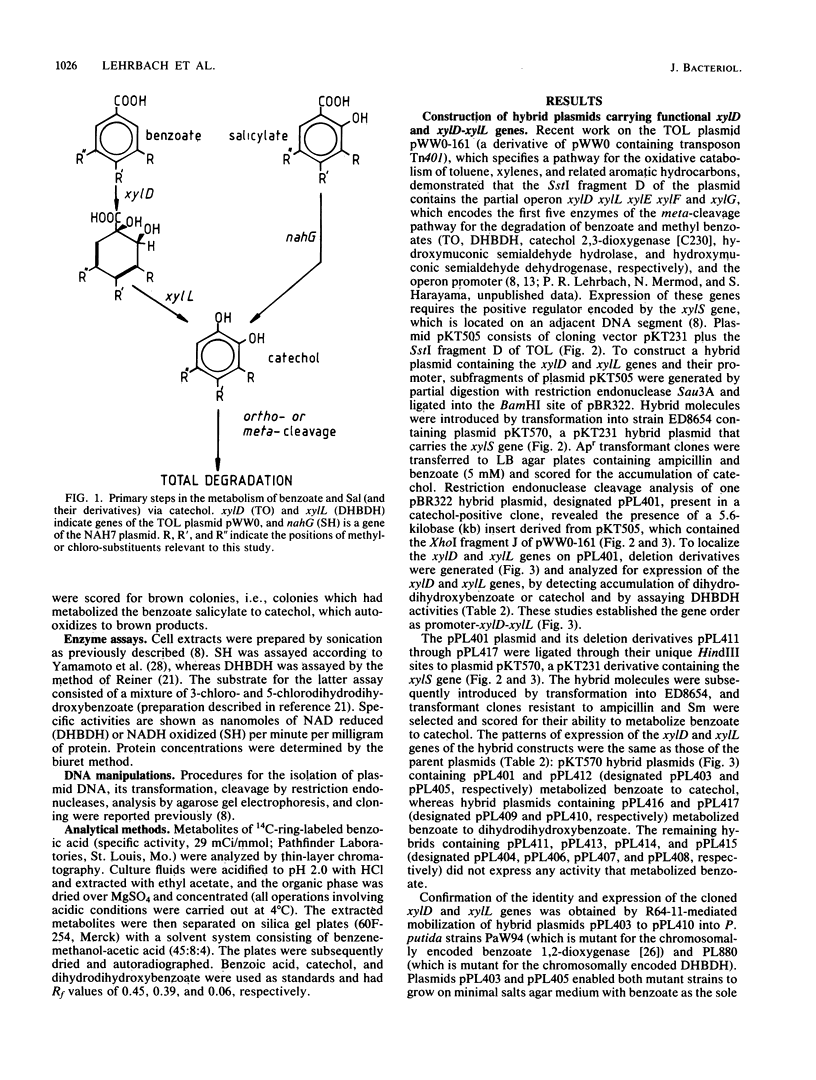
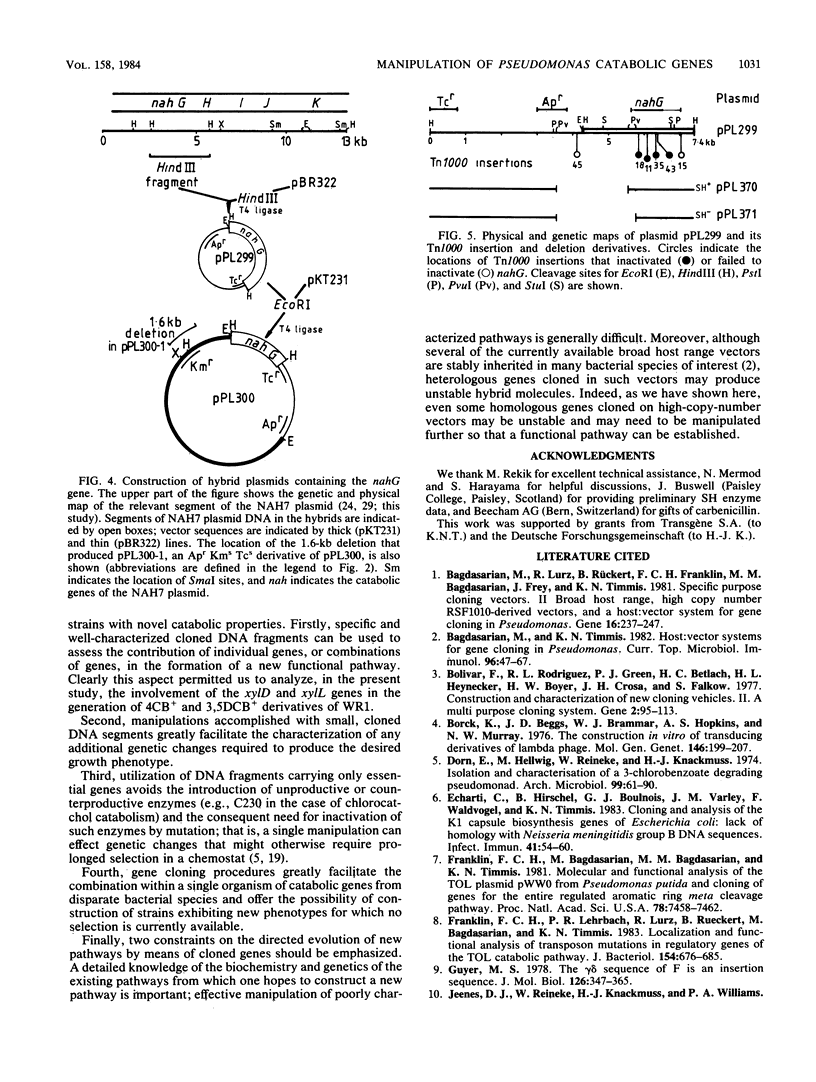
Abstract
DNA fragments containing the xylD and xylL genes of TOL plasmid pWW0 -161 of Pseudomonas putida, which code for the catabolic enzymes toluate 1,2-dioxygenase and dihydrodihydroxybenzoic acid dehydrogenase, respectively, and the nahG gene of the NAH plasmid NAH7 , which codes for salicylate hydroxylase, were cloned in pBR322 vector plasmid. Deletion and insertion mutagenesis were used to localize these genes with respect to crucial endonuclease cleavage sites. The pBR322-based plasmids were ligated to the broad host range cloning vector pKT231 , or derivatives of it, and the hybrid plasmids were introduced into Pseudomonas sp. B13( WR1 ), a bacterium able to degrade 3-chlorobenzoate but not 4-chlorobenzoate, 3,5- dichlorobenzoate , salicylate, or chlorosalicylates . The cloned xylD gene expanded the catabolic range of WR1 to include 4-chlorobenzoate, whereas the cloned xylD - xylL genes enabled the isolation of derivatives of WR1 that degraded 3-chlorobenzoate, 4-chlorobenzoate, and 3,5- dichlorobenzoate . The cloned nahG gene extended the catabolic range of WR1 to include salicylate and 3-, 4-, and 5- chlorosalicylate .
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Echarti C., Hirschel B., Boulnois G. J., Varley J. M., Waldvogel F., Timmis K. N. Cloning and analysis of the K1 capsule biosynthesis genes of Escherichia coli: lack of homology with Neisseria meningitidis group B DNA sequences. Infect Immun. 1983 Jul;41(1):54–60. doi: 10.1128/iai.41.1.54-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Lehrbach P. R., McGregor I., Ward J. M., Broda P. Molecular relationships between pseudomonas INC P-9 degradative plasmids TOL, NAH, and SAL. Plasmid. 1983 Sep;10(2):164–174. doi: 10.1016/0147-619x(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Lehrbach P. R., Timmis K. N. Genetic analysis and manipulation of catabolic pathways in Pseudomonas. Biochem Soc Symp. 1983;48:191–219. [PubMed] [Google Scholar]
