Abstract
Trimolecular interactions between the T cell antigen receptor and MHC/peptide complexes, together with costimulatory molecules and cytokines, control the initial activation of naïve T cells and determine whether the helper precursor cell differentiates into either T helper (TH)1 or TH2 effector cells. We now present evidence that regulatory CD8+ T cells provide another level of control of TH phenotype during further evolution of immune responses. These regulatory CD8+ T cells are induced by antigen-triggered CD4+ TH1 cells during T cell vaccination and, in vitro, distinguish mature TH1 from TH2 cells in a T cell antigen receptor Vβ-specific and Qa-1-restricted manner. In vivo, protection from experimental autoimmune encephalomyelitis (EAE) induced by T cell vaccination depends on CD8+ T cells, and myelin basic protein-reactive TH1 Vβ8+ clones, but not TH2 Vβ8+ clones, used as vaccine T cells, protect animals from subsequent induction of EAE. Moreover, in vivo depletion of CD8+ T cells during the first episode of EAE results in skewing of the TH phenotype toward TH1 upon secondary myelin basic protein stimulation. These data provide evidence that CD8+ T cells control autoimmune responses, in part, by regulating the TH phenotype of self-reactive CD4+ T cells.
To regulate the immune response and dampen the potential for autoimmunity, the immune system has evolved several mechanisms to down-regulate and control the outgrowth and differentiation of activated CD4+ T cells. One level of control, mediated during the initial interaction of the CD4+ T cell with MHC/peptide complexes on the surface of antigen-presenting cells, determines whether T cell activation, anergy, or apoptosis will ensue (1–3). A second level of control, mediated by cytokines, regulates the growth and differentiation of activated CD4+ T cells. Different cytokines secreted by CD4+ or CD8+ T cells either stimulate or inhibit CD4+ T cell proliferation and determine whether a naïve T helper (TH) precursor cell differentiates as an IFN-γ-producing TH1 cell or as an IL-4- and IL-10-producing TH2 cell (4–6). A third level of control resides in the regulatory T cells including both CD4+ (7) and CD8+ (8) T cell populations. For example, ample data demonstrate the ability of CD8+ T cells to regulate CD4+ T cell responses (9–13). These effects of CD8+ T cells have been mostly attributed, in recent years, to the CD8+ T cells' secretion of cytokines (14).
In addition to identifying cytokines as potential effectors of immune regulation by CD8+ T cells, other studies have identified specific cognate interactions between regulatory CD8+ T cells and activated CD4+ T cells. For example, during antigen- or superantigen-driven CD4+ T cell responses in vivo, CD8+ T cells emerge that specifically regulate CD4+ T cells in a T cell antigen receptor (TCR) Vβ-specific manner (15, 16). These CD8+ T cells preferentially recognize antigen-activated CD4+ T cell clones expressing certain TCR Vβ molecules and are restricted by the class I-b MHC molecule Qa-1. Unlike conventional MHC class I-a molecules, Qa-1 molecules are expressed only at low levels on resting T cells but are increased after antigen activation (17). These data are consistent with a model of specific immunoregulation in which after antigen activation CD4+ T cells express membrane Qa-1/TCRVβ motifs that are recognized by the αβ TCR expressed by precursor regulatory CD8+ T cells. These CD8+ T cells are induced to differentiate and down-regulate CD4+ T cells expressing the particular Qa-1/TCRVβ motifs.
A prediction of this model is that Qa-1-restricted, Vβ-specific regulatory CD8+ T cells will be induced by “vaccination” of animals with antigen-activated CD4+ T cells, using T cell vaccination (TCV) protocols known to prevent autoimmune disease in animal models (18, 19). In this regard, we have shown that Qa-1-restricted, Vβ-specific CD8+ cytotoxic T cell lines are induced by TCV (16). Moreover, we isolated a CD8+ T hybridoma clone from a T cell-vaccinated mouse that preferentially recognizes CD4+Vβ8+ but not CD4+Vβ6+ myelin basic protein (MBP)-reactive clones in a Qa-1-restricted fashion (16). In this current study, we further confirmed this prediction by investigating an experimental autoimmune encephalomyelitis (EAE) model system in which a MBP-specific encephalogenic CD4+ Vβ8+ TH1 clone, termed 1AE10, was used to induce EAE. EAE is induced in virtually all mice within 10 days, and TCV using irradiated 1AE10 cells gives complete protection from EAE. More importantly, depletion of CD8+ T cells in vivo abolished the protection.
We also demonstrate that these CD8+ T cell hybridoma cells preferentially recognize CD4+ MBP-reactive Vβ8+ TH1 but not CD4+ MBP-reactive Vβ8+ TH2 clones. The possible in vivo consequences of this differential recognition of CD4+ TH subsets is suggested by experiments showing that protection of EAE by TCV is CD8+ T cell dependent, and only MBP-reactive TH1 Vβ8+ clones, but not MBP-reactive TH2 Vβ8+ clones, protect animals from subsequent induction of EAE. We further show that CD8+ T cells control the TH phenotype of 1–9Nac MBP-reactive CD4+ T cells during the evolution of EAE in vivo. Thus, in mice depleted of CD8+ T cells during the induction of EAE, the 1–9Nac MBP-reactive TH1 CD4+ T cells are dramatically increased upon secondary antigen stimulation in vivo. In contrast, intact EAE recovered mice (possessing regulatory CD8+ T cells) showed significantly decreased TH1 cells.
Materials and Methods
Animals.
Female B10PL mice were purchased from The Jackson Laboratory and kept in our germ-free animal facilities.
Antibodies and Flow Cytometry.
Anti-Qa-1a and anti-Qa-1b antisera were kindly provided by Lorraine Flaherty (Wadsworth Center, New York State Department of Health, Albany). Antibodies to CD4 (GK1.5), CD8 (53–6.72), Vβ8.1–3 (F23.1), Vβ8.2 (F23.2), and Vβ8.1,2 (KJ16) were purified from cultured supernatants and conjugated to either fluorescein or biotin. Biotinylated antibodies were revealed by using FITC or phycoerythrin-conjugated streptavidin (Calbiochem). Antibody to Vβ6 (RR4–8) was purchased from PharMingen. Analysis of stained cells was performed by using a FACScan flow cytometer and cellquest software (Becton Dickinson) as described (16).
