the western lowland gorilla, including putative refugia in Gabon (sites 6, 11, and 14) and a new western gorilla population in Cameroon (site 2) (59). Generally only one sample per nest site was sequenced, whereas multiple nests per site were genotyped at microsatellite loci. DNA extraction, PCR protocols, and primers were carried out as previously described (22). Approximately 260 bp of the first hypervariable domain (HV1) of the mitochondrial control region were amplified from 185 gorilla samples. PCR products from 55 of these samples were cloned into the TA cloning vector (Invitrogen) before sequencing. All data from this study were then combined with sequence data published elsewhere (20, 22, 48, 60–62).
The complete database of sequences (n = 249) was aligned in Clustal X (63), and a hypervariable polycytosine stretch of the HV1 domain was deleted from the alignment. Model Test v.3.06 (64) was used to estimate the nucleotide substitution model that best approximated the data. By using previously described methods (65), candidate nuclear translocations (n = 51), in vitro PCR recombinants (n = 6), and cloned singletons (48) or variants differing by 2–5 bp outside the polyC (n = 6) were removed. GenBank sequences without geographic information were also removed (n = 20), leaving 166 gorilla HV1 sequences for analysis. Phylogenetic analysis was carried out by using PAUP 4.0 (66). Maximum likelihood estimation of phylogeny was carried out on unique mitochondrial haplotypes (n = 60). Bootstrap consensus trees were constructed with 500 replicates.
A spatial analysis of molecular variance (SAMOVA) was used to estimate the number of geographically cohesive groups of populations that were maximally differentiated from one another (67). Only populations with three or more samples (n = 20) were considered for spatial and demographic analyses. By using SAMOVA, populations were partitioned into regionally defined haplogroups (n = 7), and molecular diversity estimates were obtained by using ARLEQUIN v.3.0 (68). ARLEQUIN was also used to assess signatures of demographic expansion by using mismatch distribution profiles (69), Tajima's D (70), and Fu's F (71) statistic. Maximum likelihood estimates of the growth parameter (g) and θ within regional haplogroups was estimated by using FLUCTUATE (42). An initial θ value of 1.0 was used with a random starting tree and transition–transversion ratio of 10. Mantel tests (72) for association between geographic and genetic distances among western gorilla populations (n = 16) were performed by using the software IBD v.1.52 (73). Mitochondrial genetic distance matrices were based on either ΦST values (74) or Reynolds's transformation (75). The geographic distance matrix was constructed from either the Euclidian distance between sites or a “cost” value representing the distance required to circumnavigate intervening riverine barriers to gene flow. This cost matrix was also used as an indicator matrix in partial Mantel tests of the association between genetic and Euclidian distance.
Geospatial processing and analyses were carried out by using ArcEditor and the Spatial Analyst extension of ArcGIS 9.0. To construct the cost matrix, digital elevation data (DEMs), processed to remove No Data voids, were obtained from the Consultative Group on International Agricultural Research Consortium for Spatial Information Archives. DEMs were merged into a single raster, clipped to the boundary of the study area, and then resampled to a resolution of 0.002°. Sinks in the resulting raster were filled, and a flow accumulation raster was produced. Cells with a flow accumulation value ≥100 were extracted and converted to a stream network raster. The stream raster was used to calculate stream order (1–8) which was then used to produce a cost raster, where streams of orders 6–8 were designated as impenetrable barriers (SI Fig. 6) and those of orders 5, 4, and 3 were assigned a cost of 50, 40, and 30, respectively. Streams of order 1 or 2 and areas with a flow accumulation of <100 were assigned a cost of 1. An alternative cost matrix was also constructed, where only stream orders 7 and 8 were considered impenetrable, and stream order 6 was assigned a cost of 60.
