Abstract
The structure recognized by regulatory T cells that enables them to discriminate self from nonself in the periphery is one of the central issues of regulatory T cell biology. A link between immunoregulation and self–nonself discrimination has emerged from experiments showing that Qa-1-restricted CD8+ T cells selectively down-regulate target T cells activated by the intermediate avidity of their own T cell antigen receptor–ligand interactions. Because the peripheral self-reactive T cell repertoire is devoid of high-avidity T cells compared with the foreign-reactive repertoire, as a result of thymic negative selection, the selective down-regulation of intermediate but not high-avidity T cells enables the immune system to suppress autoimmunity without damaging the ongoing immune response to foreign pathogens. However, the molecular mechanism delineating how avidity of T cell activation is perceived by the regulatory T cells has not been elucidated. Here we show that a heat shock peptide (Hsp60sp), coupled with the MHC class Ib molecule Qa-1, is a surrogate target structure that is preferentially expressed at a higher level on the intermediate avidity T cells and specifically recognized by the Qa-1-restricted CD8+ T cells. The biological significance of this observation was confirmed by the ability of Hsp60sp-loaded relevant dendritic cells to induce a Qa-1-restricted CD8+ T cell-mediated protection from autoimmune encephalopathy in the experimental allergic encephalomyelitis model. Thus, perceiving the avidity of T cell activation can be translated into peripheral T cell regulation to discriminate self from nonself in the periphery to maintain self-tolerance.
Keywords: autoimmune diseases, Qa-1/HLA-E-restricted regulatory CD8+ T cells, self–nonself discrimination
How the immune system achieves self–nonself discrimination remains a central conundrum in the study of immunology. Intrathymic deletion of high-avidity, self-reactive T cell clones generates a truncated peripheral self-reactive repertoire composed of mainly intermediate- and low- but devoid of high-avidity clones compared with the foreign-reactive repertoire, which possesses T cells with a full range of avidity. It is likely that potentially pathogenic self-reactive T cells are included in the pool of the “intermediate-avidity” thymic escapees that could be activated in the periphery to initiate autoimmune diseases (1–4). It is thus inevitable that one of the major functions of peripheral regulatory mechanisms is to selectively down-regulate immune response to self-antigens without damaging the ongoing immune response to foreign pathogens (5). The distinctive composition of peripheral T cell repertoires to self vs. to foreign antigens provides a unique opportunity for the immune system to discriminate self from nonself in the periphery by a unified mechanism of selectively down-regulating intermediate-avidity T cells to both self and foreign antigens. Selective down-regulation of intermediate-avidity T cell populations containing the potentially pathogenic self-reactive T cells enables the immune system to specifically control autoimmune diseases without damaging the ongoing antiinfection immunity, which is largely mediated by high-avidity T cells specific to the foreign pathogens (5–7).
The experimental evidence supporting this conceptual framework of the “avidity model” comes from the observation that Qa-1-restricted regulatory CD8+ T cells inhibit the immune response to a conventional antigen, hen egg lysozyme (HEL), when it functions as a self-antigen in HEL transgenic mice but that Qa-1-restricted regulatory CD8+ T cells enhance the immune response to the same antigen when it functions as a foreign antigen in wild-type mice. This in vivo self–nonself discrimination is accomplished by CD8+ T cells that selectively down-regulate target T cells activated by the intermediate avidity of their own TCR-ligand interactions (7). The Qa-1-restricted CD8+ T cells had been identified initially as a regulatory population that mediates the resistance to autoimmune experimental allergic encephalomyelitis (EAE) induced by the first episode of the disease (8, 9). More severe symptoms of EAE develop in a much less controllable fashion during the relapse in CD8 knockout mice (9) or the reinduction of EAE in Qa-1 knockout mice (10), indicating that Qa-1-restricted CD8+ T cells play an important role in maintaining peripheral self-tolerance.
However, the cognitive molecular and cellular mechanisms that enable the Qa-1-restricted CD8+ T cells to perceive avidity of T cell activation to selectively down-regulate intermediate-avidity T cells have not been elucidated. In the current studies, we provide evidence that a signal peptide derived from the leader sequence of heat shock protein 60 (Hsp60sp), coupled with the MHC class Ib molecule Qa-1, is a surrogate target structure that is preferentially expressed at a higher level on the intermediate-avidity T cells and specifically recognized by the Qa-1-restricted CD8+ T cells. Moreover, Qa-1-restricted CD8+ T cells can be induced by vaccinating animals with Hsp60sp-loaded dendritic cells (DCs) to protect animals from the subsequently induced EAE. Thus, the cognitive mechanism in which preferential expression of particular surrogate target structures, such as Qa-1/Hsp60sp, recognized by the regulatory T cells on the surface of activated T cells of intermediate avidity enables the immune system to discriminate self from nonself in the periphery by perceiving the avidity of T cell activation.
Results
To understand the molecular mechanisms of the cognitive recognition involved in the specific T–T cell interaction between the Qa-1-restricted CD8+ T cells and the target T cells, it is crucial to identify the Qa-1-binding peptide(s) that renders the activated T cells susceptible to down-regulation by these CD8+ T cells. In this regard, it has been established that the predominant peptide bound to the MHC class Ib molecule Qa-1 is Qdm, a hydrophobic peptide derived from the leader sequence of MHC class Ia molecules (11, 12). This peptide binds with high affinity and accounts for the majority of the peptides associated with Qa-1. Qa-1/Qdm interacts with CD94/NKG2A on natural killer (NK) cells and inhibits NK activity (13, 14). We have classified the Qdm or Qdm-like peptides as “type A” peptides. However, Qa-1 can also bind other self-peptides, including those derived from heat shock proteins (15) and preproinsulin leader sequences (16). In addition, human studies have shown that a signal peptide derived from the leader sequence of a stress protein Hsp60 (Hsp60sp) is capable of competing with the B7sp peptide, the human counterpart of Qdm, for occupancy of HLA-E, the human counterpart of Qa-1 (17). Interestingly, the resultant HLA-E/Hsp60sp complex does not interact with CD94/NKG2A and therefore does not inhibit the NK activity. We have classified Qa-1-binding peptides that do not interact with CD94/NKG2A when coupled with Qa-1 as “type B” peptides. We hypothesized that some of the type B Qa-1-binding peptides that are capable of competing with Qdm for binding to Qa-1, such as Hsp60sp, may be preferentially expressed on the intermediate-avidity T cells and serve as a specific target for the Qa-1-restricted CD8+ T cells.
