Abstract
Previously, we have reported on successful imaging of colon, rectal, and pancreatic carcinomas in patients by using a radiolabeled all-human monoclonal antibody, COU-1, directed against modified cytokeratin. To further develop this antibody for use as an immunoconjugate, COU-1 was cloned by phage display selection and the human Fab fragment was expressed in bacteria. Analysis by confocal laser scanning microscopy demonstrated that COU-1 bound in a uniform punctate pattern to the surface of viable carcinoma cells stained at 4°C, and binding increased significantly when cells were cultured on fibronectin, laminin, or collagen IV. In the case of fibronectin, COU-1 staining was particularly enhanced at intercellular junctions. When carcinoma cells were cultured with COU-1 at 37°C for 6 hr, the antibody was found in large perinuclear vesicles and the punctate surface staining was significantly reduced. Similar results were obtained using intact IgM COU-1 and the recombinant Fab fragment. Immunohistological studies indicated that COU-1, in contrast to murine monoclonal antibodies against normal cytokeratin 8 and 18, could differentiate between malignant and normal colon epithelia, and between colon cancer metastasis in the liver and surrounding normal hepatocytes. Within biopsies of malignant tissue, COU-1 exhibited membrane-associated staining of proliferating cells, while resting cells had a filamentous pattern. Thus, modified cytokeratin at the surface of carcinoma cells may represent a new target for immunoconjugates and may explain the promising results of the phase I/II clinical study.
Keywords: COU-1 antibody, internalization, phage display, cancer, cytokeratin
We previously established a human monoclonal IgM antibody (HumAb), COU-1, by fusion of a human B lymphoblastoid cell line with lymphocytes from a mesenteric lymph node of a patient with colon cancer (1). The antigen recognized by COU-1 migrates as three bands in SDS/PAGE with molecular masses of approximately 43 kDa, representing modified forms of cytokeratin 8 and 18 (2). Immunohistochemical analysis shows that the antibody reacts with malignant tissues of epithelial origin, such as carcinomas of the colon, ovary, pancreas, and breast and, to a lesser extent, with normal tissues (2–4). In a phase I/II clinical trial involving intravenous administration of 131I-labeled COU-1 in patients with suspected colorectal cancer, primary tumor or recurrence was successfully detected in seven of nine patients (5). Metastasis localized to the liver was also visualized in one of three patients and, in the other two patients, “field-of-interest” analysis demonstrated increased accumulation of radioactivity in the liver compared with patients with no liver metastasis. Since COU-1 is directed against cytokeratin, and such molecules are predominantly localized intracellularly, the mechanism for the observed tumor localization was unclear. The distribution of COU-1 within the surgically removed tumors was determined after the samples were dissected into 5-mm cubic pixels and the amount of radioactivity in each pixel was correlated with morphology (5). The highest amount of radioactivity was found in viable tumor tissue compared with necrotic tumor tissue or surrounding connective tissue, indicating that accumulation of COU-1 within the tumor was due to antigen binding rather than nonspecific accumulation.
Although a clinical trial demonstrated that COU-1 effectively detected tumors in patients, IgM antibodies are likely to penetrate tissues less well than smaller molecules with the same specificity. To address this, we used chemical reduction and alkylation to generate monomeric and half-monomeric fragments containing two and one antigen-binding site(s), respectively (6). These fragments retained antigen-binding activities, although with decreased avidities (6). When tested in tumor-bearing nude mice, the fragments, especially the half-monomeric form, had a favorable cancer-to-normal tissue ratio compared with the intact IgM antibody (7). However, generation of the fragments by chemical means while retaining antigen-binding activity was not as straightforward as one might expect, and only a fraction of the reduced antibody had the desired molecular weight (6). Consequently, we have now produced the recombinant Fab fragment of COU-1 by using the phage display technology, which allows molecular cloning of human antibodies, including those already immortalized by the hybridoma technique (8). A significant advantage is that cloning the antibody variable genes into a vector allows affinity maturation of the antibody (9, 10), grafting of different Fc regions onto it (11), as well as genetic engineering of immunotoxins (12) and immunomodifiers (13). In addition, the manufacturing cost may be less for recombinant human antibodies than for human hybridoma antibodies.
In this study, we report the molecular cloning of COU-1 by phage display, bacterial expression, and characterization of the Fab fragment. The intact COU-1 as well as the recombinant Fab fragment were found to bind to the surface of colon cancer cells, an observation that provides more evidence for the presence of modified cytokeratin molecules on the surface of some malignant epithelial cells (14–16). Both the intact COU-1 and the recombinant Fab fragment were internalized by cells at 37°C and found within endocytic vesicles. We have also extended our investigation of the tissue distribution of the antigen recognized by COU-1. In ethanol-fixed tissue biopsy samples containing malignant and normal colon epithelia, or colon liver metastasis and normal liver tissue, COU-1 was able to discriminate normal from malignant tissue. In contrast, two mAbs directed against normal cytokeratins 8 and 18 stained colon carcinomas, normal colon epithelia, and most hepatocytes with similar intensities.
