Abstract
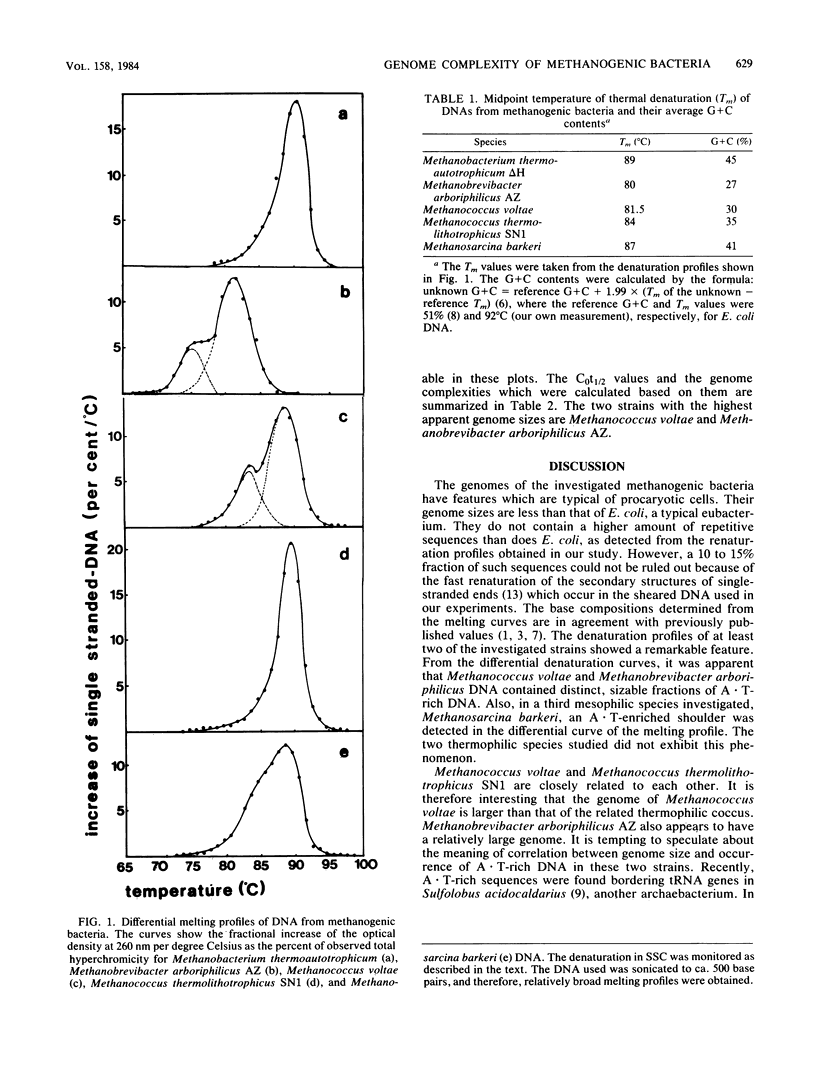
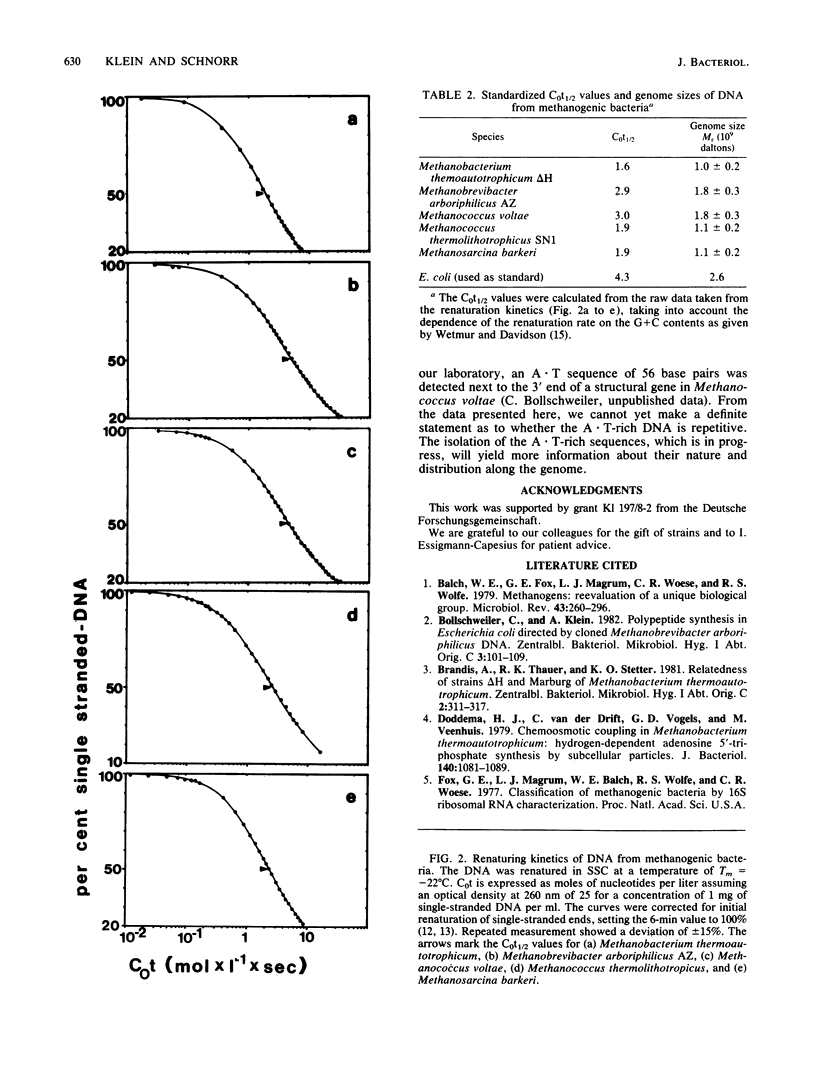
The genome complexities of different methanogenic bacteria were investigated by using an optical method to study renaturation kinetics of single-stranded DNA. The observed genome sizes ranged from 1.0 X 10(9) to 1.8 X 10(9) daltons, which is a typical range for procaryotic cells. Melting profiles of the DNA of three methanogenic species from different families show fractions which have a higher A . T content than the average DNA of that species.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H. J., van der Drift C., Vogels G. D., Veenhuis M. Chemiosmotic coupling in Methanobacterium thermoautotrophicum: hydrogen-dependent adenosine 5'-triphosphate synthesis by subcellular particles. J Bacteriol. 1979 Dec;140(3):1081–1089. doi: 10.1128/jb.140.3.1081-1089.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: rate of hybridization of excess RNA with DNA, compared to the rate of DNA renaturation. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1020–1023. doi: 10.1073/pnas.74.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine B. P., Gupta R., Woese C. R. Putative introns in tRNA genes of prokaryotes. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3309–3312. doi: 10.1073/pnas.80.11.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O. Archaebakterien und Phylogenie der Organismen. Naturwissenschaften. 1981 Apr;68(4):183–192. doi: 10.1007/BF01047198. [DOI] [PubMed] [Google Scholar]
- Mitchell R. M., Loeblich L. A., Klotz L. C., Loeblich A. R., 3rd DNA organization of Methanobacterium thermoautotrophicum. Science. 1979 Jun 8;204(4397):1082–1084. doi: 10.1126/science.377486. [DOI] [PubMed] [Google Scholar]
- Rau D. C., Klotz L. C. A unified theory of nucleation-rate-limited DNA renaturation kinetics. Biophys Chem. 1978 Mar;8(1):41–51. doi: 10.1016/0301-4622(78)85021-2. [DOI] [PubMed] [Google Scholar]
- Vosberg H. P., Hoffmann-Berling H. DNA synthesis in nucleotide-permeable Escherichia coli cells. I. Preparation and properties of ether-treated cells. J Mol Biol. 1971 Jun 28;58(3):739–753. doi: 10.1016/0022-2836(71)90037-4. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]