Generation and Analysis of CD4+ T Cell Clones.
CD4+ Vβ8+ or Vβ6+ MBP-reactive B10.PL clones were derived by limiting dilution cloning of purified CD4+ T cells obtained from the spleens or lymph nodes from EAE-recovered or MBP-immunized mice in the presence of 5 μg/ml 1–9NacMBP peptide, 2,500-R irradiated syngeneic splenocytes (16). T cells were maintained by biweekly stimulation with antigen and 2,500-R irradiated syngeneic splenocytes and expanded in recombinant human IL-2 (Proleukin, Chiron) at a concentration of 10 units of IL-2 activity/ml. Cytokine content of supernatants was assayed by capture ELISA (Pharmingen).
Generation and Assay of CD8+ T Cell Hybridomas.
The generation of the lacZ-positive CD8+ T cell hybridomas has been described (16). Briefly, B10PL mice were injected i.v. with 2.5–5 × 106, antigen-activated, 3,000-R irradiated 1AE10 cells (CD4+ Vβ8+ TH1). Seven days later, a CD8+ T cell-enriched splenic population was incubated with irradiated antigen-activated 1AE10 cells and irradiated syngeneic spleen cells and IL-2. Cells were fused on the fourth day of culture with the CD8+, lacZ-inducible fusion partner BWZ.36 (20). In the standard assay of hybridoma responses, 5 × 104 CD4+ T cells and 5 × 104 hybridomas were cocultured for 18 h in triplicate and then assayed colorimetrically for lacZ expression. In the preculture experiments, 3.5 × 105 CD4+ T cells were incubated for 24 h in the indicated cytokines plus 10 units/ml IL-2 or, in mixing experiments, 3.5 × 105 of each of the clones were incubated together for 24 h in IL-2. At the end of the preculture, 5 × 104 hybridoma cells were added to each well and assayed 18 h later as above. In all assays of CD8+ T cell function, CD4+ T cells were used on days 4–7 after activation.
TCR Cloning and Sequencing.
RNA was prepared either by the LiCl method (21) or by using a MicroFastTrack kit (Invitrogen). The mRNAs were reverse-transcribed to cDNA by using Superscript RT (Life Technologies, Gaithersburg, MD) and oligo(dT) primers. TCRB cDNAs were amplified by PCR using the TCRBV8-specific oligodeoxynucleotide 5′-CATGGAGGCTGCAGTCACCC-3′ and the TCRBC-specific oligo 5′-ACCCTCAGGCGGCCGCTCAG-3′. The PCR products were either T/A-cloned into PCR2.1-TOPO (Invitrogen) and sequenced or were purified by using QIAquick (Qiagen, Chatsworth, CA) columns and sequenced. Sequences were obtained by using an internal TCRBC oligo 5′- GATGGCTCAAACAAGGAGAC-3′.
The EAE Model and the TCV Procedure.
To establish an EAE model induced by encephalitogenic clones, different groups of mice were sublethally irradiated (350 R), and after 24 h were injected i.v. with 8–10 × 106 cells of three different 1–9Nac MBP-specific, CD4+Vβ8+, TH1 clones: 1AE10, 3AC3, and 5B2. These CD4+Vβ8+ T clones were activated by 1–9Nac MBP for 4 days before injection. Pertussis toxin also was injected at 0.1 μg/mouse i.v. 24 and 72 h later. Animals were monitored daily, and EAE was evaluated as described (10). Both 1EA10 and 3AC3, but not 5B2, effectively induced the clinical syndrome of EAE in B10PL mice with 100% efficiency. Because 1AE10 was also the most rapidly growing clone, it was chosen to be the EAE-inducing clone for the remainder of the experiments. TCV was performed by injecting mice with 2–2.5 × 106 irradiated cloned CD4+ T cells i.v. 1 week before EAE was induced. Animals were monitored daily, and EAE was evaluated as described (10). Vaccine T cells were used on days 7–10 after activation by 1–9Nac MBP. The T cell clones used as vaccine T cells were: Vβ8.2+,TH1 clones, 1AE10, 2–1D9, 2–5F6 and 3–3D11; Vβ8.2+,TH2 clones, 3AD2 and 3BG3; and Vβ6+,TH1 clones, 3AF9 and 3AB3.
In Vivo Assessment of TH Phenotype of 1–9Nac MBP-Specific CD4+ T Cells in the Periphery of EAE Mice.
EAE was induced by immunizing B10PL mice with 1–9Nac MBP as described (10). Some mice were depleted of CD8+ T cells in vivo by using anti-CD8 mAb 53–6.72 3 days before induction of EAE with 1–9Nac MBP (10). All mice were allowed to recover from EAE for 8–12 weeks. The two groups of animals then were immunized with 1–9Nac MBP in complete Freund's adjuvant at 100 μg/mouse s.c. and 7 days later, CD4+ T cells were purified from draining lymph nodes, using MACS beads (Miltenyi Biotec, Auburn, CA). Briefly, the lymph node cells were incubated with antimurine CD4-conjugated beads at 10 × 106 cells/10 μl beads, and the CD4+ and CD4- population were isolated by using a separation column exposed to a magnetic field according to the manufacturer's protocol. The purity of the CD4+ T cells was >95%. CD4+ T cells were restimulated in vitro with 1–9Nac MBP and antigen-presenting cells. At different time points after activation, the CD4+ T cells were assayed for TH phenotype by cytoplasmic cytokine staining. Briefly, the cultured cells were stimulated in vitro by phorbol ester (0.02 μg/ml) and ionomycin (0.4 μM/ml) for 1 h, and Brefeldin A was added for an additional 4 h to block the secretion of cytokines. The cells were permealized, stained for intracellular INF-γ and IL-5 using a kit (PharMingen), and analyzed by FACS.
Results
The CD8+ T Cell Hybridoma 21–5A9 Preferentially Recognizes MBP-Reactive Vβ8+TH1 Clones.