MATERIALS AND METHODS
Antibodies.
The human monoclonal IgM antibody COU-1 is secreted by the hybridoma cell line B9165, derived by fusing the human lymphoblastoid cell line WI-L2–729-HF2 and lymphocytes obtained from mesenteric lymph nodes of a patient with colon cancer (1). The hybridoma cell line was grown in protein-free medium: RPMI 1640 medium (GIBCO) supplemented with SSR3 serum replacement (Medicult, Copenhagen). The COU-1 antibody was purified from cell culture supernatants by affinity chromatography on Sepharose-coupled murine monoclonal anti-human μ-chain antibody (HB57, American Type Culture Collection, Rockville, MD) and eluted with 0.1 M diethylamine, pH 10.5, followed by fractionation by an ion-exchange FPLC (Pharmacia). IgM purified from normal human serum (Cappel) was used as a control. The human monoclonal IgM antibody 16.88 (17), which was a generous gift from R. McCabe (Organon Teknika, Biotechnology Research Institute, Rockville, MD), has been used successfully for tumor imaging in humans (18–20) and was studied for comparison. Two murine mAbs, M20, directed against normal cytokeratin 8, and CY-90, directed against normal cytokeratin 18, were obtained from Sigma.
PCR Amplification and Cloning of the Variable Heavy and Light Chain Genes.
Total RNA was prepared from the B9165 hybridoma cell line by the guanidinium method. After reverse transcription, the μ (Fd region) and κ chains were amplified by the polymerase chain reaction (PCR) using a set of family-specific primers as described (21). The subsequent construction of an IgM/κ Fab library using the pComb3 M13 surface display system (kindly provided by C. F. Barbas III, The Scripps Research Institute, La Jolla, CA) has been described previously (22, 23).
Enrichment of Antigen-Binding Phage Through Panning.
Panning of the COU-1 library was carried out as described previously (22). In brief, microtiter wells were coated overnight with ultrasonicated lysate of a colon cancer cell line (colo137) in 0.1 M sodium bicarbonate buffer, pH 8.6, at 4°C (7). After blocking with PBS containing 3% BSA for 1 hr at 37°C, a 50-μl phage suspension in PBS was added to each well and incubated for 2 hr. Unbound phage were removed by vigorous washing 10 times with PBS containing 0.05% Tween 20 (PBS-Tween)(Merck, Darmstadt, Germany). Bound phage, enriched for those bearing antigen-binding Fabs, were eluted with 0.2 M glycine⋅HCl, pH 2.2. The eluted phage were amplified by infection of Escherichia coli XLI-Blue (Stratagene) and recovered by superinfection with VCS-M13 helper phage. The panning procedure was carried out twice. Phagemid DNA was isolated from the last round of panning, cut with NheI and SpeI, and religated. This step excised the cpIII gene, resulting in a vector producing soluble Fab fragments.
ELISA Analysis of Fab COU-1 and Intact COU-1 Antibody.
Fabs were prepared as bacterial supernatants through a freeze–thawing procedure and purified by affinity chromatography, as reported earlier (22–24), with minor modifications. A goat antibody against human IgG F(ab′)2 (Pierce) crosslinked to protein G Gammabind matrix (Pharmacia) was used for the purification. The column was washed with PBS, and bound Fab was eluted with 0.2 M glycine⋅HCl, pH 2.2, and immediately neutralized with 1 M Tris⋅HCl, pH 9.0. To assess specificity, supernatants and purified Fabs were screened by ELISA for binding to ultrasonicates of colon cancer cells (colo137), BSA (30 mg/ml; Sigma), ovalbumin (20 μg/ml, Sigma), recombinant HIV-1 gp120 (2 μg/ml, IIIB) (Intracel, Issaquah, WA), and human placental DNA (2 μg/ml, Sigma). ELISA wells were coated with antigen overnight at 4°C in 0.1 M bicarbonate buffer, pH 8.6. DNA in PBS was dried on the ELISA wells at 37°C. The wells were washed twice with PBS, blocked by filling the wells with 3% BSA in PBS for 1 hr at 37°C, and incubated with human Fab samples or intact human IgM antibody for 2 hr at 37°C. Plates were washed 10 times with PBS-Tween, and bound Fab was detected with alkaline phosphatase (AP)-labeled goat anti-human IgG F(ab′)2 (Pierce) diluted 500-fold in PBS or AP-labeled rabbit anti-human κ chain (Sigma) diluted 1,000-fold in PBS. Bound antibody was visualized with para-nitrophenyl phosphate [Sigma; 1 mg/ml in 1 mM MgCl2/10% (wt/vol) diethanolamine, pH 9.6] and read at 405 nm.
Analysis by Confocal Laser Scanning Microscopy.
