Abstract
Many cancers and leukemias are associated with strong dominant oncogenic mutations that activate tyrosine kinases and other classes of molecules, including transcription factors and antiapoptotic mechanisms. Some of these events can be targeted with small molecules or antibody-based therapeutics, but many remain intractable. In addition, cancer-related enzyme targets can often mutate, and drug-resistant variants are selected. Therapies directed at the mRNA encoding dominant oncogenes could provide a more global set of technologies for cancer treatment. To test this concept, we have used the model of transformation of hematopoietic cells by the chimeric Bcr-Abl oncogene, a highly activated tyrosine kinase. Our results show that tandem arrays of miRNA mimics, but not single miRNA mimics, directed against the Abl portion of the mRNA and introduced by lentiviral vectors can effectively alter the leukemogenic potency when the degree of suppression of expression of Bcr-Abl is reduced >200-fold from control levels. Only methods capable of such dramatic sustained reduction in the level of expression of highly activated kinase oncogenes are likely to be effective in controlling malignant cell populations.
Keywords: oncogenesis, gene regulation, lentivirus
Many molecular events responsible for the initiation and progression of human leukemias have been defined over the last 30 years (1). Chromosome translocations, point mutations, and other mechanisms that activate cellular genes to become dominant active oncogenes are key targets for the development of new therapeutics (2–4). The recent success of imatinib for the inhibition of the Abl, Kit, and PDGF tyrosine kinases in a variety of blood and solid cancers (3, 5–7) has heralded a new era in targeted small-molecule therapeutics.
Human chronic myelogenous leukemia (CML) and acute lymphocytic leukemia (ALL) caused by the highly active tyrosine kinase of the Bcr-Abl oncogene (the chimeric translocation product of the Philadelphia chromosome) (8) are responsive to imatinib (4, 9) and related drugs (10–15). Daily treatment can lead to long remissions with suppression of the leukemic cell population in the blood and bone marrow. Most patients with Bcr-Abl-positive ALL will progress on treatment, but many patients with CML have survived for long periods with chronic suppression of their disease on imatinib therapy. In some studies, drug therapy has been halted without recurrence for a subset of CML patients (4). However, outgrowth of resistant populations of leukemic cells associated with either mutations in Bcr-Abl that render the kinase insensitive to imatinib and other mechanisms is a common problem. Also, nonproliferating Bcr-Abl+ progenitor cells are insensitive to imatinib treatment (16–18), and infrequently cardiotoxicity has been observed in some patients (19). New Abl tyrosine kinase inhibitors, such as dasatinib (10–14), can inhibit many but not all of the imatinib-resistant forms of Bcr-Abl.
Dramatic advances in understanding the control of mRNA production, stability, and translation by natural small RNAs have come from the study of worms, flies, and mammalian cells (20–25). These findings have led to the development of systematic approaches for the experimental regulation of gene expression using short hairpin RNA (shRNA) or microRNA (miRNA mimics) delivered by a variety of strategies into cultured cells, whole organisms, and even human patients for some disease indications (26, 27).
Several groups have demonstrated that shRNA directed to the Bcr-Abl junction sequences could reduce protein expression in cultured cell lines; but the degree of suppression, measured by Western blotting and protein kinase levels, was not tested for effects on leukemogenesis in vivo (28–32). One group demonstrated that chronic expression of a shRNA directed to the mRNA junction of the related Tel-PDGF chimeric tyrosine kinase oncogene could suppress in vivo leukemogenicity of targeted cell preparations for several weeks, but eventually all test animals died (33).
Our preliminary evaluation of shRNA directed to the Bcr-Abl junction to suppress in vivo leukemogenic activity showed that significant levels of Bcr-Abl kinase activity were still present and led to only modest delay in death from leukemia (data below and J.M. and D.C., unpublished observations). Some improvement in gene suppression was observed when combinations of small interfering RNA (siRNA) directed against Abl sequences were transfected into the K562 cell line in vitro (34).
As a general test of using small RNAs to regulate oncogene expression, we have used highly selected miRNA mimics directed to several sites within the Abl-coding sequences to suppress the expression of the Bcr-Abl oncoprotein. Individual anti-Abl miRNAs and double and triple combinations introduced by lentiviral vectors were evaluated for their ability to suppress Bcr-Abl expression and downstream substrates and pathways used by this kinase in an aggressive pre-B leukemia model. Our results show that introduction of a triple combination of miRNA mimics from a single lentiviral vector was sufficient to suppress oncogene expression and kinase pathway activation to a low enough level to prevent regrowth of leukemic cells both in vitro and in vivo. Such approaches may find eventual clinical application in modifying hematopoietic stem and progenitor cell populations for autologous transplantation in a variety of leukemias.
Results
Screening and Selection of miRNA Mimics to Suppress Bcr-Abl Expression.