Qa-1/Hsp60sp Is a Specific Target Recognized by the Qa-1-Restricted CD8+ T Cells.
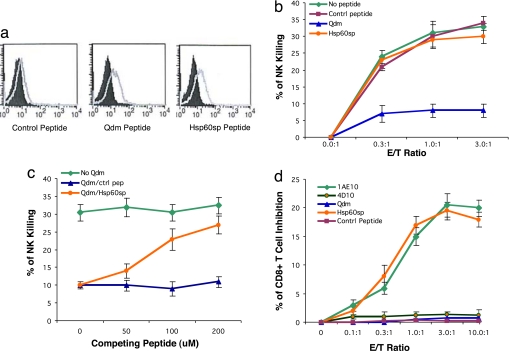
To test this hypothesis, we extended our experimental system from Qa-1b strains into the EAE-susceptible Qa-1a strain B10PL to better understand the biology of Qa-1-restricted CD8+ T cells at a molecular level in the context of an autoimmune disease model. In this regard, it has been established that most of the differences between Qa-1a and Qa-1b are located peripherally to the peptide-binding cleft, so that these two molecules associate with structurally similar peptides (18). We therefore were able to use the Qdm peptide as a model type A peptide in the studies to evaluate the relationship and function of a type B peptide, Hsp60sp, in the Qa-1a strain B10PL mice. We established Qa-1a-expressing cells by transfecting the human B cell line C1R with recombinant murine Qa-1a cDNA. In this regard, C1R transfected with Qa-1 has been widely used to identify Qa-1-binding peptide and to study the biology of Qa-1, including its use as an artificial target for NK or T cells (19, 20). One of the Qa-1a-positive transfectants detected by RT-PCR, 3F4, was capable of expressing surface Qa-1a when the cells were passively sensitized with exogenous Qa-1-binding peptides at 26°C (Fig. 1a). 3F4 was thus chosen as a Qa-1-binding peptide-presenting cell to test whether Hsp60sp served as specific target for the Qa-1-restricted CD8+ T cells. Evaluation was conducted according to three in vitro criteria, one ex vivo criterion, and one in vivo criterion, as described below.
Fig. 1.
Hsp60sp peptide is a specific target for Qa-1-restricted CD8+ T cells. (a) Hsp60sp peptide is capable of binding to Qa-1. 3F4 cells were incubated with Hsp60sp, Qdm, and control peptides at 26°C and 37°C for 18 h, stained with anti-Qa-1a serum, revealed by goat anti-mouse–PE, and analyzed by FACS. The shaded curves represent the Qa-1 staining of samples loaded with peptide at 37°C, and the light curves represent the Qa-1 staining of samples loaded with peptide at 26°C. (b) Hsp60sp peptide does not inhibit NK killing when coupled with Qa-1, indicating that Qa-1/Hsp60sp does not interact with CD94/NKG2A on the NK cells. 3F4 cells loaded with Hsp60sp, Qdm, and control peptides were used as peptide-presenting cells in a standard NK assay. The results are representative of four separate experiments. (c) Hsp60sp peptide is capable of competing with Qdm for binding to Qa-1. Hsp60sp and control peptide were loaded, together with Qdm (20 μM), to the 3F4 cells for 18 h at 26°C and tested in a standard NK assay (E/T ratio 2:1). The results are representative of four separate experiments. (d) Hsp60sp peptide renders Qa-1-expressing cells susceptible to the down-regulation by the Qa-1-restricted CD8+ T cells. 3F4 cells loaded with Hsp60sp, Qdm, and control peptides were used as peptide-presenting cells in a standard CD8+ T cell inhibition assay. The results are representative of four separate experiments.
The capacity of the peptides to bind to Qa-1 was first assessed by their ability to stabilize Qa-1 surface expression on 3F4 cells (15, 17, 21). As shown in Fig. 1a, the binding of both the Qdm and Hsp60sp at 26°C caused augmentation in the intensity of Qa-1a surface staining analyzed by FACS. Interestingly, although the Qa-1 surface expression level was relatively modest, which is consistent with reported observations of HLA-E surface expression in human studies (17, 22), it was sufficient to function as peptide-presenting molecule to NK cells and to CD8+ T cells (see below).
In human studies, it has been shown that the HLA-E/Hsp60sp complex does not interact with the CD94/NKG2A receptor on NK cells. It is important to determine whether Qa-1/Hsp60sp interacts with CD94/NKG2A on relevant cells in mice. The ability of Qa-1/Hsp60sp interacting with CD94/NKG2A was thus assessed in a standard functional NK assay, in which 3F4 cells loaded with Qdm, Hsp60sp, or nonbinding control peptide were tested for their susceptibility to NK killing. As shown in Fig. 1b, 3F4 cells loaded with control peptide were susceptible targets for NK killing. In contrast, 3F4 cells loaded with Qdm, but not Hsp60sp, were resistant to the NK killing, consistent with prior reports that Qa-1/Qdm (13, 14), but not Qa-1/Hsp60sp (17), interacts with CD94/NKG2A, leading to the inhibition of NK activity. This is also compatible with the notion that two types of Qa-1/HLA-E-binding peptides could be distinguished by their ability to interact with CD94/NKG2A in both humans (17) and mice.