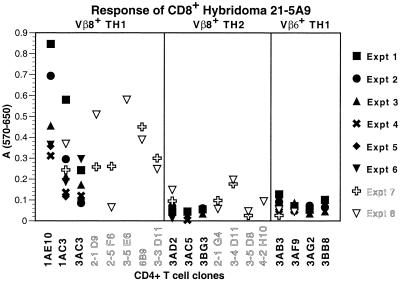
The CD8+ T cell hybridoma, 21–5A9, was derived from a B10PL mouse that had been vaccinated with the irradiated syngeneic, MBP-reactive, encephalitogenic CD4+Vβ8+TH1 clone, 1AE10. In a previous report, this hybridoma was shown to respond to 1AE10 and other CD4+Vβ8+TH1 clones but not to any CD4+Vβ6+TH1 clones. Moreover, its response was inhibited by anti-Qa-1a antisera but not by either anti-Qa-1b antisera or by monoclonal anti-MHC class I-a molecules (16). To investigate the CD4+ TH subset specificity of this hybridoma, we assayed the hybridomas' response to eight MBP-specific Vβ8+CD4+TH1 clones, seven MBP-specific Vβ8+CD4+TH2 clones, and four MBP-specific Vβ6+CD4+TH1 clones. As shown in Fig. 1, the 21–5A9 hybridoma is preferentially stimulated by the MBP specific Vβ8+CD4+TH1 clones. In contrast, the MBP-specific Vβ8+TH2 clones and Vβ6+TH1 clones did not induce significant responses. We would emphasize that the CD4+ clones were at a comparable level of activation and proliferation and expressed comparable levels of surface Qa-1 before assay. Although there was some experiment-to-experiment variability in the response of the 21–5A9 hybridoma to the CD4+ clones the preferential response of the 21–5A9 hybridoma to MBP-specific CD4+TH1 compared with TH2 clones was highly statistically significant with a P value of <0.0000000002.
Figure 1.
Qa-1-restricted Vβ8-specific CD8+ T cell hybridoma 21–5A9 differentially recognizes TH1 Vβ8+CD4+ T cells but not TH2 Vβ8+CD4+ T cells. The CD8+ T cell hybridoma was cultured with each of the MBP-specific CD4+ clones for 18 h and then assayed for the induction of β-galactosidase activity as described in Materials and Methods. Each data point represents the means of triplicate values minus background.
The Specific Recognition of CD4+ T Cells by the CD8+ T Hybridoma Is Mediated by the αβ TCR in a CD8- and Qa-1-Dependent Manner.
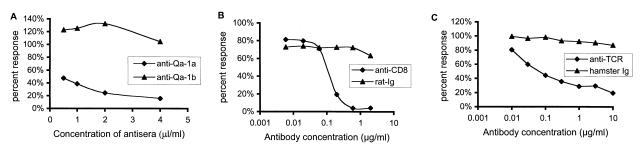
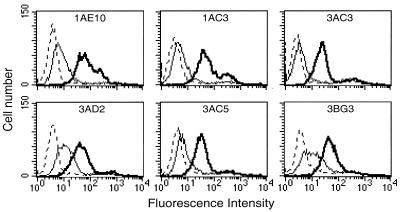
To directly establish that the αβ TCR is involved in target recognition, the antimurine pan αβ TCR antibody (H57) and an antimurine CD8 antibody (53–6.72) were used to block the specific recognition of the CD4+Vβ8+TH1 cells by the CD8+ 21–5A9 hybridoma (Fig. 2 b and c). Hamster and murine Ig served as the negative controls. As shown, the triggering of the 21–5A-9 hybridoma by CD4+Vβ8+TH1 target 1AE10 was specifically blocked by both the anti-TCR and the anti-CD8 antibodies but not control Ig. Moreover, the specific blocking of the hybridoma reactivity by the anti-Qa-1a sera was confirmed (Fig. 2a). Furthermore, to investigate whether the preferential response of the 21–5A9 CD8+ T cell hybridoma clone to TH1 compared with the TH2 CD4+ T cells was due to differential Qa-1 expression, we stained and assayed by FACS the Vβ8+CD4+ TH1 and TH2 cell clones described above with anti-Qa-1a antisera. We assayed Qa-1 expression daily after MBP activation. The Qa-1 surface expression peaks between days 4 and 7 after activation, and we observed no significant difference of Qa-1 expression between TH1 and TH2 clones at any time point. Representative FACS data on 6 days after activation is shown by Fig. 3.
Figure 2.
Anti-Qa-1a antiserum, antimurine CD8 mAb, and antimurine TCR mAb block the specific recognition of TH1 Vβ8+CD4+ T cell clone 1AE10 by the CD8+ T cell hybridoma 21–5A9. In these experiments, varying amount of either (a) murine anti-Qa-1a or control anti-Qa-1b sera; (b) rat anti-mouse CD8 mAb (53–6.72) or control rat Ig; and (c) hamster anti-mouse TCR mAb (H57) or control hamster Ig were added to the coculture of 1AE10 and 21–5A9. Reactivity of 21–5A9 was assayed as described in Fig. 1. Inhibition of 21–5A9 reactivity was assessed as a decrease in the percent response compared with the response of the 21–5A9 hybridoma without antibody or control Ig.
Figure 3.
TH1 Vβ8+CD4+ clones and TH2 Vβ8+CD4+ clones express similar levels of Qa-1 on their surface. TH1 (Upper) and TH2 (Lower). CD4+ T cell clones were stained with antisera to Qa-1a (bold solid lines), Qa-1b (thin solid lines), or with normal B10.PL serum (dotted lines) as described in Materials and Methods. CD4+ T cell lines were stained on day 6 after activation.
The CD8+ T Cell Hybridoma Can Distinguish Between TH1 and TH2 CD4+ T Cells Expressing Identical TCRs.