Human colon cancer cell lines (H3619 and colo137) and breast cancer cell lines (MCF-7 and H3396) were grown in Iscove’s modified Dulbecco’s medium containing 10% fetal bovine serum (FBS) and allowed to adhere to chambered coverslips (Nunc) for 48 hr at 37°C under a 5% CO2/95% air atmosphere to form monolayers. Experiments were performed using the primary antibodies COU-1, Fab COU-1, murine anti-cytokeratin 8, murine anti-cytokeratin 18, and HumAb 16.88, as indicated below. All antibodies were tested at 10 μg/ml except Fab COU-1 (30 μg/ml).
Intracellular antibody binding.
H3619 and colo137 cells were permeabilized with methanol at −20°C for 5 min, blocked with normal goat serum, and incubated with primary antibodies at room temperature for 1 hr. The cells were then washed three times with culture medium and incubated with fluorescein isothiocyanate (FITC)-labeled goat anti-human κ-chain antibody (Southern Biotechnology Associates) or FITC-labeled goat anti-mouse IgG (BioSource International, Camarillo, CA) diluted 1:100 and 1:50, respectively, in PBS for 1 hr at room temperature.
Antibody binding to cell surface.
Live H3619 cells were incubated with COU-1 at 4°C for 2 hr, washed three times with cold culture medium, and incubated with secondary FITC-labeled antibody at 4°C for 1 hr. In addition, H3619 cells were cultured on different extracellular matrices; fibronectin, laminin, or collagen IV (all Boehringer Mannheim). Chamber slides were coated with 100 μl of collagen (2 mg/ml), 400 μl of fibronectin (50 μg/ml), or 400 μl of laminin (50 μg/ml). Different volumes had to be used to achieve complete coating of the slides. After 45 min, excess substrate was removed (fibronectin, laminin) or air dried (collagen IV), cells were added, and the slides were incubated for 48 hr at 37°C.
Antibody internalization.
Live H3619 and colo137 cells were incubated with COU-1 or Fab COU-1 at 37°C for 6 hr, followed by washing three times and permeabilization with methanol at −20°C for 5 min. Cells were blocked with normal goat serum and incubated with secondary FITC-labeled antibody at room temperature for 1 hr. For all experiments, after primary and secondary antibody incubations, the cells were washed, fixed with 2% paraformaldehyde in PBS for 15 min at room temperature, washed twice, and mounted in anti-fading reagent (30 mM dithioerythritol/PBS/glycerol, 2:9:1 vol/vol). Staining of cells was evaluated by confocal laser scanning microscopy. As a control, all experiments were duplicated with omission of the primary antibody.
Immunohistochemical Analysis.
Tissue specimens were obtained from colorectal cancer patients undergoing surgical resection. Normal colon tissue was taken from the resectate approximately 15 cm away from the site of the tumor. Tissues were fixed in 96% ethanol for 6 hr at 4°C, embedded in paraffin, and cut into 5-μm sections. Sections were deparaffinized in xylol, rehydrated through graded ethanol, and washed in PBS-Tween. Sections were incubated for 2 hr at room temperature in a humidified chamber with 100 μl of murine mAb, HumAb, or normal polyclonal human IgM, all at 0.5–10 μg/ml. The slides were washed and incubated with AP-labeled rabbit anti-human IgM (Dako), horseradish peroxidase (HRP)-labeled rabbit anti-human IgM (Dako), or HRP-labeled rabbit anti-mouse IgG (Dako) diluted in PBS with 10% (wt/vol) BSA for 1 hr at room temperature. After washing, the HRP was visualized by development with chromogenic substrate (0.6 mg of diaminobenzidine per ml of PBS with 0.01% H2O2) and AP with 0.2 mg of naphthol-AS-MX phosphate (Sigma), 1 mg of fast red TR salt (Sigma), and 20 μg of dimethylformamide per ml of 0.1 M Tris⋅HCl/1 M levamisole, pH 8.2. The sections were counterstained with Mayer’s hematoxylin, dehydrated in xylene, and mounted in Aquamount (Gurr, Poole, England). The staining intensity was graded as follows: −, no staining; +, weak staining; ++, moderate staining; and +++, strong staining.
RESULTS
Phage Display Expression and Sequencing of HumAb COU-1.
RNA was extracted from the B9165 cell line and the heavy (μ, Fd region) and light (κ)-chain genes from the corresponding cDNA were amplified by PCR using 3′ family specific primers and a 5′ constant primer. The light and heavy chain products were then sequentially cloned into the M13 phage surface expression vector pComb3 to generate a library of 2 × 106 members. The phage library was selected twice on an ultrasonicate of the COU-1 antigen-positive colon cancer cell line (colo137). DNA was prepared from the last round of selection and gene III fragment was removed by NheI/SpeI digestion and ligation. The reconstructed phagemids were used to transform E. coli cells to produce clones secreting soluble Fab fragments. Supernates of 3 of the 80 single Fab expression clones tested by ELISA bound to colo137 lysate and not to ovalbumin.