Because both Qdm and Hsp60sp peptides, which are equally capable of binding to Qa-1, can be generated in activated T cells, it is crucial to evaluate whether Hsp60sp can compete with Qdm for binding to Qa-1. The ability of the Hsp60sp to compete with Qdm for binding to Qa-1 was further tested in a functional NK assay. As shown in Fig. 1c, Hsp60sp peptide but not control peptide abrogated the protection of NK killing of 3F4 mediated by Qdm in a dose-dependent manner. This result (i) provides evidence that Hsp60sp is capable of competing with Qdm for binding to Qa-1; (ii) is consistent with the observations shown in human studies that Hsp60sp is capable of competing with the signal peptide of the HLA class Ia leader sequence B7sp for binding to HLA-E (17); and (iii) provides a biological basis for the possibility that, in certain activated, T cells a higher level of Hsp60sp vs. Qdm can lead to higher expression of Qa-1/Hsp60sp vs. Qa-1/Qdm.
We next tested directly whether Qa-1/Hsp60sp could serve as a functionally relevant target for the regulatory CD8+ T cells. In this regard, we have demonstrated previously that the CD8+ T cells selectively down-regulate intermediate-avidity T cell clones (7) and that, in a variety of experimental settings, the suppression mediated by the CD8+ T cells is Qa-1-restricted (2, 7, 10, 23, 24). In the current studies, the Qa-1-expressing 3F4 cell was used as a Qa-1-peptide-presenting cell to test whether Hsp60sp is preferentially recognized by the regulatory CD8+ T cells in a CD8+ T cell inhibition assay. The 1-9NacMBP (myelin basic protein)-specific, pathogenic, intermediate-avidity CD4+ T cell clone 1AE10, a physiological target cell of the Qa-1-restricted CD8+ T cells (2, 24), served as a positive control, and a 1-9NacMBP-specific, nonencephalitogenic, low-avidity clone 4D10 and 3F4 cells loaded with control non-Qa-1-binding peptide or Qdm served as negative controls. In this regard, it is known that 1AE10 not only is capable of inducing EAE in vivo but also can be used as an effective vaccine to prime Qa-1-restricted CD8+ T cells, which protect animals from subsequent induced EAE (2, 24). As shown in Fig. 1d, the 1AE10 clone but not 4D10 was efficiently down-regulated by the CD8+ T cells isolated from EAE-recovered mice. These mice are known to be resistant to the reinduction of EAE and have been shown to possess Qa-1-restricted regulatory CD8+ T cells (2, 8). Interestingly, only 3F4 cells loaded with Hsp60sp, but not Qdm or control peptide, rendered the 3F4 cells susceptible to the down-regulation by the CD8+ T cells. CD8+ T cells isolated from naïve mice had no effect. Thus, Hsp60sp is indeed a specific target recognized by the regulatory CD8+ T cells when presented by Qa-1.
The Expression Ratio of Qa-1/Hsp60sp Versus Qa-1/Qdm Is Significantly Higher in the Intermediate-Avidity Clones Than in the High- and Low-Avidity Clones.
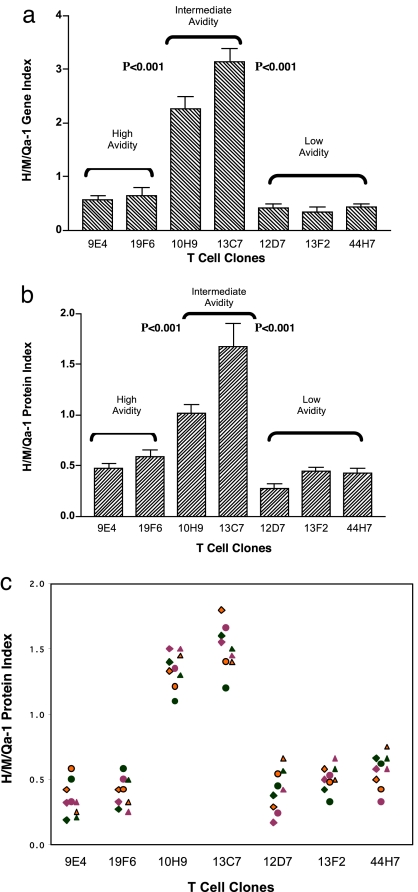
The observation that Qa-1/Hsp60sp is a specific target structure recognized by the Qa-1-restricted CD8+ T cells allowed us to further explore whether preferential expression of Qa-1/Hsp60sp on intermediate-avidity T cells is a function of T cell activation that enables these T cells to be susceptible to the down-regulation by the CD8+ T cells. We compared the expression of Qa-1 as well as the Hsp60 and MHC class Ia (H-2Dd) molecules at both mRNA and protein levels in a panel of activated T cell clones. In this regard, we took advantage of 28 HEL-specific T cell clones established from the Qa-1b strain BALB/c mice and identified in our previous studies. These T cell clones represent a range of avidity responding to HEL and have different susceptibility to the down-regulation by the Qa-1-restricted CD8+ T cells (7). The representative results from seven clones are presented in Fig. 2, supporting information (SI) Tables 3 and 4, and SI Fig. 3. The experimental design was based on the fact that Hsp60 and MHC class Ia (H-2Dd) proteins are able to generate the Qa-1-binding peptides Hsp60sp (17) and Qdm (25) peptides, respectively, in activated T cell clones. We measured the ratio of the expression of Hsp60 vs. MHC class Ia in relation to the expression of Qa-1 as a function of avidity of T cell activation and calculated the H/M/Qa-1 gene or protein indexes to reflect the relative expression of Qa-1/Hsp60sp vs. Qa-1/Qdm in each T cell clone tested.
Fig. 2.
