Abstract
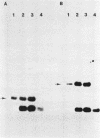
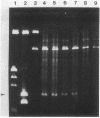
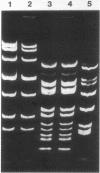
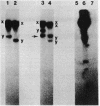
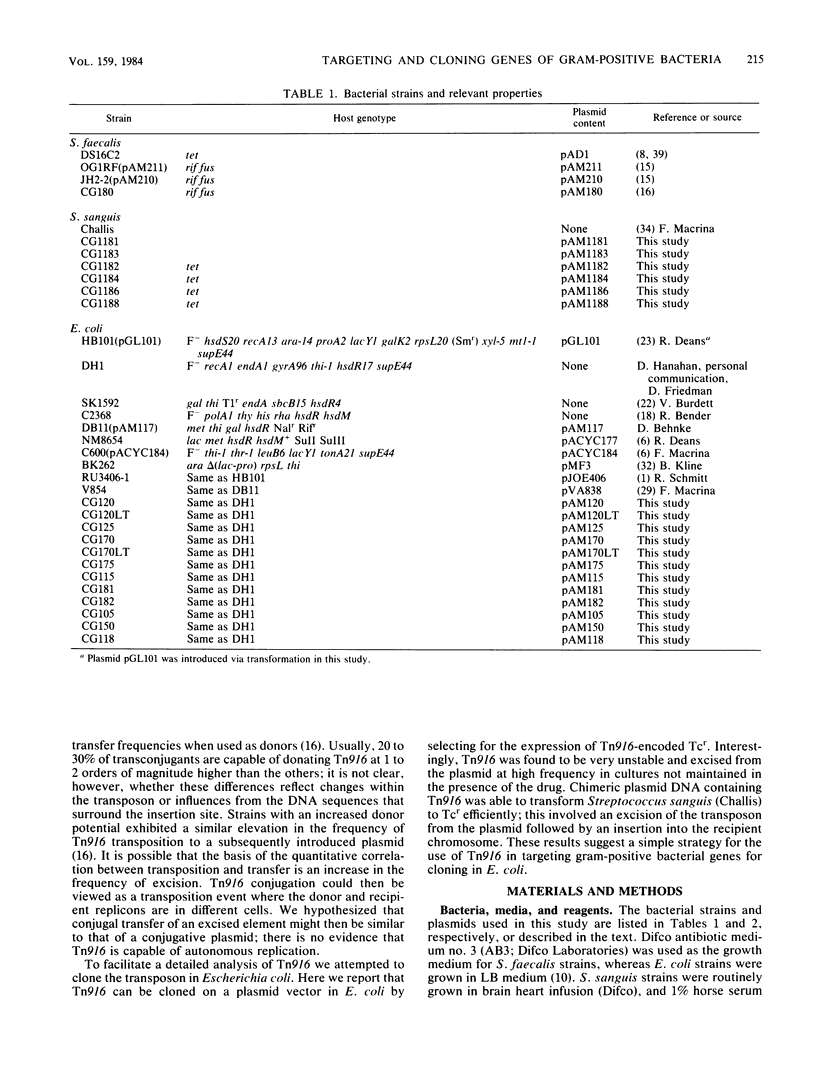
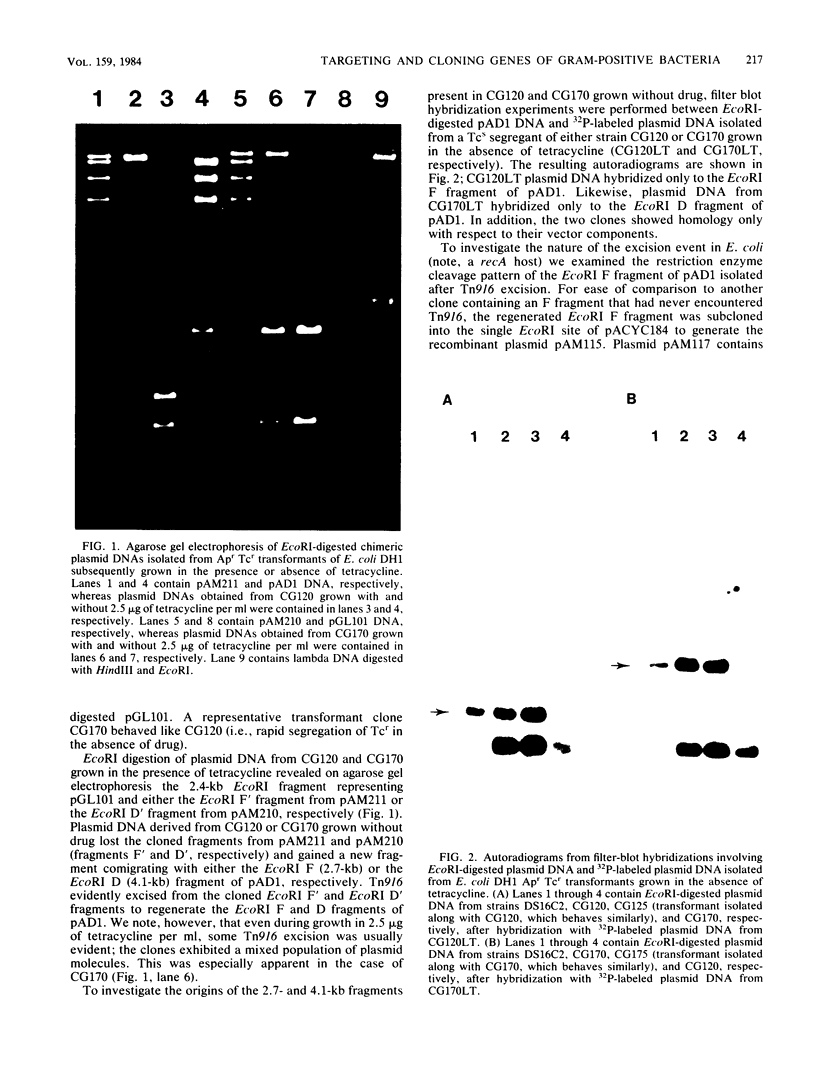
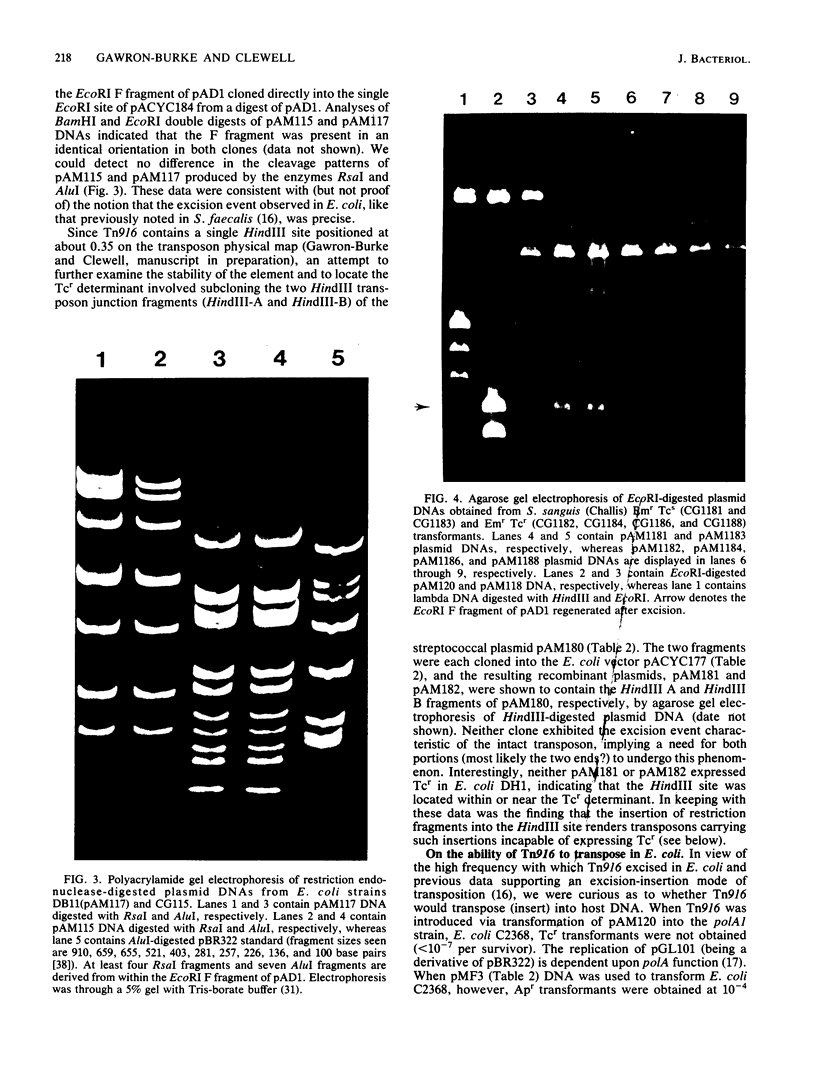
The conjugative transposon Tn916 (15 kilobases), originally identified in Streptococcus faecalis DS16, has been cloned as an intact element on the pBR322-derived vector pGL101 in Escherichia coli. The EcoRI F' (EcoRI F::Tn916) fragment of pAM211 (pAD1::Tn916) was cloned into the single EcoRI site of pGL101 to form the chimera, pAM120, by selecting for the expression of Tn916-encoded tetracycline resistance (Tcr). Interestingly, in the absence of continued selection for Tcr, Tn916 excised from pAM120 at high frequency. This excision event resulted in a plasmid species consisting of the pGL101 vector and a 2.7-kilobase restriction fragment comigrating with the EcoRI F fragment of pAD1 during agarose gel electrophoresis. Filter blot hybridization experiments showed the 2.7-kilobase fragment generated as a result of Tn916 excision to be homologous with the EcoRI F fragment of pAD1. Analogous results were obtained with another chimera, pAM170, generated by ligating the EcoRI D' (EcoRI D::Tn916) fragment of pAM210 (pAD1::Tn916) to EcoRI-digested pGL101. Comparison of the AluI and RsaI cleavage patterns of the EcoRI F fragment isolated after Tn916 excision with those from an EcoRI F fragment derived from pAD1 failed to detect any difference in the two fragments: data in support of a precise Tn916 excision event in E. coli. Subcloning experiments showed that an intact transposon was required for Tn916 excision and located the Tcr determinant near the single HindIII site on Tn916. Although excision occurred with high frequency in E. coli, Tn916 insertion into the E. coli chromosome was a much rarer event. Tcr transformants were not obtained when pAM120 DNA was used to transform a polA1 strain, E. coli C2368.