The coalescent simulation package in MESQUITE 1.05 (76) was used to compare hypotheses of minimum divergence times between eastern and western gorillas. The steps described here were modified from previous work (77) and aim to estimate the minimum divergence time required to achieve reciprocal monophyly between eastern and western gorillas by using a model of nucleotide substitution and an effective population size (Ne) estimated from the data. A simple two-refuge model was constructed, where branch lengths were varied from between 1,333 and 40,666 generations. Assuming an average generation time for gorillas of 15 years (49), these branch lengths correspond to divergence times dating from the Wisconsin glacial ≈20,000 years ago to a mid-Pleistocene interglacial period of 610,000 years, respectively (77). For all simulations, the branch length of the root was set at 5,000 generations. Estimates of θ were derived for all data and for eastern and western populations separately by using Watterson's method (40). Upper (30,000) and lower (13,400) estimates of ancestral Ne were obtained from the relationship θ = 2Neμ by using control region mutation estimates of either 0.075 × 10−6 or 0.165 × 10−6 substitutions per site per year, respectively (78), and a generation time of 15 years. For each divergence scenario, 100 gene matrices were simulated by using a model constrained to fit the two-refuge hypothesis. Sequences were simulated by using a general time-reversible model of nucleotide substitution selected by ModelTest with rates (1.0, 14.5442, 0.0, 0.0, 14.5442, 1.0), a proportion of invariant characters set to 0.0, and a gamma distribution with shape parameter of 0.3655 and four rate categories. From these simulated gene matrices, consensus trees were estimated by using the maximum likelihood method implemented in PAUP under the same model parameters, except that 0.0 rate category was adjusted to 0.0001 because of program constraints. Slatkin and Maddison's S value (79) was then estimated from each consensus tree to generate a null distribution of these values. This distribution was then compared with the value for the observed genealogy (S = 1), as is the case for reciprocal monophyly between eastern and western populations. The two-refuge model for a given divergence times was accepted if S of 1 values from the coalescent simulations were observed in at least five or more cases (P ≥ 0.05). East–west divergence times were also estimated from MDIV (80) by using five Markov chains of 2,000,000 cycles and a burn in time of 500,000 cycles. The population divergence time (t) was estimated from the highest posterior probability value of the parameter T, an effective population size of 30,000, and a generation time of 15 years.
Microsatellite multilocus genotypes based on seven human loci (D1S550, D4S1627, D5S1457, D7S817, D8S1179, D21S11, FGA) and a sex-specific amelogenin locus (81) were amplified in two separate quadriplexes by using hair samples collected from site 1 (n = 16), site 12 (n = 72), site 23 (n = 40), site 24 (n = 16), site 25 (n = 32), and site 28 (n = 18). Because of the risk of allelic dropout and false alleles, each sample was genotyped four times. Heterozygotes were accepted if both alleles were typed twice (82), whereas homozygotes were only accepted if replicated three times. Although the number of replicates is less than recommended (82), allelic dropout rates estimated from these loci indicate that only three replicates are required to give a 95% or greater probability of a homozygote being typed correctly (83). Cumulative probabilities of identity across all loci assuming either Hardy–Weinberg frequencies or full-sibling relationships were calculated for each population by using GENECAP (84). To exclude the risk of sampling the same individual more than once, redundant genotypes were removed where there was an identical match between multilocus genotypes. Tests for deviations from Hardy–Weinberg, pair-wise FST calculations, and population differentiation were conducted in ARLEQUIN. Goldstein's (δμ)2 (85) and Cavalli-Sforza and Edwards' chord (86) distances between populations were calculated by using the program MSA3.0 (87). Multidimensional scaling of this distance matrix was carried out by using the NCSS v.6 statistical package (NCSS).
Supplementary Material
ACKNOWLEDGMENTS.
We thank all of the collectors who provided samples for this and earlier work (22): M. Bermejo (University of Barcelona, Barcelona, Spain), M. Charpentier (Centre International de Recherches Médicales de Franceville), E. Dimoto (Centre International de Recherches Médicales de Franceville), J. T. Dinkagadissi (Centre International de Recherches Médicales de Franceville), M. Goldsmith (Tufts University, Boston, MA), J. Groves (Wildlife Conservation Society, Limbe, Cameroon), D. Idiata (Direction de la Favre et de la Chasse, Libreville, Gabon), S. Latour (Wildlife Conservation Society, Libreville, Gabon), F. Maisels (Wildlife Conservation Society, Libreville, Gabon), K. McFarland (City University of New York, New York), Y. Mihindou (Wildlife Conservation Society, Monitoring of Illegal Killing of Elephants, Libreville, Gabon), E. Nwufoh (Cross River National Park, Nigeria), J. Oates (City University of New York), J. Okouyi (Institut de Recherche en Ecologie Tropicale), I. Omari (Institute Congolais pour la Conservation de la Nature, Kinshasa, Democratic Republic of Congo), R. Parnell (Wildlife Conservation Society, Libreville, Gabon), P. Peignot (Centre International de Recherches Médicales de Franceville), M. E. Rogers (University of Edinburgh, Edinburgh, U.K.), and E. Williamson (University of Stirling). This work was funded by Darwin Initiative Grant 08/044 and the University of New Orleans.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU305296–EU305397).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704816105/DC1.