Previous work had defined that siRNA directed against the Bcr-Abl junction delivered as oligonucleotides or as shRNA via viral vectors could suppress expression in cultured cell lines sufficient to induce significant apoptosis (30, 32, 35). Little data were available to test whether this degree of oncogene suppression was sufficient to prevent growth of the targeted cell lines as leukemias upon in vivo transfer in rodent models. We evaluated Bcr-Abl junction-specific shRNA delivered by lentiviral vector into susceptible cell lines and observed up to 90% suppression at the protein level in vitro, but very limited effect on leukemogenic potential (J.M. and D.C., unpublished observations and additional data shown below). Because Bcr-Abl is so highly active, we reasoned that even modest amounts of residual kinase activity would be sufficient to drive the malignant growth.
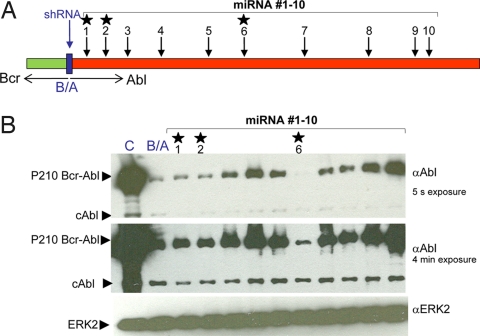
To block the expression of Bcr-Abl more stringently, we turned to miRNA technology (23, 24). We directed our attention to the sequences of the Abl gene because ample evidence from the use of imatinib and related drugs has shown that inhibition of the cellular Abl kinase in adult animals or man has limited deleterious side effects. Using currently available criteria to select sequences likely susceptible to miRNA blockade (see Materials and Methods), we evaluated 10 miRNA constructs by cotransfection with Bcr-Abl onto 293T cells (Fig. 1). For comparison, shRNA against the Bcr-Abl junction was used. The results demonstrate that there is wide variation in efficiency of suppression of p210 Bcr-Abl expression. The shRNA expressed from a polymerase III (pol III) H1 promoter (36, 37) within a lentiviral construct could suppress ≈90% of the Bcr-Abl protein production when comparing different exposure lengths of Western blots detecting the level of p210 Bcr-Abl. The degree of suppression by the 10 miRNA constructs expressed by a CMV promoter within a plasmid expression vector varied widely, but two (marked 1 and 2 in Fig. 1) were equally efficient compared with the junction specific shRNA, and one (marked 6 in Fig. 1) was remarkably efficient, blocking Bcr-Abl protein production by >99%.
Fig. 1.
Efficient suppression of p210 Bcr-Abl expression by selected cAbl-directed miRNA mimics. Numbers 1–10 represent the positions on the cAbl 1b sequence to which the miRNAs are directed. The exact sequences of all miRNAs are in Materials and Methods. B/A, shRNA Bcr-Abl junction. Five micrograms of MSCV-p210-IRES-EYFP was transfected either alone (C, control) or together with 5 μg of LV H1 shRNA Bcr-Abl junction or 5 μg of pcDNA 6.2-GW/EmGFP-miRNA Abl 1–10 onto 293T cells. Forty-eight hours after transfection, cells were lysed in extraction buffer as described in Materials and Methods, and 5 μg of protein was run per lane. The Western blot was probed with either 400 ng/ml anti-Abl or 1:1,000 anti-ERK2 as described in Materials and Methods. The stars indicate three selected miRNAs used in subsequent experiments.
Alternative Activated Forms of the Abl Oncogene Are Effectively Suppressed by Selected miRNA Mimics.
Alternative Bcr-Abl mRNAs and proteins, such as p210 and p185, are produced dependent on the precise location of the chromosomal breakpoint within the Bcr gene (38). In addition, alternative chromosomal partners, like Tel, can participate with Abl in the formation of chimeric oncogenes such as P180 Tel-Abl (39, 40). Recent clinical studies have demonstrated a high rate of selection for many imatinib-resistant forms of Bcr-Abl, such as mutations at Thr-315 (13, 41, 42). To evaluate the generality of the utility of Abl-directed miRNAs, we compared the ability of selected forms to suppress alternative members of the Abl oncogene family (Fig. 2) and demonstrated that each could be effectively suppressed by targeting Abl sequences.
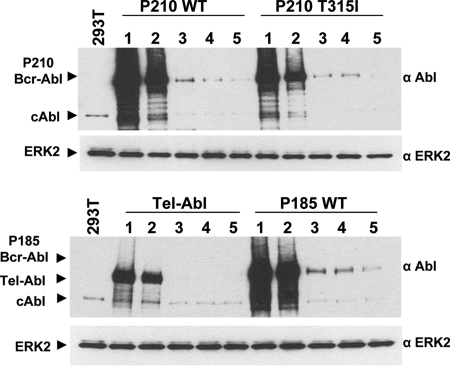
Fig. 2.