The H/M/Qa-1 indexes are higher in intermediate avidity T cell clones. (a) H/M/Qa-1 gene indexes are significantly higher in intermediate-avidity T cell clones than in high- and low-avidity clones. The real-time PCR was performed at 60 h after the T cell clones were activated by 10 μM HEL (7). The results summarize three separate experiments. The H/M/Qa-1 gene index is calculated as: [Hsp60 gene expression index/MHC class Ia (H-2Dd) gene expression index] × Qa-1 gene expression index, which represents the ratio of Hsp60 vs. MHC class Ia (H-2Dd) normalized to Qa-1 at the gene expression level. The gene expression index is the ratio of gene expression between a given gene and β-actin in the same cells. ED50 of the clones are as follows: 9E4 and 19F6, <1 μM; 10H9, 3 μM; 13C7, 10 μM; 12D7, 13F2, and 44H7, >20 μM. (b) H/M/Qa-1 protein indexes are significantly higher in intermediate-avidity T cell clones than in high- and low-avidity clones. The Western blotting assay was performed at 72 h after the T cell clones were activated by 10 μM HEL (7). The data summarize three separate experiments. The H/M/Qa-1 protein index is calculated as: [Hsp60 protein expression index/MHC class Ia (H-2Dd) protein expression index] × Qa-1 protein expression index, which represents the ratio of Hsp60 vs. MHC class Ia (H-2Dd) normalized to Qa-1 at the protein expression level. The protein expression index is the ratio of protein expression between a given protein and β-actin in the same cells. (c) There were no significant differences among the H/M/Qa-1 protein indexes of T cell clones with known ED50 activated by different doses of antigen HEL ranging from 1 to 50 μM. The results summarize three separate experiments. When H/M/Qa-1 protein indexes were compared for each clone stimulated with three doses (1, 10, and 50 μM), there were no significant differences for each clone tested (P > 0.05), whereas significant differences between intermediate vs. high- and low-avidity groups at all three doses were shown (P < 0.001). The triangle, circle, and diamond symbols represent H/M/Qa-1 protein indexes of T cell clones from three separate experiments. The orange, green, and purple colors represent H/M/Qa-1 protein indexes of T cell clones stimulated with different doses of HEL (1, 10, and 50 μM) in each experiment.
As shown in SI Table 3 and Fig. 2a, in a real-time PCR assay, the H/M/Qa-1 gene index was significantly higher in the two intermediate-avidity clones, 10H9 and 13C7, compared with the two high-avidity clones, 9E4 and 19F6 (P < 0.001), as well as the three low-avidity clones, 12D7, 13F2, and 44H7 (P < 0.001). Similarly, at the level of protein expression, the H/M/Qa-1 protein index was also significantly higher in the intermediate-avidity clones than in the high- and low-avidity clones according to a Western blot assay, as shown in SI Table 4, Fig. 2b, and SI Fig. 3. The higher H/M/Qa-1 gene and protein indexes of intermediate-avidity T cell clones reflect a higher expression ratio between Qa-1/Hsp60sp and Qa-1/Qdm in these T cell clones. This higher expression ratio in intermediate-avidity T cell clones is consistent with the particular pattern of T cell regulation observed in our previous in vivo and in vitro studies demonstrating that Qa-1-restricted CD8+ T cells selectively down-regulate T cells of intermediate but not high or low avidity (7). To test whether the dose of antigen used to activate T cell clones with known avidity, measured by ED50, could influence this particular consequence of T cell activation, we activated the panel of T cell clones with various doses of the antigen HEL, ranging from 1 to 50 μM, under standard T cell culture conditions. As shown by Fig. 2c, there were no significant differences among H/M/Qa-1 protein indexes obtained when the same clones were activated with different doses of antigen, ranging from 1 to 50 μM. Taken together, the observations support the hypothesis that a higher expression ratio between Qa-1/Hsp60sp and Qa-1/Qdm is a function of the intermediate-avidity activation of T cells, which determines their susceptibility to the down-regulation by the Qa-1-restricted CD8+ T cells.
Vaccination of Animals with Hsp60sp-Loaded Relevant DCs Induced a Qa-1-Dependent CD8+ T Cell-Mediated Significant Protection from EAE.
We further tested, in an EAE model, the biological significance that selective down-regulation of intermediate-avidity T cells by the Qa-1-restricted CD8+ T cells, via specific recognition of Qa-1/Hsp60sp, is an important mechanism that the immune system employs to control pathogenic autoimmunity in vivo. In these experiments, Hsp60sp was assessed for its potential use as a “vaccine” to activate the Qa-1-restricted CD8+ T cells in vivo to protect animals from EAE that is mediated by certain 1-9NacMBP-reactive CD4+ T cell clones (1, 2, 8, 9, 26). In this particular EAE model, Anderton et al. have shown that the encephalitogenic T cell clones that are responsible for clinical EAE are in the 1-9NacMBP reactive repertoire of relatively “low avidity” (1). In the same experimental setting, we have shown that the potentially pathogenic clones are included in the T cell pool with higher growth potential in response to 1-9NacMBP isolated from EAE mice compared with nonpathogenic 1-9NacMBP-reactive T cell clones isolated from EAE-resistant mice (2). These two sets of data are consistent with the notion that encephalitogenic T cell clones with low avidity to 1-9Nac MBP (1), which mediate clinical EAE in this model, are the same self-reactive T cell clones that we referred to in our studies as intermediate avidity, which are presumably at the low end of the intermediate-avidity range (2, 5–7). This notion provides the biological as well as the experimental basis to determine whether protection from EAE in vivo is attributable to selective down-regulation of intermediate-avidity self-reactive T cell population containing the potentially encephalitogenic T cell clones. Naïve B10PL mice were thus injected with bone-marrow-derived dendritic cells (DCs) loaded with Hsp60sp or with control Qdm peptide at least 1 week before the induction of EAE. The DCs used for vaccination expressed Qa-1 on their surface but did not express CD8. Vaccination with DCs loaded with Hsp60sp, but not Qdm, significantly protected animals from EAE compared with the control, unvaccinated group (Tables 1 and 2 and SI Table 5). Importantly, the protection was CD8+ T cell-dependent because depletion of CD8+ T cells in vivo abolished the protection. The fact that the DCs used for vaccination in this experimental setting did not express CD8 excludes the possibility that the protection itself was mediated by the CD8+ DCs. Thus, Qa-1-restricted CD8+ T cells, which are capable of down-regulating 1-9NacMBP-reactive encephalitogenic CD4+ T cells (2, 8, 9), can be specifically induced in vivo by DCs loaded with Hsp60sp, but not Qdm, to protect animals from the disease.