We next investigated whether the TH1 and TH2 cells differed in their expression of TCR molecules. There was no difference in the level of TH1 and TH2 cell TCR expression as determined by staining with anti-TCRVβ8 antibodies or anti-CD3 antibodies. To determine whether there was a structural difference in TCR expression, we cloned and sequenced TCRB cDNAs from the original series of CD4+ T cell clones by PCR. Alignment of the nucleotide and deduced polypeptide sequences demonstrated that the TH1 and TH2 cells express similar TCRβ chains (data not shown). In fact, there are two pairs of TH1 and TH2 cells (1AE10 and 3AD2, 1AC3 and 3BG3) that express TCRβ chains with identical amino acid sequence. In the case of 1AC3 and 3BG3, there were three silent nucleotide differences between the cDNAs. Interestingly, clones 1AE10 and 3AD2 were shown to have identical TCRB nucleotide sequences, which was confirmed by repeated cloning of all of the TCR cDNAs in parallel using new aliquots of cells and new reagents. Additional sequence analysis demonstrated the presence of identical TCRA mRNAs in the 1AE10 and 3AD2 clones, indicating that, on both chromosomes, these cells contain identically rearranged TCRA genes. Thus, the 1AE10 and 3AD2 cells express identical TCRαβ chains and are derived from the same progenitor Th precursor cell. Nonetheless, the 21–5A9 CD8+ T cell hybridoma responds to the TH1 clone 1AE10 but not to the TH2 clone 3AD2. These data do not rule out the possibility that the TH1 and TH2 cells may present distinct Qa-1 bound TCR peptides that are distinguished by the CD8+ regulatory cells.
Cytokines Produced by TH Cells Do Not Account for Their Different Abilities to Stimulate the CD8+ Hybridomas.
Because the phenotypic hallmark that distinguishes the TH subsets is the production of distinct cytokines, we considered the possibility that particular cytokines are required to activate the CD8+ T cell hybridoma response or that TH2 cytokines might be inhibitory. To address this issue, we first sought to determine whether CD8+ T hybridoma activation required actual contact with the Vβ8+TH1 cells or whether soluble products from TH1 cells could activate the hybridoma. In a series of two-chamber experiments in which the CD8+ hybridoma cells and CD4+ cells were cultured either together or separated by a 0.4 μM membrane (Costar) we demonstrated that CD8+ T hybridoma cell activation required actual cell-cell contact because 1AE10 cells lost the capacity to stimulate 21–5A9 when separated from 21–5A9 by the membrane.
We next investigated whether this contact-dependent CD8+ T cell activation could be modulated by soluble TH cell products. We first asked whether the coculture of CD4+Vβ8+ TH1 and TH2 cells in various cytokines either induced, augmented, or inhibited their ability to activate the CD8+ hybridoma. CD4+ T cells were preincubated for 24 h in the presence of: (i) IFN-γ at two different concentrations, (ii) IL-4 at two different concentration, (iii) 25% (vol/vol) supernatants from 1AE10 cells harvested at 48 or 96 h after antigen activation, or (iv) 25% (vol/vol) supernatants from 3AD2 cells harvested at 96 h after activation. In addition, we precultured mixtures of the various cells in different combinations for 24 h to further investigate the abilities of TH1 or TH2 cytokines to induce or inhibit the CD8 anti-Vβ response. IL-2 was included in all of the preculture wells to maintain viability of the CD4+ T cell clones. At the end of the preincubation, 21–5A9 hybridoma cells were added to each well for an additional 18 h culture. As a further set of controls, fresh stimulator cells were incubated with 21–5A9 hybridoma cells for 18 h as we had done for the previous experiments. As shown in Table 1, none of these cytokine modulations altered the ability of the CD4+ T cell clones to stimulate the 21–5A9 CD8+ hybridoma. For example, after preculture of the 3AD2 (Vβ8+TH2) clone in IL-2 plus IFN-γ, IL-2 plus IL-4, or IL-2 plus supernatants from 1AE10, the 3AD2 cells were no more stimulatory for the hybridoma than were fresh 3AD2 cells from the same culture. Similarly, 3AD2 cultured for 24 h with the Vβ6+TH1 clone, 3AF9, remained nonstimulatory for the Vβ8-specific CD8+ hybridoma. As a control, it was noted that the coculture of 3AF9 with the 1AE10 cell did not inhibit the ability of 1AE10 to stimulate the CD8+ hybridoma. Moreover, the culture of 1AE10 with the various cytokines or with the 3AD2 cell line did not inhibit its ability to stimulate the 21–5A9 hybridoma. These results indicate that cytokines or other soluble mediators produced by the CD4+ T cells do not serve either to stimulate or inhibit the hybridoma cells.
Table 1.
Cytokines or other soluble TH products do not influence the TH specificity of the 21-5A9 CD8+ T cell hybridoma
| Stimulator cells
|
|||
|---|---|---|---|
| 1AE10 Vβ8 TH1
|
3AF9 Vβ6
TH1
|
3AD2 Vβ8 TH2
|
|
| Response of 21-5A9 (A570–A650) | |||
| Experiment A* | |||
| IL-2 starved | 0.157 | 0.073 | 0.064 |
| IL-2 10 units/ml | 0.602 | 0.099 | 0.067 |
| IL-2 + IFN-γ 200 units/ml | 0.650 | 0.108 | 0.075 |
| IL-2 + IFN-γ 20 units/ml | 0.590 | 0.107 | 0.074 |
| IL-2 + IL-4 200 units/ml | 0.602 | 0.091 | 0.079 |
| IL-2 + IL-4 20 units/ml | 0.596 | 0.106 | 0.081 |
| IL-2 + 48 h TH1 sups | 0.740 | 0.104 | 0.033 |
| IL-2 + 96 h TH1 sups | 0.523 | 0.086 | 0.058 |
| Fresh cells (control) | 0.460 | 0.049 | 0.039 |
| Experiment B† | |||
| IL-2 alone | 0.314 | 0.028 | 0.026 |
| IL-2 + 96 h TH1 sups | 0.373 | 0.071 | 0.076 |
| IL-2 + 96 h TH2 sups | 0.352 | 0.054 | 0.065 |
| Fresh cells (control) | 0.357 | 0.044 | 0.033 |
| Cell mixing experiments† | |||
| 1AE10 + 3AF9 | 0.297 | ||
| 1AE10 + 3AD2 | 0.374 | ||
| 3AD2 + 3AF9 | 0.062 | ||
Representative of three separate experiments.
Representative of two separate experiments.
In Vivo Evidence that Regulatory CD8+ T Cells Are Specifically Induced by TH1 But Not TH2 MBP-Reactive CD4+ T Cells to Protect Mice from EAE.