Efficient knockdown of multiple chimeric forms of cAbl using single, double, and triple miRNA mimics. Five micrograms of MSCV-IRES-EYFP expressing either p210 Bcr-Abl WT, p210 Bcr-Abl T315I (ref. 41), Tel-Abl (ref. 39), or p185 Bcr-Abl WT (ref. 61) were transfected alone (lane 1) or cotransfected with 5 μg of either miRNA scrambled (lane 2), miRNA Abl single 2 (lane 3), miRNA Abl double 6/2 (lane 4), or miRNA Abl triple 6/2/1 (lane 5) onto 293T cells. All miRNAs were in the pcDNA 6.2-GW/EmGFP vector. Forty-eight hours after transfection, cells were lysed in extraction buffer as described in Materials and Methods, and 5 μg of protein was run per lane. Blots were probed with 400 ng/ml anti-Abl and 1:1,000 anti-ERK2 as described in Materials and Methods.
A specific advantage of miRNAs is their ability to be expressed in tandem arrays within a single vector (26, 43, 44). We constructed a double combination (6/2) and a triple tandem combination (6/2/1). Each of the activated Abl genes was expressed via transfection in the 293T cell line, and a set of miRNAs expressing plasmids including a scrambled sequence control (lane 2), a single (2, in lane 3), double (6/2, in lane 4), and triple (6/2/1, in lane 5) were introduced by transfection. The results show that the most effective suppression resulted from the introduction of the triple combination of miRNAs (Fig. 2, lane 5 in each set).
Effective Blockade of Downstream Tyrosine Kinase Signaling Pathways by Abl miRNA Mimics.
Bcr-Abl and related kinases activate a number of key downstream signals to promote cell growth and block apoptosis (2, 45, 46). To evaluate in detail the effects of miRNA blockade on susceptible cells, we infected a Ba/F3 cell line expressing a luciferase marker (47) with a construct expressing the p210 Bcr-Abl gene and an enhanced yellow fluorescent protein (EYFP) marker (see vectors details in Materials and Methods). This cell line was IL-3-independent and highly leukemogenic (see below). To ensure that all cells in our subsequent analyses contained the specific sequences we desired, we infected the Ba/F3-Fluc-p210 IRES-EYFP cells with lentiviral preparations expressing single, double, or triple miRNA combinations anticipated to give a graded levels of gene suppression, the shRNA, and scrambled controls. Each construct contains an EGFP marker expressed from a CMV promoter. Cells were cultured for at least 2 days in IL-3-containing medium, sorted for EYFP and EGFP double-positive cells, cultured again in IL-3-containing medium, and sorted again to ensure that a high fraction of cells had received each of the desired vectors. Populations prepared for further analysis expressed the EGFP marker and presumably the desired miRNA or combinations (data not shown).
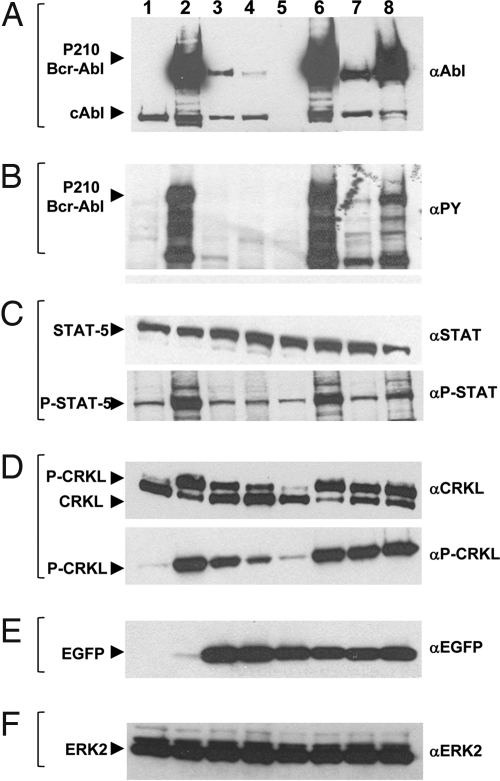
Western blot analysis showed that single, double, or triple miRNA-expressing cells were superior to shRNA for suppression of the Bcr-Abl protein level (Fig. 3A) and total phosphotyrosine-modified proteins (Fig. 3B). Several downstream phosphorylation events have been highly correlated with Bcr-Abl kinase activity and biological effects (2, 45, 46). The levels of phospho-STAT5 (Fig. 3C) and phospho-CRKL were most efficiently suppressed by the triple miRNA combination (Fig. 3D). Fig. 3E demonstrates the production of either EGFP or EYFP from the small RNA-expressing vectors and the Bcr-Abl vector, respectively. Fig. 3F is the level of cellular ERK that serves as a loading control for equal cell numbers analyzed.
Fig. 3.
Increasing knockdown of Bcr-Abl and of downstream targets STAT5 and CRKL in Ba/F3 p210-expressing cells transduced with single, double, and triple miRNA mimics. Ba/F3 Fluc-Neo cells (lane 1) expressing p210 IRES-EYFP (lanes 2–8) and grown in the presence of 10% IL-3-containing medium were infected with lentivirus expressing either miRNA Abl single 2 (lane 3), miRNA Abl double 6/2 (lane 4), miRNA Abl triple 6/2/1 (lane 5), miRNA scrambled (lane 6), shRNA Bcr-Abl junction (lane 7), or shRNA scrambled (lane 8). After sorting, cells were lysed for total protein in 2× sample buffer, and 5 × 104 cell equivalents were run per lane. Western blots were probed with 400 ng/ml anti-Abl (A), 1:1,000 anti-phosphotyrosine (B), 1:1,000 anti-STAT5 or 1:1,000 anti-phospho-STAT5 (C), 1:1,000 anti-CRKL or 1:1,000 anti-phospho-CRKL (D), 200 ng/ml anti-GFP (E), and 1:1,000 anti-ERK2 (F). Antibodies are described in Materials and Methods.