Table 1.
Mice vaccinated with DCs loaded with Hsp60sp but not Qdm are significantly protected from the subsequent induction of EAE
| Group | Incidence, n (%) | Severity | Mean days |
|
|---|---|---|---|---|
| Onset | Maximum duration | |||
| Control | 15/16 (94) | 2.6 ± 0.63 | 25 ± 3.6 | 21 ± 3.2 |
| DC w/Hsp60sp | 4/18 (22) | 0.6 ± 0.66 | 53 ± 7.9 | 3 ± 2.9 |
| DC w/Qdm | 13/16 (81) | 2.3 ± 0.87 | 32 ± 8.8 | 17 ± 4.4 |
| CD8−/DC w/Hsp60sp | 10/16 (63) | 2.1 ± 0.59 | 32 ± 2.6 | 16 ± 1.6 |
Unless indicated otherwise, values are means ± SD. Control, mice induced to develop EAE without vaccination; DC w/Hsp60sp, mice injected with DCs loaded with Hsp60sp peptide before EAE induction; DC w/Qdm, mice injected with DCs loaded with Qdm peptide before EAE induction; CD8−/DC w/Hsp60sp, mice depleted of CD8+ T cells by injection of anti-CD8 mAb 53-6.72, 1 week after DC vaccination and 3 days before EAE induction. The data in Table 1 summarize four separate experiments shown in SI Table 5.
Table 2.
Comparison of mouse vaccination groups
| Group comparison | Incidence, P | Severity, P | Mean days, P |
|
|---|---|---|---|---|
| Onset | Maximum duration | |||
| DC w/Hsp60sp vs. control | <0.003 | <0.001 | <0.001 | <0.001 |
| DC w/Hsp60sp vs. DC w/Qdm | <0.01 | <0.01 | <0.01 | <0.002 |
| DC w/Hsp60sp vs. CD8−/DCw/Hsp60sp | <0.02 | <0.01 | <0.002 | <0.002 |
Discussion
We have proposed in the avidity model that Qa-1-restricted CD8+ T cells perceive the avidity of T cell activation to guide their regulatory functions in vivo (5–7). Here we provide a molecular and cellular mechanism suggesting that selective down-regulation of intermediate-avidity T cells can be accomplished via the specific recognition of Qa-1/Hsp60sp on intermediate-avidity T cells by the Qa-1-restricted CD8+ T cells. Hsp60sp was not only capable of competing with Qdm for binding to Qa-1 (Fig. 1 a–c) but was also a specific target structure recognized by the Qa-1-restricted CD8+ T cell when coupled with Qa-1 (Fig. 1d). More importantly, regulatory CD8+ T cell could be directly induced by Qa-1-expressing DCs loaded with Hsp60sp in vivo to protect animals from the subsequently induced EAE (Tables 1 and 2 and SI Table 5). The central issue of our current studies was to understand the biology of why and how Qa-1/Hsp60sp expressed in intermediate- but not high- and low-avidity T cells render the activated T cells susceptible to the CD8+ T cell regulation. In this regard, it already has been established from previous elution and mass spectroscopic studies that Qa-1-binding peptides from both Hsp60 and H-2D can be eluted from cells and transfectants (19, 27). The key finding from our current studies is the observation that “quantitative” but not “qualitative” differences between Qa-1/Hsp60sp vs. Qa-1/Qdm are a function of avidity of T cell activation and to determine the susceptibility of the target T cells to down-regulation by the CD8+ T cells. We showed that the intermediate-avidity T cells have a significantly higher ratio of Qa-1/Hsp60sp vs. Qa-1/Qdm than the ratio of high- and low-avidity T cells, a finding that is reflected by the differential expression of Hsp60 protein and the Qdm-containing MHC class Ia protein H-2Dd (Fig. 2). This led to more Qa-1/Hsp60sp, a specific target for the regulatory CD8+ T cells (Fig. 1d) on the surface of intermediate-avidity T cells with or without coexpression of Qa-1/Qdm. In this regard, we consistently observed that among the three molecules, Hsp60 is the one that shows the greatest variation in expression level, whereas the expression levels of both the Qa-1 and the H-2Dd are relatively constant between intermediate- vs. high- and low-avidity clones. Although Qdm could be generated by both H-2Dd and H-2Ld (28), data obtained from H-2Dd should be sufficient to represent the relative level between Hsp60sp and Qdm between intermediate vs. high and low clones because of the likely relatively constant expression of total H-2d among intermediate-, high-, and low-avidity T cell clones.
Moreover, a higher Qa-1/Hsp60sp vs. Qa-1/Qdm ratio in intermediate-avidity T cells may decrease the level of Qa-1/Qdm on the cell surface, which might increase their susceptibility to the NK killing (13, 14). This may lead to another possible mechanism guided by the conceptual framework of the avidity model to discriminate self from nonself, linking the regulation of adaptive immunity to innate immunity, which should be further explored.
It is known that in addition to Hsp60sp, three potential Qa-1/HLA-E-binding peptides could be generated from the Hsp60 protein (17, 27). It is also known that these three peptides either do not efficiently bind to Qa-1/HLA-E (Hsp60.2 and 3 peptides) (17) or are unable to compete with Qdm for occupancy of Qa-1 (Hsp60.4 peptide) (20). It is thus unlikely that these peptides would interfere with the overall biological outcome of the predominant expression of Qa-1/Hsp60sp on activated intermediate-avidity T cells. In this regard, Hsp60sp peptide may represent one example of a type B Qa-1-binding peptide capable of competing with Qdm or Qdm-like type A peptide(s) for occupancy of Qa-1 and serving as a specific target structure. Our studies did not exclude the possibility that other Hsp60sp-like type B peptide(s) may exist that function as targets for the Qa-1-restricted regulatory CD8+ T cells.