In the next series of experiments we asked whether the capacity of CD8+ T cells to distinguish TH1 versus TH2 CD4+ T cells in vitro has biological consequences in vivo. Because the Vβ8-specific and Qa-1-restricted CD8+ T cell hybridoma, 21–5A9, which preferentially recognizes TH1 cells, originally was generated from mice vaccinated with the MBP-reactive CD4+Vβ8+TH1 clone, 1AE10, we first asked whether the process of TCV, using 1AE10, induces regulatory CD8+ T cells in vivo, which protect animals from EAE. We then asked whether the TH2 clone, 3AD2, which expresses the identical TCR VA and VB chains as 1AE10, is also capable of inducing CD8+ T cell-dependent protection from EAE.
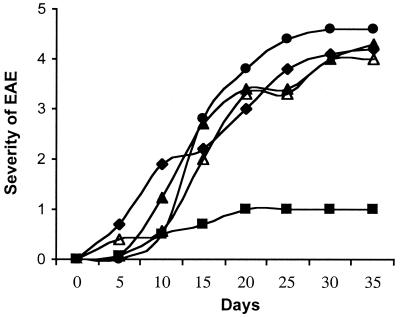
EAE was induced by the TH1 Vβ8+ encephalitogenic clone 1AE10, which induces EAE in virtually all B10PL mice with clinical symptoms emerging within 7–14 days. In this model, we first showed that TCV using 2–2.5 × 106 irradiated 1AE10 T cells administered 7 days before disease induction effectively protected animals from subsequently induced EAE by 1AE10 (Fig. 4). We then asked whether the protection induced by TCV depended on CD8+ T cells in vivo. Thus, one group of mice were depleted of CD8+ T cells before TCV using the antimurine CD8 mAb 53–6.72 as described (10). Control mice either received rat Ig before TCV or received no TCV before the induction of EAE. The effectiveness of the antimurine CD8 mAb in depleting CD8+ T cells was assayed by FACS analysis of peripheral T cells at days 3, 7, 10, and 14. The CD8+ T cells were markedly reduced (<1%) from days 3 to 10 and gradually reappeared after day 10. The TCV was performed on day 3, and the EAE induction was performed on day 10 of the CD8+ T cell depletion. This time window is sufficient to reveal the effect of CD8+ T cells on the protection of EAE induced by TCV. The results from multiple experiments show that depletion of CD8+ T cells before TCV completely abolished the protection from EAE induced by TCV (Fig. 4).
Figure 4.
TCV-induced protection in EAE is CD8+ T cell dependent and TCR Vβ and TH type specific. EAE was induced by the encephalitogenic clone 1AE10 as described. TCV was performed 1 week before EAE induction with different T cell clones: One group of mice were treated with either rat Ig (■) or anti-CD8 (●) 3 days before TCV with 1AE101 (Vβ8.2+, TH1); other groups received TCV with 3AD2 (Vβ8.2+, TH2) (▵) or 3AF9 (Vβ6+, TH1) (▴). Mice in the control group did not receive TCV (⧫). Results are representative of four separate experiments with four mice per group per experiment.
To further define the specificity of the TCV-induced protection from EAE, we next compared the protection induced by 1AE10 with three additional 1–9Nac MBP-specific TCR Vβ8+ TH1 clones (data not shown) as well as two 1–9Nac MBP-specific TCR Vβ6+ TH1 clones. The three 1–9Nac MBP-specific Vβ8+ TH1 clones protected animals from EAE induced by 1AE10 to some degree but not as efficiently as 1AE10. In contrast, vaccination with the two Vβ6+ TH1 clones did not protect animals. These results show that the protective effect of CD8+ T cells in vivo by TCV in this model is induced preferentially by several CD4+Vβ8+ clones in a TCR Vβ-specific manner and does not depend on clonal identity between the vaccine T cells and the EAE-inducing clone.
In addition, we asked whether TH phenotype of the vaccine T cells played an in vivo role in CD8+ T cell-dependent protection from EAE. Thus, we directly compared the CD4+Vβ8+ T cell clone 1AE10 (TH1) with the 3AD2 (TH2) for their capacity to induce protection from EAE. As noted above, both of these T cell clones are specific for the 1–9Nac MBP and express identical TCR αβ chains. Thus, different groups of mice were vaccinated with either 1AE10 or 3AD2 (2–2.5 × 106 cells/mouse) 7 days before induction of EAE with 1AE10 cells. The TH1 clone 1AE10 completely protected mice from subsequent induction of EAE. In marked contrast, the TH2 clone 3AD2 did not protect at all (Fig. 4). In addition, another Vβ8+ TH2 clone 3BG3 was used as vaccine T cells in TCV experiments. This clone, like 3AD2, failed to protect animals from EAE (data not shown). Because the protection afforded by Vβ8+ TH1 clones is CD8+ T cell-dependent, these data are consistent with the model that Vβ8+ TH1 cells, but not Vβ8+ TH2 cells, are able to induce the regulatory CD8+ T cells, which specifically down-regulate TH1 cell-induced disease in vivo.
CD8+ T Cells Control the TH Phenotype of 1–9Nac MBP-Specific CD4+ T Cells in the Periphery of EAE Mice.