References
- 1.Bush MB. J Biogeogr. 1994;21:5–17. [Google Scholar]
- 2.Haffer J. Biodiv Conserv. 1997;6:451–476. [Google Scholar]
- 3.Haffer J. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- 4.Endler JA. In: Biological Diversification in the Tropics. Prance GT, editor. New York: Columbia Univ Press; 1982. pp. 641–657. [Google Scholar]
- 5.Patton JL, da Silva MNF. In: Endless Forms: Species and Speciation. Howard DJ, Berlocher SH, editors. New York: Oxford Univ Press; 1998. pp. 202–216. [Google Scholar]
- 6.Moritz C, Patton JL, Schneider CJ, Smith TB. Annu Rev Ecol Syst. 2000;31:533–563. [Google Scholar]
- 7.Lessa EP, Cook JA, Patton JL. Proc Natl Acad Sci USA. 2003;100:10331–10334. doi: 10.1073/pnas.1730921100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silva MNF, Patton JL. Mol Phylogenet Evol. 1993;2:243–255. doi: 10.1006/mpev.1993.1023. [DOI] [PubMed] [Google Scholar]
- 9.Schneider CJ, Smith TB, Larison B, Moritz C. Proc Natl Acad Sci USA. 1999;96:13869–13873. doi: 10.1073/pnas.96.24.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugall A, Moritz C, Mousalli A, Stanisic J. Proc Natl Acad Sci USA. 2002;99:6112–6117. doi: 10.1073/pnas.092538699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond AW, Hamilton AC. J Zool. 1980;191:379–402. [Google Scholar]
- 12.Mayr E, O'Hara RJ. Evolution (Lawrence, Kans) 1986;40:55–67. doi: 10.1111/j.1558-5646.1986.tb05717.x. [DOI] [PubMed] [Google Scholar]
- 13.Sosef MSM. Wageningen Agricultural University Papers, 94-1: Studies in Begoniaceae V. Wageningen, The Netherlands: Wageningen Agricultural University; 1994. [Google Scholar]
- 14.Rietkirk M, Ketner P, De Wilde JJFE. In: The Biodiversity of African Plants. van der Maesen LJG, editor. Dordrecht, The Netherlands: Kluwer; 1996. pp. 608–623. [Google Scholar]
- 15.Maley J. Proc R Soc Edinburgh. 1996;104B:31–73. [Google Scholar]
- 16.Leal ME. Wageningen, The Netherlands: Wageningen University; 2004. PhD thesis. [Google Scholar]
- 17.Roy M. Proc R Soc London Ser B. 1997;264:1337–1344. [Google Scholar]
- 18.Smith TB, Holder K, Girman D, O'Keefe K, Larison B, Chan Y. Mol Ecol. 2000;9:1505–1516. doi: 10.1046/j.1365-294x.2000.01032.x. [DOI] [PubMed] [Google Scholar]
- 19.Bowie RCK, Fjeldsa J, Hackett SJ, Bates JM, Crowe TM. Mol Phylogenet Evol. 2006;38:171–188. doi: 10.1016/j.ympev.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Jensen-Seaman MI, Kidd KK. Mol Ecol. 2001;10:2241–2247. doi: 10.1046/j.0962-1083.2001.01365.x. [DOI] [PubMed] [Google Scholar]
- 21.Telfer PT, Souquiere S, Clifford SL, Abernethy KA, Bruford MW, Disotell TR, Sterner KN, Roques P, Marx PA, Wickings EJ. Mol Ecol. 2003;12:2019–2024. doi: 10.1046/j.1365-294x.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- 22.Clifford SL, Anthony NM, Bawe-Johnson M, Abernethy KA, Tutin CEG, White LTJ, Bermejo M, Goldsmith ML, McFarland K, Jeffery K, et al. Mol Ecol. 2004;13:1551–1567. doi: 10.1111/j.1365-294X.2004.02140.x. [DOI] [PubMed] [Google Scholar]
- 23.Muloko-Ntoutoume N, Petit RJ, White L, Abernethy K. Mol Ecol. 2000;9:359–363. doi: 10.1046/j.1365-294x.2000.00859.x. [DOI] [PubMed] [Google Scholar]