Besides its role in providing the Hsp60sp peptide involved in immune regulation mediated by the Qa-1-restricted CD8+ T cells, the Hsp60 protein itself has multiple other effects on T cell functions, including enhancement of CD4+ regulatory T cells function through innate signaling via TLR2 (29). Moreover, Hsp60, like CD25, is also expressed as a target for ergotypic regulation, which is not directly involved self–nonself discrimination (30).
We also observed that there were no significant differences among the ratio of Hsp60 vs. MHC class Ia (H/M/Qa-1 protein indexes) obtained when the same clones were activated with varying doses of the antigen HEL ranging from 1 to 50 μM under standard T cell culture conditions (Fig. 2c). This observation further indicates that one of the biological consequences of T cell activation, the differential expression of Qa-1/Hsp60sp vs. Qa-1/Qdm, is predominantly dependent on the avidity of TCR on T cells within a wide range of antigen doses used to activate T cells in an environment providing relatively constant costimulation. This observation suggested that variation of this particular consequence of peripheral T cell activation determined by the avidity of TCR on activated T cells is quite limited, in vivo, within a biological range of antigen presented because somatic hypermutation is rare in TCR after thymic selection in the periphery (31). This is important because the observation suggested a necessary functional stability of the pathway to selectively down-regulate intermediate-avidity self-reactive T cells in vivo, which could be activated under various conditions and become potentially pathogenic. However, to what extent the extremely high and low doses of antigens, the drastic change of intensity and duration of costimulation as well as the possible altered peptide–ligand interactions may influence this particular biological outcome of activation of T cells with fixed TCR avidity (affinity and density of TCR expressed on each T cell) is unknown and requires further investigation.
In summary, perceiving the avidity of T cell activation can be translated into peripheral T cell regulation to discriminate self from nonself is the essence of the conceptual framework of the avidity model and allows an alternative intellectual theme to understand the development and control of immunologically relevant clinical problems. Manipulation of the common target structures recognized by Qa-1-restricted CD8+ T cells is the basis for potential therapeutic interventions to specifically enhance or block this regulatory pathway in vivo. In this regard, the potential application of such interventions in humans is based on the evidence that the human homolog of Qa-1, HLA-E, can function as a restricting element for human regulatory CD8+ T cells (22, 32).
Materials and Methods
Animals.
WT Balb/C and B10PL mice (The Jackson Laboratory) were housed in the pathogen-free animal facility associated with the Columbia University Department of Comparative Medicine. The Institutional Animal Care and Use Committee at Columbia University provided approval for all animal studies.
Reagents.
See the SI Text for details.
Real-Time PCR.
RNA was isolated by using the RNEasy system (Promega). Reverse transcription and real-time PCR were performed by using the avian myeloblastosis virus reverse transcriptase (Promega) and LightCycler Faststar DNA Master SYBR Green I Systems on a Roche LightCycler (Roche). The cycling parameters were 40 cycles of the following: 95°C, 10 min; 95°C, 10 s; 55–61°C (55°C, H-2Dd; 59°C, Qa-1b; 61°C, Hsp60), 8 s; 72°C, 13–20 s (13 s, Hsp60; 13 s, Qa-1b; 20 s, H-2Dd). Sequences of primers are shown in the SI Text. All cDNA samples were amplified in triplicate wells in each PCR assay. The PCR products for Qa-1b, mHsp60, and H-2Dd were each subcloned into the plasmid pCR2.1 (Invitrogen Life Technologies) and used to create standard curves, allowing calculation of the copy number.
SDS/PAGE and Western Blot Analysis.
SDS/PAGE and Western blotting were conducted according to standard procedures. HEL-specific CD4+ clones with different avidity to HEL were stimulated with irradiated splenic cells (antigen-presenting cells) and HEL (1–50 μM) for 72 h. The Abs used were as follows: anti-actin, anti-Hsp60, anti-MHC class Ia pan mAb M1/42 or H-2d-specific mAb 34–2-12 (33), and anti-Qa-1b 6A8.6F10.1A6, followed by incubation with the secondary Ab rabbit anti-mouse HRP or rabbit anti-Rat HRP. Target proteins were detected by using the ECL detection kit (Amersham Biosciences). All blots were densitometrically quantitated by using ChemiDoc XRS Imager Quantity One 4.5.0 software (Bio-Rad).
Qa-1a Transfectants and T Cell Clones.
The cDNA encoding full-length Qa-1a was isolated from the T cells from B10PL mice by RT-PCR by using the following primers: forward, CATGGTGAGGATGTTGCTTTTTGCCCACTTGCTCCAGC T G C T G G T C AGCG; and reverse, AGAACATGAGCATAGCATCCTTT. The Qa-1a cDNA was subcloned into the mammalian expression vector pCDNA3.1 (Invitrogen) and transfected by electroporation (Gene-Pulser, Bio-Rad) into the human B cell line C1R, following which stably transfected clones were isolated by limiting dilution under G418 selection (Invitrogen Life Technologies).
Qa-1-Binding Assay.
Selected Qa-1a-PcDNA3.1-positive transfectants detected by RT-PCR were assayed for their Qa-1 surface expression after loading with known Qa-1-binding peptides. In brief, Qa-1a transfectants were incubated at 26°C for ≈18 h with Hsp60sp or Qdm peptides and control peptide 1-9Nac MBP (12, 15, 21). The surface expression of Qa-1a was assessed by staining with anti-Qa-1a sera followed by PE–goat anti-mouse Ig and analyzed on a FACScan flow cytometer and CellQuest software (Becton Dickinson) (7). One of the transfectants, 3F4, which expresses Qa-1a on the surface when exogenously loaded with Hsp60sp and Qdm at 26°C, was chosen to further perform the subsequent functional assays. All peptides were used at 50 μM unless specifically indicated.