The sequences of these three clones were identical. Sequence analysis showed that the COU-1 light chain belongs to the VκIII family and exhibits 97% (269/276) nucleotide identity to L6 as the closest germ line (Fig. 1). The COU-1 light chain contained an extra serine inserted corresponding to codon 30. The light-chain J segment showed 95% (36/38) nucleotide identity to the germ-line Jκ5 segment. Further sequence analysis showed that the heavy chain belongs to the VHI family, exhibiting 98% nucleotide identity to the heavy-chain germ line DP-7. The heavy-chain J segment showed 96% (53/55) nucleotide identity to the germ-line JH6b segment. The D segment of COU-1 showed closest homology to the D2 germ-line D segment, with a 16 nucleotide stretch of complete identity. The deduced amino acid sequence of the COU-1 heavy and light chains, together with the closest germ-line homologues, are shown in Fig. 1.
Figure 1.
Deduced amino acid sequence of the variable heavy and light chain of HumAb COU-1 compared with the closest known germ-line sequences. FR, framework region; CDR, complementarity-determining region.
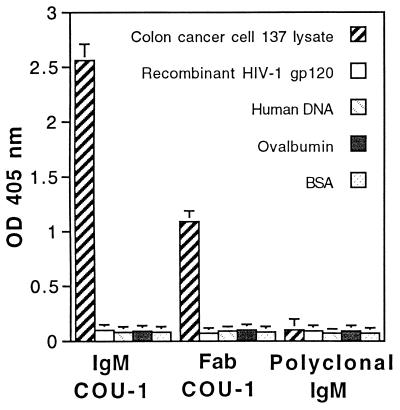
Purified recombinant Fab fragment of COU-1 (Fab COU-1) was tested in parallel with the intact COU-1 and normal polyclonal IgM for binding to lysate of colon cancer cells (colo137) and irrelevant antigens in ELISA. As depicted in Fig. 2, the Fab COU-1 and COU-1 exhibited strong binding to colo137 lysate, but not to a panel of other antigens, including BSA, ovalbumin, human DNA, and HIV-1 gp120. In contrast, normal human IgM did not bind to any of these antigens. The concentration needed for saturation was significantly higher for the Fab (20 μg/ml) than for the intact antibody (1 μg/ml), and it was similar to that previously measured for chemically derived half-monomeric fragments, exhibiting a Ka of 2 × 106 M−1 (6).
Figure 2.
Binding of HumAb IgM COU-1 (0.5 μg/ml), recombinant Fab fragment of COU-1 (10 μg/ml), and normal human IgM (10 μg/ml) to a panel of solid-phase antigens tested by ELISA.
COU-1-Defined Cytokeratins: Localization, Induction by the Extracellular Matrix, and Internalization.
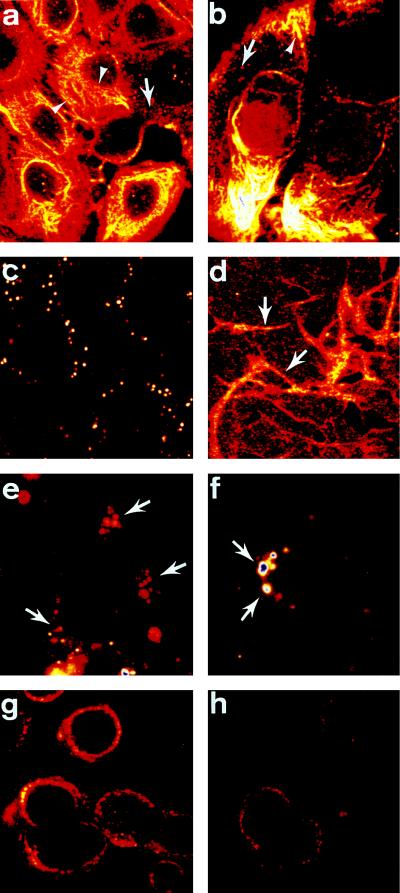
Cells from the human colon carcinoma lines H3619 (Fig. 3a) and colo137 (Fig. 3b) were fixed, permeabilized, and stained with COU-1 (Fig. 3a) or Fab COU-1 (Fig. 3b). Intermediate filaments were heavily labeled, as expected. The staining pattern for murine mAbs directed against cytokeratins 8 and 18 and the HumAb 16.88 was similar to that observed with COU-1 (data not shown). The breast carcinoma cell lines MCF-7 and 3396 gave comparable results. In addition, COU-1 localized to vesicles (Fig. 3 a and b; arrows) within the cytoplasm, which were more abundant in colo137 cells (Fig. 3b).
Figure 3.