- 24.Morin PA, Moore JJ, Chakraborthy R, Jin L, Goodall J, Woodruff D S. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 25.Gonder MK, Oates JF, Disotell TR, Forstner MRJ, Morales JC, Melnick DJ. Nature. 1997;388:337–338. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg T, Ruvolo M. Mol Biol Evol. 1998;14:976–984. doi: 10.1093/oxfordjournals.molbev.a025841. [DOI] [PubMed] [Google Scholar]
- 27.Roca AL, Georgiadis N, Pecon-Slattery J, O'Brien SJ. Science. 2001;293:1473–1477. doi: 10.1126/science.1059936. [DOI] [PubMed] [Google Scholar]
- 28.Eggert SL, Rasner CA, Woodruff DS. Proc R Soc London Ser B. 2002;269:1993–2006. doi: 10.1098/rspb.2002.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klicka J, Zink RM. Science. 1997;277:1666–1669. [Google Scholar]
- 30.Knapp S, Mallet J. Science. 2003;300:71–72. doi: 10.1126/science.1083007. [DOI] [PubMed] [Google Scholar]
- 31.Hewitt GM. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- 32.Wallace AR. Proc Zool Soc Lond. 1849;20:107–110. [Google Scholar]
- 33.Ayres JM, Clutton-Brock TH. Am Nat. 1992;140:531–537. doi: 10.1086/285427. [DOI] [PubMed] [Google Scholar]
- 34.Aleixo A. Evolution (Lawrence, Kans) 2004;58:1303–1317. doi: 10.1111/j.0014-3820.2004.tb01709.x. [DOI] [PubMed] [Google Scholar]
- 35.Peres CA, Patton JL, da Silva MNF. Folia Primatol. 1996;67:113–124. doi: 10.1159/000157213. [DOI] [PubMed] [Google Scholar]
- 36.Kingdon J. The Kingdon Field Guide to African Mammals. Princeton: Princeton Univ Press; 1995. pp. 1–450. [Google Scholar]
- 37.Gonder MK, Disotell TR, Oates JF. Int J Primatol. 2006;27:1103–1127. [Google Scholar]
- 38.Eriksson J, Hohmann G, Boesch C, Vigilant L. Mol Ecol. 2004;13:3425–3435. doi: 10.1111/j.1365-294X.2004.02332.x. [DOI] [PubMed] [Google Scholar]
- 39.Tutin CEG, White LJT, Mackanga-Missandzou A. Conserv Biol. 1997;11:1190–1203. [Google Scholar]
- 40.Watterson GA. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 41.Tajima D. In: Mechanisms of Molecular Evolution. Takahata N, Clark AG, editors. Tokyo/Sinauer, Sunderland, MA: Japan Sci Soc Press; 1993. pp. 37–59. [Google Scholar]
- 42.Kuhner M, Yamato J, Felsenstein J. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plana V. Phil Trans R Soc London B. 2004;359:1585–1594. doi: 10.1098/rstb.2004.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colyn M, Gautier-Hion A, Verheyen W. J Biogeogr. 1991;18:403–407. [Google Scholar]
- 45.Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Ruvolo M. Mol Phylogenet Evol. 1996;5:202–219. doi: 10.1006/mpev.1996.0014. [DOI] [PubMed] [Google Scholar]
- 47.Jensen-Seaman MI, Deinard AS, Kidd K. In: Gorilla Biology: A Multidisciplinary Perspective. Taylor AB, Goldsmith ML, editors. Cambridge, UK: Cambridge Univ Press; 2003. pp. 247–268. [Google Scholar]
- 48.Thalmann O, Hebler J, Poinar HN, Pääbo S, Vigilant L. Mol Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294x.2003.02070.x. [DOI] [PubMed] [Google Scholar]
- 49.Thalmann O, Fischer A, Lankester F, Pääbo S, Vigilant L. Mol Biol Evol. 2007;24:146–158. doi: 10.1093/molbev/msl160. [DOI] [PubMed] [Google Scholar]
- 50.Hamilton AC, Taylor D. Clim Change. 1991;19:65–78. [Google Scholar]
- 51.Bonnefille R, Roeland JC, Guiot J. Nature. 1990;346:347–349. [Google Scholar]
- 52.deMenocal PB. Science. 1995;270:53–58. doi: 10.1126/science.270.5233.53. [DOI] [PubMed] [Google Scholar]
- 53.Vrba ES. In: Palaeoclimate and Evolution, with Emphasis on Human Origins. Vrba ES, Denton GH, Partridge TC, Burckle LH, editors. New Haven, CT: Yale Univ Press; 1995. pp. 385–424. [Google Scholar]