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbuchner J., Schmitt R. Transposon Tn1721: site-specific recombination generates deletions and inversions. Mol Gen Genet. 1983;190(2):300–308. doi: 10.1007/BF00330655. [DOI] [PubMed] [Google Scholar]
- Barany F., Tomasz A. Genetic transformation of Streptococcus pneumoniae by heterologous plasmid deoxyribonucleic acid. J Bacteriol. 1980 Nov;144(2):698–709. doi: 10.1128/jb.144.2.698-709.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182(3):490–497. doi: 10.1007/BF00293940. [DOI] [PubMed] [Google Scholar]
- Burdett V., Inamine J., Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982 Mar;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Tomich P. K., Gawron-Burke M. C., Franke A. E., Yagi Y., An F. Y. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J Bacteriol. 1982 Dec;152(3):1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R. H. Multiple RNA polymerase holoenzymes exert transcriptional specificity in Bacillus subtilis. Arch Biochem Biophys. 1982 Apr 1;214(2):772–781. doi: 10.1016/0003-9861(82)90084-4. [DOI] [PubMed] [Google Scholar]
- Eccles S., Docherty A., Chopra I., Shales S., Ball P. Tetracycline resistance genes from Bacillus plasmid pAB124 confer decreased accumulation of the antibiotic in Bacillus subtilis but not in Escherichia coli. J Bacteriol. 1981 Mar;145(3):1417–1420. doi: 10.1128/jb.145.3.1417-1420.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. DNA cloning in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1433–1436. doi: 10.1073/pnas.75.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for conjugal transfer of a Streptococcus faecalis transposon (Tn916) from a chromosomal site in the absence of plasmid DNA. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):77–80. doi: 10.1101/sqb.1981.045.01.014. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982 Nov 18;300(5889):281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- Goebel W., Schrempf H. Replication of the minicircular DNA of E. coli 15 is dependent on DNA polymerase I but independent of DNA polymerase 3. Biochem Biophys Res Commun. 1972 Oct 17;49(2):591–600. doi: 10.1016/0006-291x(72)90452-4. [DOI] [PubMed] [Google Scholar]
- Harding N. E., Cleary J. M., Smith D. W., Michon J. J., Brusilow W. S., Zyskind J. W. Chromosomal replication origins (oriC) of Enterobacter aerogenes and Klebsiella pneumoniae are functional in Escherichia coli. J Bacteriol. 1982 Dec;152(3):983–993. doi: 10.1128/jb.152.3.983-993.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Craig R. A., White B. A., Yagi Y., Clewell D. B. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft J., Bernhard K., Goebel W. Recombinant plasmids capable to replication in B. subtilis and E. coli. Mol Gen Genet. 1978 Jun 1;162(1):59–67. doi: 10.1007/BF00333851. [DOI] [PubMed] [Google Scholar]
- Lauer G., Pastrana R., Sherley J., Ptashne M. Construction of overproducers of the bacteriophage 434 repressor and cro proteins. J Mol Appl Genet. 1981;1(2):139–147. [PubMed] [Google Scholar]
- Lawson J. W., Gooder H. Growth and development of competence in the group H streptococci. J Bacteriol. 1970 Jun;102(3):820–825. doi: 10.1128/jb.102.3.820-825.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Jones K. R., Welch R. A. Transformation of Streptococcus sanguis with monomeric pVA736 plasmid deoxyribonucleic acid. J Bacteriol. 1981 May;146(2):826–830. doi: 10.1128/jb.146.2.826-830.