COU-1 localization, internalization, and influence of fibronectin on surface cytokeratin expression. Colon cancer cells [H3619 (a), colo137 (b)] were fixed, permeabilized, and stained with COU-1 (a) or Fab COU-1 (b) and FITC-labeled goat anti-human κ-chain antibody. Note intense fibrillar staining of the intermediate filament (arrowhead). In addition, vesicles (arrows) throughout the cytoplasm were routinely observed. Live H3619 cells incubated with COU-1 at 4°C gave dispersed punctate staining of the upper cell surface (c). When the H3619 cells were grown on fibronectin-coated slides, the level of punctate staining at the cell surface increased and COU-1 was enriched at intercellular junctions (arrows, d). When the colon cancer cells [H3619 (e, f), colo137 (g, h)] were incubated with COU-1 at 37°C, the dispersed punctate surface staining disappeared and COU-1 (e, f, h) or Fab COU-1 (g) localized in large cytoplasmic vesicles adjacent to the nucleus. (×1000.)
To determine whether the cytokeratins recognized by the mAbs were present on the outer cell membrane, viable H3619 cells were stained with COU-1 at 4°C to block antibody internalization. Approximately 20% of all cells showed intense punctate staining (Fig. 3c), and the fluorescence associated with the remaining 80% of the cells gave a similar pattern, but was less intense. A similar surface staining pattern was observed with the murine anti-cytokeratin 8 mAb (data not shown).
We next examined whether culturing of carcinoma cells on different extracellular matrices—i.e., fibronectin, laminin, or collagen IV—could affect the distribution of the cell surface cytokeratin. When cells were grown on laminin or collagen IV and stained by incubation with COU-1 at 4°C for 1 hr, there was a significant increase in the number of cells with intense punctate cell surface staining, and, in addition, cell surface staining corresponding to the intercellular junctions was observed (data not shown). The distribution of COU-1 was further altered when cells were cultured on fibronectin. The cytokeratin was highly enriched at intercellular junctions (Fig. 3d, arrow), whereas punctate cell surface staining was reduced. The fact that adhesion to different extracellular matrix proteins can alter the cell surface distribution of cytokeratin is a further indication that cytokeratin is present on the outer plasma membrane.
Since we observed COU-1-positive intracellular vesicles when we used permeabilized cells (Fig. 3 a and b; arrows), we investigated whether cell surface cytokeratin might be internalized into an endocytic compartment. H3619 (Fig. 3 e and f) and colo137 (Fig. 3 g and h) cells were cultured with COU-1 (Fig. 3 e, f, and h) or Fab COU-1 (Fig. 3g) for 6 hr at 37°C, fixed, and prepared for immunofluorescence. The dispersed punctate staining typical of cytokeratins localized at the cell surface was not observed. Instead, both COU-1 and Fab COU-1 accumulated in large perinuclear vesicles, which may be lysosomes. Since the distributions of Fab COU-1 and COU-1 were identical, the data indicate that antibody internalization was not due to multivalent crosslinking and imply that cytokeratins may cycle between the cell surface and the endocytic compartment.
COU-1 Binds Preferentially to Carcinoma Cells.
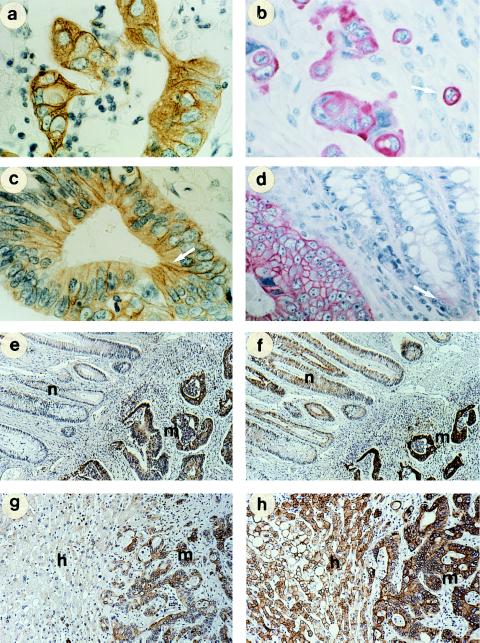
The subcellular localization of the antigen recognized by COU-1 in tissue biopsies of colon and rectal adenocarcinomas was studied by using indirect immunoperoxidase and AP techniques. At high magnification, we observed a distinct fibrillar staining of intermediate filaments by COU-1 (Fig. 4a). In small clusters or individual cells, there was intense staining at the cell periphery, possibly associated with the cell surface (Fig. 4b). In addition, enhanced staining associated with the junctional zone between adjacent cells was seen (Fig. 4c, arrow). No staining was observed in adjacent crypt epithelial cells from normal colon in five of eight colon or rectal cancers (Fig. 4e). In the remaining three cancers, however, there was weak staining of a few individual cells from normal colon, surrounded by negative cells, while the cancer tissue was stained strongly (Fig. 4d). Although the colon epithelium in Fig. 4d looked morphologically normal, it may have contained transformed cells. Murine mAbs anti-cytokeratin 8 (Fig. 4f) and 18 (data not shown) gave intense staining of the adjacent normal colon epithelium as well as of the carcinoma tissue, while COU-1 did not (Fig. 4e). The HumAb 16.88 antibody stained colon cancer cells strongly, and some areas of the normal colon epithelium weakly, but also stained smooth muscle fibers and myoepithelium.