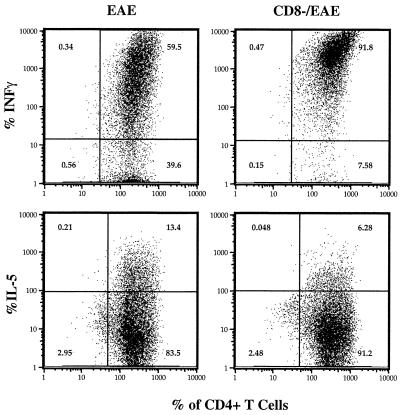
We previously showed that B10PL mice immunized with an encephalitogenic peptide of MBP develop acute EAE and recover from the first episode of disease and do not develop relapsing EAE. Importantly, the first episode of EAE renders the animals resistant to the reinitiation of EAE by secondary immunization with MBP. In contrast, animals depleted of CD8+ T cells in vivo, using anti-CD8 mAb-mediated clearance are no longer resistant to a second induction of disease with MBP (10). These data demonstrate that CD8+ T cells play a major role in the resistance to a second induction of EAE. Moreover, if CD8+ T cell depletion is instituted during the primary induction of EAE, animals recover from EAE but are not rendered resistant to reinduction of EAE. Taken together these data suggest that the regulatory CD8+ T cells need to be primed during the first episode of EAE to down-regulate encephalitogenic CD4 T cells during the second induction of EAE. These observations together with our data described above that CD8+ T cell hybridoma cells differentially recognize TH1 versus TH2 cells suggest the possibility that CD8+ T cells may control the TH phenotype of 1–9Nac MBP-reactive CD4+ T cells that emerge in the periphery during the evolution of EAE. To directly test this hypothesis, the TH phenotype of MBP-reactive CD4+ T cells in EAE recovered mice (EAE) and CD8+ T cell-depleted EAE recovered mice (CD8-/EAE) were assayed and compared. The latter group of mice were depleted of CD8+ T cells by using antimurine CD8 mAb 3 days before induction of EAE with 1–9Nac MBP. Both the CD8+ T cell-depleted and CD8+ T cell-nondepleted, EAE recovered mice were immunized with 1–9Nac MBP s.c., and 7 days later CD4+ T cells were purified from draining lymph nodes and restimulated in vitro with 1–9Nac MBP. The TH type of the MBP-specific CD4+ T cells was assayed on days 4–7 by intracellular staining of IFN-γ and IL-5 and analyzed by FACS. The experiment depicted in Fig. 5 is representative of four separate experiments. As shown, in CD8-/EAE mice, the IFN-γ-positive CD4+ T cells increased, whereas the IL-5-positive CD4+ T cells decreased upon secondary MBP stimulation, compared with the EAE recovered mice that possess the regulatory CD8+ T cells. We also assayed the TH phenotype of CD4+ T cells from draining lymph nodes at the different time points after in vitro retriggering with MBP. We consistently found an increase in the fraction of MBP-reactive CD4+ TH1 cells and a decrease in the fraction of MBP-reactive CD4+ TH2 cells at all time points in cultures derived from CD8-/EAE mice. These results show that CD8+ T cells preferentially down-regulate TH1 1–9Nac MBP-reactive CD4+ T cells in the periphery of EAE recovered mice in vivo. These data are consistent with the idea that regulatory CD8+ T cells control the function of antigen-specific CD4+ T cells in the periphery, in part, by controlling the TH phenotype of antigen-activated CD4+ T cells.
Figure 5.
CD8+ T cells control the TH phenotype of 1–9Nac MBP-reactive CD4+ T cells in EAE mice. The TH phenotype of 1–9Nac MBP-reactive CD4+ T cells in EAE recovered mice (EAE) and CD8+ T cell-depleted EAE recovered mice (CD8-/EAE) were assayed and compared. The CD4+ T cells from the EAE mice and CD8-/EAE mice were isolated and assayed for TH phenotype by intracellular staining by FACS analysis for the expression of intracellular IFN-γ and IL-5 as described. Results are representative of four separate experiments.
Discussion
In this report, we provide in vitro and in vivo evidence that CD8+ T cells regulate immune responses to the 1–9Nac MBP self-peptide, by differentially recognizing TH1 from TH2 MBP-reactive CD4+ T cells. We first further investigated the specificity of the Qa-1-restricted Vβ-specific CD8+ T hybridoma clone, 21–5A9, which previously had been shown to distinguish between Vβ8+ and Vβ6+ MBP-specific CD4+ TH1 clones in a Qa-1 restricted manner (16). This hybridoma is shown here to preferentially recognize MBP-reactive CD4+Vβ8+ TH1 but not CD4+Vβ8+ TH2 clones, even for pairs of TH1 and TH2 clones expressing identical TCR Vα and Vβ chains and identical Qa-1 molecules. The potential in vivo significance of these studies is suggested by experiments showing that TCV using TH1 but not TH2 CD4+ MBP-specific T cells as vaccine cells protect mice from EAE. Moreover, protection from EAE is CD8+ T cell-dependent, because CD8+ T cell depletion completely abrogates the protection induced by TH1 clones. Taken together, these data are consistent with the model that Vβ8+ TH1 cells but not Vβ8+ TH2 cells are able to induce regulatory CD8+ T cells, which specifically down-regulate TH1 cell-mediated disease in vivo. Moreover, additional evidence to support this model was provided in experiments showing that CD8+ T cell depletion in vivo results in a skewing of the TH phenotype in EAE mice. Thus, there are a greater percentage of TH1 cells responding to MBP stimulation during the evolution of MBP-induced EAE in CD8+ T cell-depleted mice as compared with CD8+ T cell intact mice.
In the current studies, we considered several general mechanisms for the differential recognition of TH1 and TH2 clones by the regulatory CD8+ T cells. First, although cytokines or other soluble TH products secreted by TH1 or TH2 cells may influence the specificity of the CD8+ T cells, in a series of cytokine, supernatant, and cell mixing experiments, a direct role for cytokines in this response was not observed. Instead, activation of the 21–5A9 CD8+ T cell hybridoma required CD4+/CD8+ T cell contact. These observations let us to consider the second possibility that TH1 and TH2 cells might differ in the expression of cell surface molecule(s) important in this CD8+ T cell response. We first showed that the recognition molecule used by the regulatory CD8+ T cells is a classical αβ TCR structure and that the triggering of this αβ TCR depends on Qa-1 structures expressed on the CD4+ T cells. These conclusions were firmly supported by our data that the activation of the 21–5A9 hybridoma is blocked by both anti-TCR and anti-CD8 antibodies (reactive with the CD8+ hybridoma) as well as by anti-Qa-1 antibodies (reactive with the CD4+ T cells). The possibility that TH1 and TH2 cells may differ in quantitative Qa-1 surface expression was excluded by staining and FACS analysis with the anti-Qa-1 antiserum. Furthermore, pairs of TH1 and TH2 cells were identified that expressed identical TCRβ chains but differed in their abilities to activate the CD8+ hybridomas. Moreover, in one case a pair of TH1 and TH2 clones derived from the same clone with identical TCRβ and TCRα chains were identified. Here, too, only the TH1 cells triggered the hybridoma. Together these data suggested that the differential expression of the αβ TCR or Qa-1 by the CD4+ T cells does not account for the capacity of CD8+ T cells to distinguish TH1 from TH2 cells. It remains possible that other differentially expressed cell surface molecules involved in cell-cell contact between CD8+ T cells and targets may account for the differential recognition of TH1 but not TH2 cells.