These effects were not caused by differences in the average number of small RNA-expressing genomes introduced into each of the cell populations. Analysis by Southern blotting to measure the number of integrated lentiviral genomes in the cell populations showed that each contained a very similar level of integrated viral templates [supporting information (SI) Fig. 6]. Interestingly, analysis of the level of mRNA stably expressed from the Bcr-Abl vector showed that single, double, or triple miRNAs lowered mRNA production with the same gradient of efficiency (SI Fig. 7) but left a surprising fraction (5–10% for the triple miRNA) intact. Given the more dramatic suppression of Bcr-Abl at the protein level (>100-fold; see Figs. 1–3), this finding suggests that both mRNA destruction as well as inhibition of translation may play a role in this process.
More Efficient Suppression of Bcr-Abl in Ba/F3 Cells Leads to Enhanced Apoptosis and Failure to Thrive in Vitro.
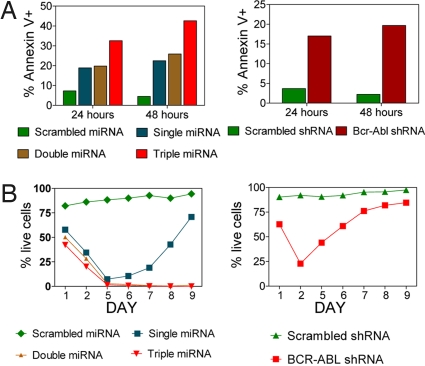
Each of the sorted populations of Bcr-Abl Ba/F3 cells described in Fig. 3 above were analyzed for survival when removed from IL-3-containing medium (Fig. 4). As expected, the scrambled shRNA and scrambled miRNA controls that continue to express high levels of Bcr-Abl showed high cell viability as determined by lack of annexin staining assayed at both 24 and 48 h after IL-3 removal (Fig. 4A). Cells expressing the shRNA directed to the Bcr-Abl junction showed a transient drop of viable cells down to ≈25% over 2 days but then quickly recovered to >80% viable cells at the end of 9 days (Fig. 4B). With the single miRNA mimic construct used, the extent of apoptotic cells was similar (Fig. 4A), and they also recovered and produced a viable and growing cell population at the end of 9 days (Fig. 4B). There was no change in the level of p210 expression in this recovered population. With the double and triple combinations tested, the extent and duration of the apoptotic effect were much greater (Fig. 4A). No viable cells were detected after 9 days (Fig. 4B).
Fig. 4.
Reduced cell viability and enhanced apoptosis induced by Bcr-Abl silencing and IL-3 removal in Ba/F3 cells expressing p210-IRES-EYFP. Ba/F3 Fluc-Neo cells expressing 210-IRES-EYFP were infected via lentivirus with miRNA-EGFP (Left) or shRNA-EGFP (Right) and sorted twice for EGFP/EYFP high signal (Materials and Methods). Cells (1 × 105) were analyzed by flow cytometry for apoptosis (A) and cell viability (B) 24 or 48 h after IL-3 removal (washed three times) and up to 9 days in the absence of IL-3 (Materials and Methods). Viable cells are quantified as annexin V- and 7AAD-double negative.
Degree of Bcr-Abl Suppression Determines Survival from Leukemia in Vivo.
Previous work has demonstrated that Bcr-Abl-expressing Ba/F3 cells are highly malignant and produce a disseminated leukemia with massive splenic, bone marrow, and nodal infiltration (47) within several weeks from an innoculum of 106 cells. We compared the same carefully selected cell populations described in Figs. 3 and 4 for their ability to transfer leukemia in vivo. Because our cells expressed the human form of Bcr-Abl and additional foreign proteins (EYFP, EGFP, and F-Luc), we used CB17.scid/scid mice as recipients to prevent any T or B cell-mediated immune rejection. To follow disease progression, we monitored the animals by CCD camera detection of luciferase activity after injection of d-luciferin and noted the time of death when the animals became moribund. The results (Fig. 5) were striking. Animals receiving Ba/F3 Bcr-Abl cells with either scrambled shRNA or scrambled miRNA controls succumbed within 14–25 days. The group receiving cells modified with the shRNA to the Bcr-Abl junction survived between 45 and 60 days. Animals in the group suppressed with a single miRNA mimic survived to between 50 and 60 days, and those with two miRNAs from 70 to 110 days. All animals tested that received the cells modified with the triple miRNA combination were alive and well at 120 days with no detectable luciferase activity. Western blot analysis from lysates of the splenocytes from the triple miRNA animals at 120 days did not show any detectable Bcr-Abl protein expression (data not shown).