Figure 4.
Comparison of the tissue distribution of COU-1 and murine anti-cytokeratin 8 antibody in ethanol-fixed tissues. Tumor cells within tissue sections bound COU-1, while surrounding normal cells were not stained (a–d). Fibrillar staining characteristic of intermediate filaments (a, arrow), membrane staining of single proliferating cells (b, arrow; note metaphase, arrowhead), and enrichment of COU-1 at intercellular junctions (arrows in c and d). In adjacent normal colon epithelia, weak staining was found only in a few cells of some biopsies (arrow, d). (e and f) Comparison of staining with COU-1 (e) and murine anti-cytokeratin 8 (M20) (f) on serial sections of malignant (m) and adjacent normal colon epithelium (n). (g and h) Adjacent sections of a colon cancer metastasis in the liver (m) and surrounding normal hepatocytes (h) incubated with COU-1 (g) and with M20 (h). (a and b, ×600; c and d, ×500; and e–h, ×200.)
The mAbs were also compared for staining of colon metastases in liver versus surrounding normal liver tissue. COU-1 gave intense staining of liver metastases, whereas hepatocytes were not stained (Fig. 4g) except for a few cells in the periportal zones, which were weakly positive. Similarly, HumAb 16.88 did not stain the majority of the hepatocytes, but stained myoepithelia, in contrast to COU-1. Both HumAbs stained the biliary ducts. The murine mAbs anti-cytokeratin 8 (Fig. 4h), and 18 (not shown) stained liver metastases as well as normal hepatocytes strongly and with equal intensity. The staining decreased toward the centrilobular area. Particularly strong staining by the murine mAbs was associated with the cell membrane of the hepatocytes (Fig. 4h).
DISCUSSION
Increasing interest has focused on cytokeratin expression in epithelial cancer and the potential of cytokeratins as tumor therapy targets. A recent report showed that a truncated cytokeratin 8 was the major protein accumulating in association with ubiquitin in colorectal cancer, but not in normal colon (25). This was observed in both early and advanced stages of colorectal cancer. The study suggested that malignant cells have cytokeratin 8 degradation pathways that do not exist in normal colon cells, resulting in the formation of cytokeratin neoepitopes. In another study, modified cytokeratin 8 was found to have plasminogen-binding activity (14), and it has been suggested that cytokeratin 8 may be a plasminogen receptor on breast cancer cells (26) and, therefore, be of importance in tumor metastasis (14).
We have used phage display technology to clone and further characterize the HumAb COU-1, which is an IgM antibody directed against modified cytokeratins 8 and 18. The Fab fragment of COU-1 used in this study was generated by phage display cloning and bacterial expression. Its binding characteristics were similar to those previously reported for half-monomeric fragments generated by chemical reduction and alkylation (6). The major advantage of the phage approach is that production of fragments by bacterial expression is more straightforward than chemical reduction and size fractionation of whole immunoglobulin. Moreover, although the existing Fab COU-1 has a lower avidity than the whole mAb, it can be improved by affinity maturation using the phage technique (9, 10). Sequence analysis showed that the variable region of the heavy and light chains had minimal somatic mutations, with 98% and 97% nucleotide identity to the closest germ-line V genes, respectively. This is in accordance with COU-1 being an IgM antibody, and indicates that substantial affinity maturation through site-directed mutagenesis is possible.
A phase I/II clinical trial with radiolabeled COU-1 showed that mAb localized to colorectal cancer in seven of nine patients (15). These findings are compatible with the view that a modified cytokeratin is expressed on the surface of colon carcinoma cells and that it can be recognized by circulating radiolabeled COU-1. To investigate this, we examined the distribution of COU-1 on the surface of live carcinoma cells exposed to the mAb at 4°C or 37°C and attached to plastic or different extracellular matrix proteins. Our results show that the distribution of COU-1 can be altered by changing temperature or by culturing on adhesive substratum, which makes it unlikely that its binding to the cell surface is due to cell damage or is an artifact of cell preparation. Recently, other results have indicated that cytokeratins may be present on the surface of some cancer cells (14–16). Interestingly, one of these studies used the HumAb 16.88 (15), which exhibits many similarities to COU-1 (27). Both antibodies recognize modified forms of cytokeratins 8 and 18, but 16.88 also recognizes an antigen with a molecular mass of 190 kDa in Western blots of melanoma cell extracts, and it crossreacts with some normal tissues in immunohistochemical analysis (27). Godfroid et al. (15) reported surface staining of breast cancer cells with 16.88, but not with a series of murine mAbs directed against normal cytokeratin 18 or 19; normal breast epithelia were not stained by 16.88. In another study, a murine antibody (M20) was shown to bind to cytokeratin at the surface (14), and we have confirmed these data. Two-dimensional gel electrophoretic separation of iodinated MCF-7 cells has also been used to demonstrate surface expression of cytokeratins (15), but this technique may detect intracellular cytokeratin, since the amount of intracellular cytokeratin so far exceeds the amount at the cell surface that a few injured cells would cause misleading results (28). Riopel et al. (28) found no labeling of cytokeratins when they used biotinylation of whole cells, and they concluded that the detection of cytokeratin at the cell surface was an artifact. A further criticism was that only the breast cancer cell line MCF-7 was used to demonstrate surface expression. Our data, collected by using other cell lines and techniques where minimal surface damage can occur, are consistent with the presence of modified cytokeratins 8 and 18 at the cell surface.
