Abstract
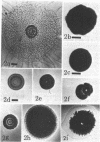
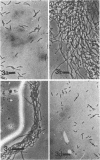
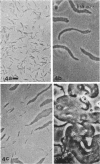
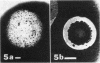
Three approaches were taken to isolate a total of 153 nonspreading mutants derived from our laboratory strain of Cytophaga johnsonae, UW101, or from its auxotrophic derivative, UW10538. Characterization of 109 of these mutants led to their placement in five general categories: (i) motile, nonspreading (MNS) mutants whose cells are motile to various degrees but whose colonies fail to spread on agar gels under any conditions of incubation; (ii) conditional nonspreading (CNS) mutants with motile cells whose colonies require more moisture to spread on agar gels than do those of wild-type cells; (iii) filamentous conditional motility (FCM) mutants whose cells grow as nonmotile filaments or as motile cells with wild-type morphology, depending on conditions of incubation; (iv) short, tumbling, nonspreading (STN) mutants with short cells that tumble constantly; and (v) truly nonmotile (TNM) mutants whose cells never move and whose colonies never spread under any conditions tested. All TNM mutants exhibited a remarkable pleiotropy not seen in the other four classes of mutants: all were resistant to 39 phages to which wild-type cells are sensitive, and all were unable to digest chitin, which is digested by wild-type cells. The correlation between ability to move and phage sensitivity was strengthened further by showing that 150 additional TNM mutants derived from UW101 and 43 TNM mutants derived from 29 independent isolates of C. johnsonae were resistant to all phages to which their parents were sensitive. Furthermore, motile revertants of TNM mutants became phage sensitive, and temperature-sensitive mutants were motile and phage sensitive at 25 degrees C and nonmotile and phage resistant at 32 degrees C. Evidence supports the conclusion that any mutation rendering cells truly nonmotile invariably alters cell surface-associated properties such as phage sensitivity and chitin digestion merely as a consequence of changing a moving cell surface to a static surface.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayusawa D., Yoneda Y., Yamane K., Maruo B. Pleiotropic phenomena in autolytic enzyme(s) content, flagellation, and simultaneous hyperproduction of extracellular alpha-amylase and protease in a Bacillus subtilis mutant. J Bacteriol. 1975 Oct;124(1):459–469. doi: 10.1128/jb.124.1.459-469.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P. Gliding motility mutants of Myxococcus xanthus. J Bacteriol. 1970 Nov;104(2):940–947. doi: 10.1128/jb.104.2.940-947.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- Dworkin M., Keller K. H., Weisberg D. Experimental observations consistent with a surface tension model of gliding motility of Myxococcus xanthus. J Bacteriol. 1983 Sep;155(3):1367–1371. doi: 10.1128/jb.155.3.1367-1371.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein J. E., Rogers H. J. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J Bacteriol. 1976 Sep;127(3):1427–1442. doi: 10.1128/jb.127.3.1427-1442.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J., Pate J. L. Isolation and characterization of gliding motility mutants of Cytophaga columnaris. Arch Mikrobiol. 1973 Nov 19;93(4):295–309. doi: 10.1007/BF00427927. [DOI] [PubMed] [Google Scholar]
- Keller K. H., Grady M., Dworkin M. Surface tension gradients: feasible model for gliding motility of Myxococcus xanthus. J Bacteriol. 1983 Sep;155(3):1358–1366. doi: 10.1128/jb.155.3.1358-1366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus I. R., Berg H. C. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982 Jul;151(1):384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. A. Bacteriophage receptors. Annu Rev Microbiol. 1973;27:205–241. doi: 10.1146/annurev.mi.27.100173.001225. [DOI] [PubMed] [Google Scholar]
- Mendelson N. H. Bacterial growth and division: genes, structures, forces, and clocks. Microbiol Rev. 1982 Sep;46(3):341–375. doi: 10.1128/mr.46.3.341-375.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller E. C., Durham N. N. Sealed aerobic slide culture for photomicrography. Appl Microbiol. 1968 Feb;16(2):439–440. doi: 10.1128/am.16.2.439-440.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEWART D. J. A selective-diagnostic medium for the isolation of pectinolytic organisms in the Enterobacteriaceae. Nature. 1962 Sep 8;195:1023–1023. doi: 10.1038/1951023a0. [DOI] [PubMed] [Google Scholar]
- Schade S. Z., Adler J., Ris H. How bacteriophage chi attacks motile bacteria. J Virol. 1967 Jun;1(3):599–609. doi: 10.1128/jvi.1.3.599-609.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y. Studies on Nonfruiting Myxobacteria: I. Cytophaga johnsonae, n.sp., a Chitin-decomposing Myxobacterium. J Bacteriol. 1947 Mar;53(3):297–315. [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y., Maruo B. Mutation of Bacillus subtilis causing hyperproduction of alpha-amylase and protease, and its synergistic effect. J Bacteriol. 1975 Oct;124(1):48–54. doi: 10.1128/jb.124.1.48-54.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]