- Reineke W., Jeenes D. J., Williams P. A., Knackmuss H. J. TOL plasmid pWW0 in constructed halobenzoate-degrading Pseudomonas strains: prevention of meta pathway. J Bacteriol. 1982 Apr;150(1):195–201. doi: 10.1128/jb.150.1.195-201.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Chemical structure and biodegradability of halogenate aromatic compounds. Substituent effects on 1,2-dioxygenation of benzoic acid. Biochim Biophys Acta. 1978 Sep 6;542(3):412–423. doi: 10.1016/0304-4165(78)90372-0. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on dehydrogenation of 3,5-cyclohexadiene-1,2-diol-1-carboxylic acid. Biochim Biophys Acta. 1978 Sep 6;542(3):424–429. doi: 10.1016/0304-4165(78)90373-2. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Hybrid pathway for chlorobenzoate metabolism in Pseudomonas sp. B13 derivatives. J Bacteriol. 1980 May;142(2):467–473. doi: 10.1128/jb.142.2.467-473.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A. M. Metabolism of aromatic compounds in bacteria. Purification and properties of the catechol-forming enzyme, 3,5-cyclohexadiene-1,2-diol-1-carboxylic acid (NAD + ) oxidoreductase (decarboxylating). J Biol Chem. 1972 Aug 25;247(16):4960–4965. [PubMed] [Google Scholar]
- Ribbons D. W., Williams P. A. Genetic engineering on microorganisms for chemicals: diversity of genetic and biochemical traits of pseudomonads. Basic Life Sci. 1982;19:211–232. doi: 10.1007/978-1-4684-4142-0_18. [DOI] [PubMed] [Google Scholar]
- Schell M. A. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 1983 Feb;153(2):822–829. doi: 10.1128/jb.153.2.822-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO S., KATAGIRI M., MAENO H., HAYAISHI O. SALICYLATE HYDROXYLASE, A MONOOXYGENASE REQUIRING FLAVIN ADENINE DINUCLEOTIDE. I. PURIFICATION AND GENERAL PROPERTIES. J Biol Chem. 1965 Aug;240:3408–3413. [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]