Fig. 5.
A triple miRNA mimic-encoding lentivirus effectively suppresses oncogenic activity of Bcr-Abl in vivo, whereas the shRNA against the junction region is less potent. CB17.scid/scid mice were injected with 1 × 106 Ba/F3-Fluc-p210-IRES-EYFP cells transduced with control shRNA (scrambled-2 mice) and shRNA targeted against the junction region (miRNA 4 mice) (A) or various miRNAs (scrambled-8 mice; single miRNA-8 mice; double miRNA-8 mice; and triple miRNA-10 mice) (B). Kaplan–Meier survival curves were plotted. Mice were imaged at the indicated time points by using a Xenogen IVIS 100 system to record bioluminescent images of luciferase activity indicating the presence of Ba/F3-Luc-p210-IRES-EYFP cells. A false color scale to demonstrate light intensity is shown. The scale is between 2.472e ± 0.06 and 2.472e ± 0.07 photons per s per cm2 per steridian.
Direct comparison with the levels of Bcr-Abl kinase inhibition seen in clinical tests with imatinib or dasatinib may not be directly comparable with the level of suppression described above. Our retroviral vector expresses Bcr-Abl at a much higher level per cell, and more stringent suppression was needed to block leukemic growth.
Discussion
The potential to inhibit specific mRNAs essential for a wide range of disease states holds much promise for new therapeutics. Clinical efficacy depends on the ability to deliver an effective dose to the right cell type in a time frame consistent with slowing or reversing the specific disease process (26, 27). The degree of gene suppression required for a meaningful clinical effect will vary with the specific functional role of the mRNA targeted and the nature of the disease process.
Our work demonstrates that selected combinations of miRNA mimics delivered by viral vectors that are directed against the Abl portion of the Bcr-Abl oncogene can effectively suppress gene function and have a dramatic therapeutic effect, if a very high percentage of the malignant cells can be forced to express the desired therapeutic miRNAs in a sustained manner. The suppressive function of this triple miRNA combination appears equally effective on all forms of activated Abl, including those associated with imatinib and dasatinib resistance mutations. The marrow, blood, and peripheral tissue involvement of Bcr-Abl-positive CML and ALL would require systemic administration of such viral vectors or alternative strategies such as targeted liposomes or nanoparticles (48). Recent advances in specific targeting of lentiviral particles by engineering of surface receptor specificity could be useful in such attempts (49). However, the extremely high numbers of leukemic stem or progenitor cells in any patient with chronic, accelerating, or blast crisis CML would be prohibitive unless the pool of cells to target could be dramatically reduced by prior treatment with agents such as imatinib, conventional cytotoxic chemotherapy, or biological response modifiers like IFN-α (50). Even with cytoreductive conditioning, the logistical issues for in vivo delivery of the therapeutic miRNAs by viral vector or other technologies remain substantial (51).
A more realistic test of the potential efficacy of such targeted therapies may come from the setting of bone marrow or hematopoietic stem cell transplantation in combination with myeloablative chemotherapy and/or irradiation for patients who progress on imatinib or other targeted therapies because of drug-resistant mutations in Bcr-Abl. Relatively few patients will have an HLA-matched donor available for allogeneic transplantation. An autologous transplantation scheme in which the patient's endogenous stem and progenitor elements, isolated by cell enrichment procedures from their bone marrow or peripheral blood, are genetically modified to chronically suppress the action of the Bcr-Abl oncogene may have value (52, 53). In this strategy, many fewer cells would need to be infected, repeat inoculums could be stored for later use, and the fraction of cells modified with the therapeutic vector could be quantitatively assessed. Such strategies may eventually find utility for the many forms of leukemia where no small-molecule-targeted chemotherapy is available but a clear target for such mRNA based therapies is known.
Materials and Methods
Generation of Retrovirus Stock and Bcr-Abl Cell Lines.
The retroviral vector MSCV-IRES-EGFP, where EGFP was replaced by EYFP, was used to generate retrovirus (54) expressing Bcr-Abl p210 wild type (WT), which was used to infect the cell line Ba/F3 Fluc-Neo (47) as described in ref. 55. Forty-eight hours after infection, cells were sorted for the EYFP+ population on the FACS Vantage (Becton Dickinson). Cells were subsequently allowed to become growth factor-independent by removal of IL-3-containing medium from the cultures.
Preparation of Lentiviral Vector FCW attR1-attR2.
The ubiquitin promoter and EGFP were removed from the FUGW lentiviral vector (56) and replaced with a linker containing the sites NheI, XbaI, HpaI, and PacI to create FlinkW. A CMV promoter was inserted into the XbaI site to create FCW. The attR1-attR2 cassette from the Gateway pLenti6/V5-DEST vector from Invitrogen was removed by EcoRV and cloned as a blunt end ligation into the FCW vector digested with HpaI. Positive clones were selected by using ampicillin and chloramphenicol.
Preparation of miRNA Mimics.