NK Assay.
NK cell lines are routinely established from B10PL mice (34). Unlabeled 3F4 cells were loaded with test peptides at 26°C overnight and mixed with an equal number of 5(6)-carboxyfluorescein diacetate, succinimidyl-ester (CFSE)-labeled human EBV-transformed lymphoblastoid cell line (LCL) cells, which are not susceptible to the killing by mouse NK cells. Graded numbers of mouse NK cells were added to the mixture of 3F4 and LCL cells and further incubated at 37°C for 2 h. 3F4 cells, which were not loaded with peptides or loaded with 1-9NacMBP, served as controls. The cell mixtures were then stained with anti-mouse H-2U mAb 3-8-3P-PE to distinguish NK cells from target cells. The change in the ratio of 3F4 to LCL cells in the presence or absence of NK cells in each group detected by FACS (7) reflects the specific NK killing: percentage of specific killing = {[killing ratio in control wells (without NK) − the killing ratio in experimental wells (with NK)]/the killing ratio of control wells} × 100%.
Peptide Competition Assay.
Peptide-unloaded 3F4 cells showed high susceptibility to NK killing [effector-to-target cell ratio (E/T), 2:1] and served as a positive control in this assay. 3F4 cells loaded with Qdm (20 μM), which were resistant to the NK killing, served as targets for competition. Competing peptides (Hsp60sp or control non-Qa-1-binding peptide) at 50, 100, and 200 μM were added to the 3F4 cells, together with 20 μM Qdm for 18 h at 26°C (17). Equal numbers of CFSE-labeled LCL cells were mixed with 3F4 cells, and NK cells were then added into the mixture of peptide-loaded 3F4 cells and LCL cells. The percentage of NK killing of 3F4 cells was evaluated. In the presence of the peptides that are capable of effectively competing with Qdm for occupancy for Qa-1, but do not interact with CD94/NKG2A, the abrogation of inhibition of 3F4 target cell lysis by NK cells in the presence of Qdm serves as a functional parameter of effective competition.
CD8+ T Cell Inhibition Assay.
CD8+ T cells were purified with CD8 MACS magnetic beads (Miltenyibiotec, Inc.) (7). 3F4 cells were passively loaded with peptides overnight at 26°C. An equal number of unlabeled 3F4 cells loaded with peptides and CFSE-labeled 3F4 cells that were not loaded with peptide were mixed, and a graded number of CD8+ T cells were added to the targets. CD8+ T cells from naïve mice served as a control, and we established that these CD8+ T cells have no effect on the activated target T cells. The 1-9Nac MBP-specific intermediate-avidity T cell clone 1AE10 and a low-avidity clone 4D10 served as positive and negative controls, respectively, and were built in all of the functional assays in B10PL mice. In addition, in all assays, the CD8+ T cells tested had no effect on C1R cells pulsed with Hsp60sp or Qdm. Four days later, the cell mixtures were stained with anti-mouse CD8–PE mAb to distinguish CD8+ T cells from target cells, and the CD8+ T cells were gated out during the analysis when the ratio between two types of targets were calculated. The ratio between the peptide-loaded (non-CFSE-labeled) 3F4 cells and unloaded (CFSE-labeled) 3F4 cells in the presence of CD8+ T cells was determined as percentage of specific inhibition (7): {[the ratio of loaded vs. unloaded 3F4 cells in control cultures (without CD8+ T cells) − the ratio in experimental cultures (with CD8+ T cells)]/the ratio in control cultures} × 100%. Because the time course of this assay was at least 4 days, we titrated the CFSE carefully and used a relatively high dose to label the target cells to ensure that by day 4 of the culture, the labeled target cells did not move into the unlabeled target cells. SI Fig. 4 illustrates a set of representative FACS plots showing that the CD8+ T cells inhibited the 3F4 cells loaded with Hsp60sp but not Qdm on day 4 of the culture.
Vaccination of Animals with DCs Loaded with Qa-1-Binding Peptides in the EAE Model.
Bone-marrow-derived DCs were generated from the femurs of B10PL mice as described (35). Briefly, the bone marrow was flushed out, erythrocytes were lysed, and hematopoietic precursors were cultured in complete RPMI medium 1640 containing 2.5% J558 cell supernatant as the source of GM-CSF for 6 days. On day 3, fresh culture media were added to the plate. On day 6, assessed by cell surface staining, these DCs routinely expressed Qa-1 on their surface but were CD8−. Day 6 DCs were loaded with either Hsp60sp or Qdm at 50 μM for 2 h at 37°C. Cells were then washed once with 50 ml of PBS and injected into naïve B10PL mice intravenously at 1 × 106 cells per mouse at least 1 week before EAE induction. EAE was induced in B10PL mice as described previously (8).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Richard Axel for discussion of the concept, Dr. Steve Canfield for critical reading of the manuscript, and Yihua Jiang for the technical help. The studies were supported by National Institutes of Health Grants R01 AI065609 (to H.J.) and U19 AI46132 (to L.C. and H.J.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709878104/DC1.