Studies by Hembrough et al. (26) have shown that normal cytokeratins 8 and 18 associate with the inner plasma membrane, and they have suggested that their presence on the outer surface of cells may be due to the projection of cytokeratin through the plasma membrane as part of a protein complex. Alternatively, it was hypothesized that the cytokeratins may translocate to the outer membrane by covalently binding to plasma membrane lipids (29). We have used a variety of techniques to demonstrate that COU-1 binds to cytokeratin exposed at the outer surface of the cell membrane. Using viable cells stained at 4°C, we found COU-1 distributed in a punctate pattern at the cell surface. This pattern was distinct from the fibrillar staining observed when the plasma membrane was disrupted under nonphysiologic conditions. We also found that the level of cell surface cytokeratin could be increased by culturing cells on fibronectin, laminin, and collagen IV. Again, this cytokeratin was not fibrillar, but instead gave an enhanced punctate staining, significantly enriched at points of cell-to-cell contact.
It is not known how modified cytokeratins 8 and 18 are associated with the cell surface. It has been suggested that this may be due to secondary bindings of cytokeratin found in the extracellular fluid (28), and cytokeratin 8 fragments have been identified in the culture media of MCF-7 cells (15, 30) and in the serum of cancer patients (31). Alternatively, overexpression of cytokeratin by transformed cells leads to excess protein accumulating at the cell surface due to inefficient incorporation into intermediate filaments. We now show that cell surface cytokeratin cycles to an intracellular endocytic compartment. This internalization of cytokeratin was not induced by crosslinking of the antibody, since Fab COU-1 and the whole IgM gave comparable results. Further, we observed some cytoplasmic vesicles stained with COU-1 in fixed preparations, implying that modified cytokeratin may cycle between the cell surface and the endocytic compartment. A possible explanation may be that excess protein is processed via the endocytic compartment and eventually degraded in lysosomes. Recycling vesicles may then shuttle some cytokeratin to the cell surface. The internalization of monovalent Fab COU-1 is consistent with this idea and is the first step in dissecting this process. In addition, factors that stimulate cell growth might further increase the level of cell surface cytokeratin if there is an imbalance between cytokeratin synthesis and their organization into filaments. We found that fibronectin, which stimulates tyrosine phosphorylation and cell proliferation (32), caused a significant increase in nonfibrillar cell surface cytokeratin.
The distribution of the antigen recognized by COU-1 in ethanol-fixed biopsies revealed that COU-1 could discriminate between malignant and normal colon epithelia, and also between colon cancer metastases and surrounding normal liver tissue. In contrast, two murine mAbs directed against normal cytokeratins 8 and 18 reacted equally well with colon cancer and normal colon epithelia and the majority of normal hepatocytes. These observations extend our previous comparative studies of four different tissue preparation methods that identified alcohol fixation as the preferred method of tissue preparation for COU-1 (3). Altmannsberger et al. (33) also found ethanol superior to formaldehyde for the preservation of intermediate filament proteins (keratin, vimentin, and desmin) in a variety of human tissues which were subsequently paraffin-embedded.
The demonstration of modified cytokeratin as a potential new tumor target at the surface of carcinoma cells combined with the promising results of the early phase I/II clinical study of radiolabeled COU-1 is very encouraging. Comparison of recombinant Fab COU-1 and the whole IgM for in vivo tumor localization and targeting of immunoconjugates warrants further investigation.
Acknowledgments
We are grateful to Karin Kejling, Helle Ladefoged, Anne Andersen, Helen Wan, Per Borup-Christensen, and Ingrid Titlestad for their contribution to this work.