Because Qa-1, like other class Ib MHC molecules, is known to present peptides to T cells (22–24), a third possibility for the differential recognition of TH1 but not TH2 clones is that TH1 and TH2 cells might differ in their processing and presentation of relevant self-peptides to regulatory CD8+ T cells. In principle, this distinction could be either at the level of the presenting molecules used or the self-peptide presented. In this regard, it is known that the predominant self-peptide presented by Qa-1 is termed Qdm (for Qa-1 determinant modifier), derived from the signal leader sequence of certain murine class Ia molecules (22, 25, 26). This peptide (AMAPRTLLL) binds with high affinity and accounts for the majority of the peptides associated with this molecule (26). However, Qa-1 also can bind other self-peptides including those derived from heat shock proteins (23) and preproinsulin leader sequences (24) as well as exogenous peptides including bacterial-derived peptides (27, 28). In addition, our data suggest that CD8+ T cells recognize TCR Vβ motifs restricted by Qa-1. For example, it is possible that although TH2 cells express Qa-1, they may use other MHC class Ib molecules as presenting molecules or they may present different self-TCR peptides or other non-Qdm self-peptides to regulatory CD8+ T cells. In either case, the MHC class Ib/TCR peptide complex generated and presented to the CD8+ T cells could differ between TH1 and TH2 cells.
Regardless of the mechanisms used by the Qa-1-restricted TCR Vβ-specific CD8+ T hybridoma cells to differentially recognize TH1 versus TH2 cells in vitro, the in vivo studies reported here provide a way of thinking about the control of TH phenotype during an immune response. It is known that the TH phenotype of an immune response is regulated at several stages during its evolution. Cognate trimolecular interactions between the TCR and MHC/peptide complexes, together with costimulatory molecules and cytokines, control the initial activation of a naïve T cell and determine whether the helper precursor cell differentiates as a TH1 or TH2 effector (6, 29–31). Many cytokine-producing cells have been shown to influence this process, including natural killer cells (32), γδ T cells (33), and the TH1 and TH2 cells themselves (4). Mechanisms for the differential regulation of mature TH1 and TH2 clones are less well understood. Some cytokines such as IL-4 and IL-12 contribute to the differential growth of the subsets. For example, IL-4 preferentially promotes the activation and growth of TH2 cells (34, 35); IL-12 stimulates TH1 cells (36). The local production of cytokines by regulatory cells at sites of inflammation may serve to effect antigen-specific immunoregulation. The data presented in this report suggest that, in addition to the control of TH phenotype during the initial antigen triggering, regulatory CD8+ T cells that distinguish TH1 from TH2 CD4+ T cells provide another level of control of TH phenotype during the further evolution of an immune response. There are two possible pathways by which CD8+ T cells could control the TH phenotype of an ongoing immune response in vivo. The CD8+ T cells might directly alter the existing TH type of antigen-activated CD4+ T cells. At the present time, there is no data to support this possibility. Alternatively, CD8+ T cells might selectively recognize and down-regulate TH1 antigen-activated CD4+ T cells in vivo. In this case, the differentiation of CD4+ T cells upon secondary antigen stimulation will be skewed toward the outgrowth of antigen-specific TH2 cells simply because of the decreased competition from down-regulated antigen reactive TH1 cells. Our experimental data support the latter possibility.
In this regard, of interest are studies documenting changes of the TH phenotype of CD4+ T cell subsets during various vaccination procedures used to ameliorate or prevent autoimmune disease. For example, Waisman et al. (37) demonstrated that vaccination of naive PL/J mice with TCR Vβ8 cDNA protected animals from subsequent induction of EAE with pAc1–20 peptide of MBP. It is significant that accompanying this protection was a skewing of the MBP-reactive CD4+ T cells from TH1 to TH2 phenotype. Although the mechanism for this shift is unknown, the protection was shown to be TCR Vβ specific. Moreover, in the NOD autoimmune murine model, Elias et al. (38) demonstrated that immunizing NOD mice with a peptide (C9) derived from the TCR CDR3 region of a T cell clone specific to a self-peptide p277 induced down-regulation of IFN-γ secreted by the anti-p277 T cells. Furthermore, analogous to the 1–9Nac MBP-induced resistance in EAE mice, immunizing NOD mice with p277 peptide clinically arrested the autoimmune diabetic process. Interestingly, the effectiveness of this peptide treatment was associated with the transient activation of anti-p277 splenic TH2 cells (39). A similar observation was also made by Chaturvedi et al. (40), when immunizing NOD mice with a different self-peptide I-Aβg754–76. The precise mechanisms for the shift in the self-reactive T cells from TH1 to TH2 after immunization are complex and currently unknown. Nevertheless, a common feature of these models is the induction of protection accompanied by a skewing of the immune response to self-antigen from TH1 to TH2. We thus would propose that the common mechanism may involve the regulatory CD8+ T cells described here, which selectively down-regulate potentially pathogenic self-reactive TH1 cells in vivo. As a consequence of this differential down-regulation, self-reactive TH2 cells are indirectly promoted by the same self-antigen.
Acknowledgments
We thank Gregory Tau, Elena Ruiz-Vazquez, and Helena Kashleva for technical assistance. The research was supported by National Institutes of Health Grants AI39630, AI30675, and AI44927 and National Multiple Sclerosis Society Grant RG2938A.