References
- 1.Anderton SM, Radu CG, Lowrey PA, Ward ES, Wraith DC. J Exp Med. 2001;193:1–11. doi: 10.1084/jem.193.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang H, Curran S, Ruiz-Vazquez E, Liang B, Winchester R, Chess L. Proc Natl Acad Sci USA. 2003;100:8378–8383. doi: 10.1073/pnas.1432871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han B, Serra P, Yamanouchi J, Amrani A, Elliott JF, Dickie P, Dilorenzo TP, Santamaria P. J Clin Invest. 2005;115:1879–1887. doi: 10.1172/JCI24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zehn D, Bevan MJ. Immunity. 2006;25:261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Chess L. N Engl J Med. 2006;354:1166–1176. doi: 10.1056/NEJMra055446. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H, Chess L. Annu Rev Immunol. 2000;18:185–216. doi: 10.1146/annurev.immunol.18.1.185. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H, Wu Y, Liang B, Zheng Z, Tang G, Kanellopoulos J, Soloski M, Winchester R, Goldstein I, Chess L. J Clin Invest. 2005;115:302–312. doi: 10.1172/JCI23879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, Zhang SI, Pernis B. Science. 1992;256:1213–1215. doi: 10.1126/science.256.5060.1213. [DOI] [PubMed] [Google Scholar]
- 9.Koh D-R, Fung-Leung W-P, Ho A, Gray D, Acha-Orbea H, Mak T-W. Science. 1992;256:1210–1213. doi: 10.1126/science.256.5060.1210. [DOI] [PubMed] [Google Scholar]
- 10.Hu D, Ikizawa K, Lu L, Sanchirico ME, Shinohara ML, Cantor H. Nat Immunol. 2004;5:516–523. doi: 10.1038/ni1063. [DOI] [PubMed] [Google Scholar]
- 11.Aldrich CJ, Waltrip R, Hermel E, Attaya M, Lindahl KF, Monaco JJ, Forman J. J Immunol. 1992;149:3773–3777. [PubMed] [Google Scholar]
- 12.Lowen LC, Aldrich CJ, Forman J. J Immunol. 1993;151:6155–6165. [PubMed] [Google Scholar]
- 13.Cotterill LA, Stauss HJ, Millrain MM, Pappin DJ, Rahman D, Canas B, Chandler P, Stackpoole A, Simpson E, Robinson PJ, et al. Eur J Immunol. 1997;27:2123–2132. doi: 10.1002/eji.1830270902. [DOI] [PubMed] [Google Scholar]
- 14.Kurepa Z, Hasemann CA, Forman J. J Exp Med. 1998;188:973–978. doi: 10.1084/jem.188.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imani F, Soloski MJ. Proc Natl Acad Sci USA. 1991;88:10475–10479. doi: 10.1073/pnas.88.23.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun T, Aldrich CJ, Baldeon ME, Kawczynski LV, Soloski MJ, Gaskins HR. Immunology. 1998;94:64–71. doi: 10.1046/j.1365-2567.1998.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaelsson J, Teixeira de Matos C, Achour A, Lanier LL, Karre K, Soderstrom K. J Exp Med. 2002;196:1403–1414. doi: 10.1084/jem.20020797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly DJ, Cotterill LA, Hederer RA, Thorpe CJ, Travers PJ, McVey JH, Dyson J, Robinson PJ. J Immunol. 1993;151:6089–6098. [PubMed] [Google Scholar]
- 19.Aldrich CJ, DeClousc A, Woods AS, Cotter RJ, Woloski MJ, Forman J. Cell. 1994;79:649–659. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 20.Gays F, Fraser KP, Toomey JA, Diamond AG, Millrain MM, Dyson PJ, Brooks CG. J Immunol. 2001;166:1601–1610. doi: 10.4049/jimmunol.166.3.1601. [DOI] [PubMed] [Google Scholar]
- 21.Lo WF, Ong H, Metcalf ES, Soloski MJ. J Immunol. 1999;162:5398–5406. [PubMed] [Google Scholar]
- 22.Li J, Goldstein I, Glickman-Nir E, Jiang H, Chess L. J Immunol. 2001;167:3800–3808. doi: 10.4049/jimmunol.167.7.3800. [DOI] [PubMed] [Google Scholar]
- 23.Jiang H, Ware R, Stall A, Flaherty L, Chess L, Pernis B. Immunity. 1995;2:185–194. doi: 10.1016/s1074-7613(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, Braunstein NS, Yu B, Winchester R, Chess L. Proc Natl Acad Sci USA. 2001;98:6301–6306. doi: 10.1073/pnas.101123098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sher BT, Nairn R, Coligan JE, Hood LE. Proc Natl Acad Sci USA. 1985;82:1175–1179. doi: 10.1073/pnas.82.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamvil S, Nelson P, Trotter J, Mitchell D, Knobler R, Fritz R, Steinman L. Nature. 1985;317:355–358. doi: 10.1038/317355a0. [DOI] [PubMed] [Google Scholar]
- 27.Davies A, Kalb S, Liang B, Aldrich CJ, Lemonnier FA, Jiang H, Cotter R, Soloski MJ. J Immunol. 2003;170:5027–5033. doi: 10.4049/jimmunol.170.10.5027. [DOI] [PubMed] [Google Scholar]
- 28.Wolf PR, Cook RG. J Exp Med. 1995;181:657–668. doi: 10.1084/jem.181.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanin-Zhorov A, Cahalon L, Tal G, Margalit R, Lider O, Cohen IR. J Clin Invest. 2006;116:2022–2032. doi: 10.1172/JCI28423. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Quintana FJ, Cohen IR. Scand J Immunol. 2006;64:205–210. doi: 10.1111/j.1365-3083.2006.01807.x. [DOI] [PubMed] [Google Scholar]
- 31.Wagner SD, Neuberger MS. Annu Rev Immunol. 1996;14:441–457. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- 32.Ware R, Jiang H, Braunstein N, Kent J, Wiener E, Pernis B, Chess L. Immunity. 1995;2:177–184. doi: 10.1016/s1074-7613(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 33.Stallcup KC, Springer TA, Mescher MF. J Immunol. 1981;127:923–930. [PubMed] [Google Scholar]
- 34.Takai T, Li M, Sylvestre D, Clynes R, Ravetch JV. Cell. 1994;76:519–529. doi: 10.1016/0092-8674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 35.Basu S, Binder RJ, Ramalingam T, Srivastava PK. Immunity. 2001;14:303–313. doi: 10.1016/s1074-7613(01)00111-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.