Using the BLOCK- iT pol II miR RNAi expression vector kit from Invitrogen and the online site www.invitogen.com/rnai, we designed 10 miRNA mimics targeted against the human cAbl 1b gene that were 21 nt in length. Numbering and sequence of miRNAs are as follows: 1, 137CTCGTTGGAACTCCAAGGAAA157; 2, 226GGAGATAACACTCTAAGCATA246; 3, 339CAACTACATCACGCCAGTCAA359; 4, 501CGAAGGGAGGGTGTACCATTA521; 5, 1047AGCCATGGAGTACCTGGAGAA1067; 6, 1226AGAGCCTGGCCTACAACAAGT1246; 7, 1705GTCTCCATTGCTCCCTCGAAA1725; 8, 2236CAGTTTGACTCGTCCACATTT2256; 9, 2777CAGTCCTGGGCGCAAAGACAA2797; 10, 2805GAGTCTGGTTGATGCTGTGAA2825. The numbers in superscript are the first and last nucleotides of the cDNA targeted sequence. These 10 miRNAs were cloned into the pcDNA 6.2-GW/EmGFP-miR vector in which the miRNA sequence is inserted into the 3′-UTR of the EmGFP gene. Single, double, and triple miRNAs were subsequently transferred to the vector pDONR 221 from Invitogen by using BP Clonase II enzyme. The miRNA cassette was transferred from the intermediate vector into the lentiviral vector FCW attR1-attR2 by using LR Clonase II to create the final lentiviral vector FC EmGFP W miRNA Abl 1–10. Additionally, we used a sequence that does not target any known gene: miRNA scrambled GTCTCCACGCGCAGTACATTT.
shRNAs Design.
Design of H1-driven shRNA cassettes was done according to ref. 57. Briefly, shRNA constructs were generated by synthesizing a long oligonucleotide that contains the entire hairpin cassette (including a pol III termination signal), 20 nt complementary to the 3′ end of the pol III H1 promoter, and a 5′ end containing a unique XbaI restriction site. Briefly, each long oligonucleotide was used in a PCR with T3 oligonucleotide (5′-CTCGAAATTAACCCTCACTAAAGGG) and a template (pH1-siGFP) to generate a fragment that contains the entire H1 promoter plus shRNA sequences; the resulting product was cut with XbaI and ligated to a NheI-digested lentivirus vector (37). PCR conditions are: initial cycle of 94°C for 3 min followed by 30 cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 50 s. We recommend using a hydrogen bond-reducing agent such as DMSO in the reaction. shRNA oligonucleotides that were used are as follows (bold indicates sense and antisense strands): siBCR-ABL, 5′-CTGTCTAGACAAAAA GCAGAGTTCAAAAGCCCTTCA TCTCTTGAA TGAAGGGCTTTTGAACTCTGC GGGGATCTGTGGTCTCATACA; siControl, 5′-CTGTCTAGACAAAAA CTTCATTGTCGGCATGGGT TCTCTTGAA ACCCATGCCGACAATGAAG GGGGATCTGTGGTCTCATACA.
The siBCR-ABL sequence was designed to target the junction between BCR and ABL, GCAGAGTTCAA-AAGCCCTTCA. siControl is a nonfunctional siRNA designed to target mouse GLUT4 (57).
Preparation of Lentivirus and Infection of Cells.
The lentivirus containing the miRNAs and shRNAs was prepared as described in ref. 58 with some modifications. Briefly, the lentiviral vector was cotransfected with the vectors pRSV-Rev, pCMV-VSV-G, and pMDL by calcium phosphate precipitation onto 293T cells. Virus was collected at 48 and 72 h, concentrated by ultracentrifugation, and titered onto 293T cells (59). The Ba/F3 Fluc-Neo p210 cells were infected at a multiplicity of infection of 20 with the lentivirus by spinduction in the presence of 10% WEHI-conditioned medium and 4 μg/ml Polybrene and maintained in WEHI unless otherwise indicated. The lentivirus-infected cells were sorted twice for EYFP/EGFP+ cells to ensure close to 100% purity on the FACS Vantage.
Annexin/Live Cell.
Cells were washed three times to remove WEHI from the medium, and 4 × 105 cells were plated per sample. Cell viability and Annexin V staining were performed as described in ref. 54 with minimum modification. Briefly, 510 nm of EGFP, 550 nm of EYFP, 7-amino-actinomycin D (7AAD) (PharMingen), and Annexin V-Cy5 (PharMingen) were measured on a FACSCanto flow cytometry (Becton Dickinson). Live cell percentage is quantified as Annexin V 7AAD-double negative, and apoptotic cell percentage is Annexin V-positive.
In Vivo Experiments and Animal Imaging.