Abbreviations
- TCR
T cell antigen receptor
- TH
T helper
- TCV
T cell vaccination
- EAE
experimental autoimmune encephalomyelitis
- MBP
myelin basic protein
References
- 1.Mueller D L, Jenkins M K, Schwartz R H. Annu Rev Immunol. 1989;7:445–480. doi: 10.1146/annurev.iy.07.040189.002305. [DOI] [PubMed] [Google Scholar]
- 2.Zheng L, Boehme S A, Critchfield J M, Zuniga-Pflucker J C, Freedman M, Lenardo M J. Adv Exp Med Biol. 1994;365:81–89. doi: 10.1007/978-1-4899-0987-9_9. [DOI] [PubMed] [Google Scholar]
- 3.Sloan-Lancaster J, Allen P M. Annu Rev Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Mosmann T R, Coffman R L. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 5.Coffman R L, Varkila K, Scott P, Chatelain R. Immunol Rev. 1991;123:189–207. doi: 10.1111/j.1600-065x.1991.tb00611.x. [DOI] [PubMed] [Google Scholar]
- 6.Seder R A, Paul W E. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 7.Shevach E M. Annu Rev Immunol. 2000;18:423–449. doi: 10.1146/annurev.immunol.18.1.423. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Chess L. Annu Rev Immunol. 2000;18:185–216. doi: 10.1146/annurev.immunol.18.1.185. [DOI] [PubMed] [Google Scholar]
- 9.Cantor H, Hugenberger J, McVay-Boudreau L, Eardley D D, Kemp J, Shen F W, Gershon R K. J Exp Med. 1978;148:871–877. doi: 10.1084/jem.148.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Zhang S I, Pernis B. Science. 1992;256:1213–1215. doi: 10.1126/science.256.5060.1213. [DOI] [PubMed] [Google Scholar]
- 11.Koh D-R, Fung-Leung W-P, Ho A, Gray D, Acha-Orbea H, Mak T-W. Science. 1992;256:1210–1213. doi: 10.1126/science.256.5060.1210. [DOI] [PubMed] [Google Scholar]
- 12.Gaur A, Ruberti G, Haspel R, Mayer J P, Fathman C G. Science. 1993;259:91–94. doi: 10.1126/science.8418501. [DOI] [PubMed] [Google Scholar]
- 13.Kumar V, Sercarz E E. J Exp Med. 1993;178:909–916. doi: 10.1084/jem.178.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloom B R, Salgame P, Diamond B. Immunol Today. 1992;13:131–136. doi: 10.1016/0167-5699(92)90110-S. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Ware R, Stall A, Flaherty L, Chess L, Pernis B. Immunity. 1995;2:185–194. doi: 10.1016/s1074-7613(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Kashleva H, Xu L X, Forman J, Flaherty L, Pernis B, Braunstein N S, Chess L. Proc Natl Acad Sci USA. 1998;95:4533–4537. doi: 10.1073/pnas.95.8.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanton T H, Carbon S. Immunogenetics. 1982;16:435–444. doi: 10.1007/BF00372102. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Nun A, Wekerle H, Cohen I R. Nature (London) 1981;292:60–61. doi: 10.1038/292060a0. [DOI] [PubMed] [Google Scholar]
- 19.Cohen I R, Weiner H L. Immunol Today. 1988;9:332–335. doi: 10.1016/0167-5699(88)91330-8. [DOI] [PubMed] [Google Scholar]
- 20.Sanderson S, Shastri N. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 21.Auffray C, Rougeon F. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 22.Aldrich C J, DeCloux A, Woods A S, Cotter R J, Soloski M J, Forman J. Cell. 1994;79:649–659. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 23.Imani F, Soloski M J. Proc Natl Acad Sci USA. 1991;88:10475–10479. doi: 10.1073/pnas.88.23.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun T, Aldrich C J, Baldeon M E, Kawczynski L V, Soloski M J, Gaskins H R. Immunology. 1998;94:64–71. doi: 10.1046/j.1365-2567.1998.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowen L C, Aldrich C J, Forman J. J Immunol. 1993;151:6155–6165. [PubMed] [Google Scholar]
- 26.Kurepa Z, Hasemann C A, Forman J. J Exp Med. 1998;188:973–978. doi: 10.1084/jem.188.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo W F, Ong H, Metcalf E S, Soloski M J. J Immunol. 1999;162:5398–5406. [PubMed] [Google Scholar]
- 28.Seaman M S, Perarnau B, Lindahl K F, Lemonnier F A, Forman J. J Immunol. 1999;162:5429–5436. [PubMed] [Google Scholar]
- 29.Kuchroo V K, Das M P, Brown J A, Ranger A M, Zamvil S S, Sobel R A, Weiner H L, Nabavi N, Glimcher L H. Cell. 1995;80:707–718. doi: 10.1016/0092-8674(95)90349-6. [DOI] [PubMed] [Google Scholar]
- 30.Constant S L, Bottomly K. Annu Rev Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- 31.Ragers P R, Croft M. J Immunol. 1999;163:1205–1213. [PubMed] [Google Scholar]
- 32.Yoshimoto T, Bendelac A, Watson C, Hu-Li J, Paul W E. Science. 1995;270:1845–1847. doi: 10.1126/science.270.5243.1845. [DOI] [PubMed] [Google Scholar]
- 33.Ferrick D A, Schrenzel M D, Mulvania T, Hsieh B, Ferlin W G, Lepper H. Nature (London) 1995;373:255–257. doi: 10.1038/373255a0. [DOI] [PubMed] [Google Scholar]
- 34.Coffman R L, Mosmann T R. Res Immunol. 1991;142:7–9. doi: 10.1016/0923-2494(91)90002-z. [DOI] [PubMed] [Google Scholar]
- 35.Huang H, Paul W E. J Exp Med. 1998;187:1305–1313. doi: 10.1084/jem.187.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germann T, Gately M K, Schoenhaut D S, Lohoff M, Mattner F, Fischer S, Jin S C, Schmitt E, Rude E. Eur J Immunol. 1993;23:1762–1770. doi: 10.1002/eji.1830230805. [DOI] [PubMed] [Google Scholar]
- 37.Waisman A, Ruiz P J, Hirschberg D L, Gelman A, Oksenberg J R, Brocke S, Mor F, Cohen I R, Steinman L. Nat Med. 1996;2:899–905. doi: 10.1038/nm0896-899. [DOI] [PubMed] [Google Scholar]
- 38.Elias D, Tikochinski Y, Frankel G, Cohen I R. Int Immunol. 1999;11:957–966. doi: 10.1093/intimm/11.6.957. [DOI] [PubMed] [Google Scholar]
- 39.Elias D, Meilin A, Ablamunits V, Birk O S, Carmi P, Konen W, Cohen I R. Diabetes. 1997;46:758–764. doi: 10.2337/diab.46.5.758. [DOI] [PubMed] [Google Scholar]
- 40.Chaturvedi P, Agrawal B, Zechel M, Lee-Chan E, Singh B. J Immunol. 2000;164:6610–6620. doi: 10.4049/jimmunol.164.12.6610. [DOI] [PubMed] [Google Scholar]