- 54.deMenocal PB, Bloemendal J. In: Paleoclimate and Evolution, with Emphasis on Human Origins. Vrba ES, Denton GH, Partridge TC, Burckle LH, editors. New Haven, CT: Yale Univ Press; 1995. pp. 262–288. [Google Scholar]
- 55.Nichol JE. Geogr J. 1999;165:79–89. [Google Scholar]
- 56.Aide TM, Rivera E. J Biogeogr. 1998;25:695–705. [Google Scholar]
- 57.Smith TB, Wayne RK, Girman DJ, Bruford MW. Science. 1997;276:1855–1857. [Google Scholar]
- 58.Fjeldsa J, Lovett JC. Biodiv Conserv. 1997;6:325–346. [Google Scholar]
- 59.Morgan BJ, Wild C, Ekobo A. Int J Primatol. 2003;24:1129–1137. [Google Scholar]
- 60.Garner KJ, Ryder OA. Mol Phylogenet Evol. 1996;6:39–48. doi: 10.1006/mpev.1996.0056. [DOI] [PubMed] [Google Scholar]
- 61.Hofreiter M, Siedel H, Vigilant L. Am J Phys Anthropol. 2003;121:361–368. doi: 10.1002/ajpa.10186. [DOI] [PubMed] [Google Scholar]
- 62.Jensen-Seaman MI, Sarmiento EE, Deinard AS, Kidd KK. Am J Primatol. 2004;63:139–147. doi: 10.1002/ajp.20047. [DOI] [PubMed] [Google Scholar]
- 63.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 65.Anthony NM, Clifford SL, Bawe-Johnson M, Abernethy KA, Bruford MW, Wickings EJ. Mol Phylogenet Evol. 2007;43:553–566. doi: 10.1016/j.ympev.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0 Beta. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 67.Dupanloup I, Schneider S, Excoffier L. Mol Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- 68.Excoffier L, Laval G, Schneider S. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 69.Rogers AR, Harpending H. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 70.Tajima D. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu Y-X. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantel N. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 73.Bohonak AJ. J Heredity. 2002;93:153–154. doi: 10.1093/jhered/93.2.153. [DOI] [PubMed] [Google Scholar]
- 74.Excoffier L, Smouse P, Quattro J. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reynolds J, Weir BS, Cockerham CC. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maddison W, Maddison D. Mesquite: A Modular System for Evolutionary Analysis. 2006 Version 1.12 http://mesquiteproject.org. [Google Scholar]
- 77.DeChaine EG, Martin AP. Evolution (Lawrence, Kans) 2006;60:1004–1013. [PubMed] [Google Scholar]
- 78.Pakendorf B, Stoneking M. Annu Rev Genom Hum Genet. 2005;6:165–183. doi: 10.1146/annurev.genom.6.080604.162249. [DOI] [PubMed] [Google Scholar]
- 79.Slatkin M, Maddison WP. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen R, Wakely J. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sullivan KM, Mannucci A, Kimpton CP, Gill P. BioTechniques. 1993;15:636–641. [PubMed] [Google Scholar]
- 82.Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeffery K. Cardiff, UK: Cardiff University; 2003. PhD thesis. [Google Scholar]
- 84.Wilberg MJ, Dreher BP. Mol Ecol Notes. 2004;4:783–785. [Google Scholar]
- 85.Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. Proc Natl Acad Sci USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cavalli-Sforza LL, Edwards AWF. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- 87.Dieringer D, Schlotterer C. Mol Ecol Notes. 2003;3:167–169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.