Ba/F3 Fluc-Neo cells (1 × 106) expressing BCR-ABL p210 and different miRNAs and shRNAs were injected into the tail vein of CB17.scid/scid mice. Starting from day 14 after injection, mice were imaged by using a bioluminescent optical camera (IVIS system; Xenogen). Mice were anesthetized with 2–4% isoflurane, and each was injected i.p. with 200 μl of 15 mg/ml d-luciferin in PBS. Exposures of 20 s were collected repeatedly after d-luciferase injection until activity started to drop off. Images were analyzed with Living Image (Igor Pro; WaveMatrics, Inc.). At days 124 (experiment I) and 126 (experiment II) mice were euthanized after final imaging. Spleens were weighed and prepared to single cells free of RBCs (ammonium chloride lysis) for GFP/YFP expression by flow cytometry and Western blotting. All animal experiments followed the guidelines of the UCLA Animal Research Committee.
Protein Analyses by Western Blotting.
Cell lysates were prepared by using extraction buffer as described in ref. 60 or by gel sample buffer. Five micrograms of protein or 5 × 104 cell equivalents were run and analyzed by immunoblotting as described in ref. 54 with the following antibodies: anti-Abl (55); anti-phosphotyrosine, clone 4G10 (Upstate Biotechnology); phospho-STAT5 (Tyr-694), STAT5, phospho-CRKL (Tyr-207) (Cell Signaling Technology); CRKL C-20 (sc319), ERK2 (sc154) (Santa Cruz Biotechnology); anti-EGFP prepared in our laboratory. Briefly, EGFP was expressed with a His tag and purified over nickel-nitrilotriacetic acid–agarose. Rabbits were immunized with purified protein (Covance), and anti-GFP antibodies were affinity-purified over a GFP–Sepharose column.
Southern Blotting.
Total DNA was isolated by using the DNeasy blood and tissue kit (Qiagen). Ten micrograms of DNA was digested with BamHI, run on a 0.8% agarose gel, and processed as described in ref. 61. The blot was probed with [α-32]ATP-labeled GFP.
Northern Blotting.
Total RNA was isolated by using TRIzol reagent (Invitrogen). Twenty micrograms of RNA was fractionated on a 1% agarose–formaldehyde gel and processed as described in ref. 61. The blot was probed with either a [α-32]ATP-labeled fragment of Abl cDNA or actin.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Shirley Quan and LaKeisha Perkins for technical assistance, Stephanie Shelly and Jenny Shu for help with animals, Mireille Riedinger (University of California, Los Angeles) for making the anti-GFP antibody, Neil Shah (University of California, San Francisco) for the Ba/F3 Fluc-Neo cell line, and Barbara Anderson for help with manuscript preparation. This work was supported in part by the Leukemia and Lymphoma Society of America and a National Institutes of Health grant (to I.M.V.). O.S. is a California Institute for Regenerative Medicine Postdoctoral Fellow. I.M.V. is an American Cancer Society Professor of Molecular Biology. O.N.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710532105/DC1.
References
- 1.Warner JK, Wang JC, Hope KJ, Jin L, Dick JE. Oncogene. 2004;23:7164–7177. doi: 10.1038/sj.onc.1207933. [DOI] [PubMed] [Google Scholar]
- 2.Wong S, Witte ON. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 3.Krause DS, Van Etten RA. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 4.Deininger M, Buchdunger E, Druker BJ. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 5.Hunter T. J Clin Invest. 2007;117:2036–2043. doi: 10.1172/JCI31691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchdunger E, Cioffi CL, Law N, Stover D, Ohno-Jones S, Druker BJ, Lydon NB. J Pharmacol Exp Ther. 2000;295:139–145. [PubMed] [Google Scholar]
- 7.Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, et al. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 8.Sawyers CL. N Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, et al. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 10.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, et al. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 11.Burgess MR, Sawyers CL. Sci World J. 2006;6:918–930. doi: 10.1100/tsw.2006.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giles FJ, Cortes J, Jones D, Bergstrom D, Kantarjian H, Freedman SJ. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 13.Azam M, Nardi V, Shakespeare WC, Metcalf CA, III, Bohacek RS, Wang Y, Sundaramoorthi R, Sliz P, Veach DR, Bornmann WG, et al. Proc Natl Acad Sci USA. 2006;103:9244–9249. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, Milanov ZV, Atteridge CE, Biggs WH, III, Edeen PT, et al. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Hare T, Pollock R, Stoffregen EP, Keats JA, Abdullah OM, Moseson EM, Rivera VM, Tang H, Metcalf CA, III, Bohacek RS, et al. Blood. 2004;104:2532–2539. doi: 10.1182/blood-2004-05-1851. [DOI] [PubMed] [Google Scholar]
- 16.Holtz M, Forman SJ, Bhatia R. Cancer Res. 2007;67:1113–1120. doi: 10.1158/0008-5472.CAN-06-2014. [DOI] [PubMed] [Google Scholar]
- 17.Holtz MS, Forman SJ, Bhatia R. Leukemia. 2005;19:1034–1041. doi: 10.1038/sj.leu.2403724. [DOI] [PubMed] [Google Scholar]