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Jones K. R., Wood P. H. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis). J Bacteriol. 1980 Sep;143(3):1425–1435. doi: 10.1128/jb.143.3.1425-1435.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Tobian J. A., Jones K. R., Evans R. P., Clewell D. B. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982 Oct;19(3):345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Wood P. H., Jones K. R. Simple method for demonstrating small plasmid deoxyribonucleic acid molecules in oral streptococci. Appl Environ Microbiol. 1980 May;39(5):1070–1073. doi: 10.1128/aem.39.5.1070-1073.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J. J., Kline B. C. Restriction endonuclease mapping and mutagenesis of the F sex factor replication region. Mol Gen Genet. 1977 Apr 29;152(3):175–182. doi: 10.1007/BF00268815. [DOI] [PubMed] [Google Scholar]
- Nida K., Cleary P. P. Insertional inactivation of streptolysin S expression in Streptococcus pyogenes. J Bacteriol. 1983 Sep;155(3):1156–1161. doi: 10.1128/jb.155.3.1156-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina J. L., Ravin A. W. Fate of homospecific transforming DNA bound to Streptococcus sanguis. J Bacteriol. 1978 Mar;133(3):1212–1223. doi: 10.1128/jb.133.3.1212-1223.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C. W., Guild W. R. Pathway of plasmid transformation in Pneumococcus: open circular and linear molecules are active. J Bacteriol. 1981 May;146(2):517–526. doi: 10.1128/jb.146.2.517-526.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich P. K., An F. Y., Damle S. P., Clewell D. B. Plasmid-related transmissibility and multiple drug resistance in Streptococcus faecalis subsp. zymogenes strain DS16. Antimicrob Agents Chemother. 1979 Jun;15(6):828–830. doi: 10.1128/aac.15.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]