- 18.Graham SM, Jorgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L, Holyoake TL. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 19.Kerkela R, Grazette L, Yacobi R, Iliescu C, Patten R, Beahm C, Walters B, Shevtsov S, Pesant S, Clubb FJ, et al. Nat Med. 2006;12:908–916. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- 20.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 22.Mello CC, Conte D., Jr Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 24.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 25.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 26.Kim DH, Rossi JJ. Nat Rev. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 27.Aagaard L, Rossi JJ. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MJ, McMahon R, Snyder DS, Yee JK, Rossi JJ. Oligonucleotides. 2003;13:401–409. doi: 10.1089/154545703322617087. [DOI] [PubMed] [Google Scholar]
- 29.Scherr M, Battmer K, Winkler T, Heidenreich O, Ganser A, Eder M. Blood. 2003;101:1566–1569. doi: 10.1182/blood-2002-06-1685. [DOI] [PubMed] [Google Scholar]
- 30.Rangatia J, Bonnet D. Leukemia. 2006;20:68–76. doi: 10.1038/sj.leu.2403999. [DOI] [PubMed] [Google Scholar]
- 31.Scherr M, Battmer K, Schultheis B, Ganser A, Eder M. Gene Ther. 2005;12:12–21. doi: 10.1038/sj.gt.3302328. [DOI] [PubMed] [Google Scholar]
- 32.Thomas M, Greil J, Heidenreich O. Acta Pharmacol Sin. 2006;27:273–281. doi: 10.1111/j.1745-7254.2006.00282.x. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Wall NR, Kocher K, Duclos N, Fabbro D, Neuberg D, Griffin JD, Shi Y, Gilliland DG. J Clin Invest. 2004;113:1784–1791. doi: 10.1172/JCI20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohba H, Zhelev Z, Bakalova R, Ewis A, Omori T, Ishikawa M, Shinohara Y, Baba Y. Cancer. 2004;101:1390–1403. doi: 10.1002/cncr.20468. [DOI] [PubMed] [Google Scholar]
- 35.Wilda M, Fuchs U, Wossmann W, Borkhardt A. Oncogene. 2002;21:5716–5724. doi: 10.1038/sj.onc.1205653. [DOI] [PubMed] [Google Scholar]
- 36.Tiscornia G, Singer O, Verma IM. Nat Protocols. 2006;1:234–240. doi: 10.1038/nprot.2006.36. [DOI] [PubMed] [Google Scholar]
- 37.Tiscornia G, Singer O, Ikawa M, Verma IM. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong S, Witte ON. Oncogene. 2001;20:5644–5659. doi: 10.1038/sj.onc.1204638. [DOI] [PubMed] [Google Scholar]
- 39.Golub TR, Goga A, Barker GF, Afar DE, McLaughlin J, Bohlander SK, Rowley JD, Witte ON, Gilliland DG. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Million RP, Aster J, Gilliland DG, Van Etten RA. Blood. 2002;99:4568–4577. doi: 10.1182/blood-2001-12-0244. [DOI] [PubMed] [Google Scholar]
- 41.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 42.Corbin AS, La Rosee P, Stoffregen EP, Druker BJ, Deininger MW. Blood. 2003;101:4611–4614. doi: 10.1182/blood-2002-12-3659. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. Nat Rev Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- 45.Ren R. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 46.Deininger MW, Goldman JM, Melo JV. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 47.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 48.Heidel JD, Yu Z, Liu JY-C, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Proc Natl Acad Sci USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, Bailey L, Baltimore D, Wang P. Proc Natl Acad Sci USA. 2006;103:11479–11484. doi: 10.1073/pnas.0604993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MWN, Silver RT, Goldman JM, Stone RM, et al. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 51.Koldehoff M, Steckel NK, Beelen DW, Elmaagacli AH. Clin Exp Med. 2007;7:47–55. doi: 10.1007/s10238-007-0125-z. [DOI] [PubMed] [Google Scholar]
- 52.An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao SH, Bonifacino A, Krouse AE, Darlix JL, Baltimore D, et al. Proc Natl Acad Sci USA. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amado RG, Mitsuyasu RT, Rosenblatt JD, Ngok FK, Bakker A, Cole S, Chorn N, Lin LS, Bristol G, Boyd MP, et al. Hum Gene Ther. 2004;15:251–262. doi: 10.1089/104303404322886101. [DOI] [PubMed] [Google Scholar]
- 54.Wong S, McLaughlin J, Cheng D, Zhang C, Shokat KM, Witte ON. Proc Natl Acad Sci USA. 2004;101:17456–17461. doi: 10.1073/pnas.0407061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong S, McLaughlin J, Cheng D, Witte ON. Blood. 2003;101:4088–4097. doi: 10.1182/blood-2002-11-3376. [DOI] [PubMed] [Google Scholar]
- 56.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 57.Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Nat Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 58.Xin L, Ide H, Kim Y, Dubey P, Witte ON. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiscornia G, Singer O, Verma IM. Nat Protocols. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 60.Lim YM, Wong S, Lau G, Witte ON, Colicelli J. Proc Natl Acad Sci USA. 2000;97:12233–12238. doi: 10.1073/pnas.210253497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McLaughlin J, Chianese E, Witte ON. Mol Cell Biol. 1989;9:1866–1874. doi: 10.1128/mcb.9.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.