ABBREVIATIONS
- HumAb
human monoclonal antibody
- BSA
bovine serum albumin
- AP
alkaline phosphatase
- FITC
fluorescein isothiocyanate
- HRP
horseradish peroxidase
Footnotes
References
- 1.Borup-Christensen P, Erb K, Jensenius J C, Nielsen B, Svehag S-E. Int J Cancer. 1986;37:683–688. doi: 10.1002/ijc.2910370507. [DOI] [PubMed] [Google Scholar]
- 2.Erb K, Borup-Christensen P, Ditzel H, Chemnitz J, Haas H, Jensenius J C. Hybridoma. 1992;11:121–134. doi: 10.1089/hyb.1992.11.121. [DOI] [PubMed] [Google Scholar]
- 3.Ditzel H, Erb K, Borup-Christensen P, Nielsen B, Jensenius J C. Hum Antibodies Hybridomas. 1991;2:135–141. [PubMed] [Google Scholar]
- 4.Borup-Christensen P, Erb K, Ditzel H, Nielsen B, Larsen J K, Svehag S-E, Jensenius J C. AMPIS. 1996;98:674–684. doi: 10.1111/j.1699-0463.1990.tb04988.x. [DOI] [PubMed] [Google Scholar]
- 5.Ditzel H, Rasmussen J W, Erb K, Borup-Christennsen P, Titlestad I, Lassen E, Fenger C, Kronborg O, Jensenius J C. Cancer Res. 1993;53:5920–5928. [PubMed] [Google Scholar]
- 6.Ditzel H, Erb K, Leslie G, Jensenius J C. Hum Antibodies Hybridomas. 1993;4:86–93. [PubMed] [Google Scholar]
- 7.Ditzel H, Rasmussen J W, Erb K, Jensenius J C. Eur J Nucl Med. 1992;19:409–417. doi: 10.1007/BF00177367. [DOI] [PubMed] [Google Scholar]
- 8.Siegel D L, Silberstein L E. Blood. 1994;83:2334–2344. [PubMed] [Google Scholar]
- 9.Barbas C F, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry R M, Nara P L, Burton D R. Proc Natl Acad Sci USA. 1994;91:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W-P, Gren K, Pinz-Sweeney S, Briones A T, Burton D R, Barbas C F. J Mol Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 11.Brandtzaeg P, Rognum T O. Pathol Res Pract. 1984;179:250–266. doi: 10.1016/S0344-0338(84)80142-9. [DOI] [PubMed] [Google Scholar]
- 12.Kuan C-T, Pastan I. Proc Natl Acad Sci USA. 1996;93:974–978. doi: 10.1073/pnas.93.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker J C, Pancook J D, Gillies S D, Mendelsohn J, Reisfeld R A. Proc Natl Acad Sci USA. 1996;93:2702–2707. doi: 10.1073/pnas.93.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hembrough T A, Vasudevan J, Allietta M M, Glass W F, Gonias S L. J Cell Sci. 1995;108:1071–1082. doi: 10.1242/jcs.108.3.1071. [DOI] [PubMed] [Google Scholar]
- 15.Godfroid E, Geuskens M, Dupressoir T, Parent I, Szpirer C. J Cell Sci. 1991;99:595–607. doi: 10.1242/jcs.99.3.595. [DOI] [PubMed] [Google Scholar]
- 16.Donald P J, Cardiff R D, He D, Kendall K. Otolaryngol Head Neck Surg. 1991;105:781–787. doi: 10.1177/019459989110500603. [DOI] [PubMed] [Google Scholar]
- 17.Haspel M V, McCabe R P, Pomato N, Janesch N J, Knowlton J V, Peters L C, Hoover H C, Hannah M G. Cancer Res. 1985;45:3951–3961. [PubMed] [Google Scholar]
- 18.Steis R G, Carrasquillo J A, McCabe R, Bookman M A, Reynolds J C, Larson S M, Smith J W, Clark J W, Dailey V, Del Vecchio S, Shuke N, Pinsky C M, Urba W J, Haspel M, Perentesis P, Paris B, Longo D L, Hanna M G. J Clin Oncol. 1990;8:476–490. doi: 10.1200/JCO.1990.8.3.476. [DOI] [PubMed] [Google Scholar]
- 19.Boven E, Haisma H J, Bril H, Martens H J, van Lingen A, den Hollander W, Kessel M A, DeJager R L, Roos J C. Eur J Cancer. 1991;27:1430–1436. doi: 10.1016/0277-5379(91)90025-9. [DOI] [PubMed] [Google Scholar]
- 20.Rosenblum M G, Levin B, Rohn D, Hohn D, McCabe R, Thompson L, Cheung L, Murray J L. Cancer Immunol Immunother. 1994;39:397–400. doi: 10.1007/BF01534427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson M A, Caothien R H, Burton D R. Proc Natl Acad Sci USA. 1991;88:2432–2436. doi: 10.1073/pnas.88.6.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burton D R, Barbas C F, Persson M A A, Koenig S, Chanock R M, Lerner R A. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbas C F, Kang A S, Lerner R A, Benkovic S J. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ditzel H J, Binley J M, Moore J P, Sodroski J, Sullivan N, Sawyer L S W, Hendry R M, Yang W-P, Barbas C F, Burton D R. J Immunol. 1995;154:895–908. [PubMed] [Google Scholar]
- 25.Nishibori H, Matsuno Y, Iwaya K, Osada T, Kubomura N, Iwamatsu A, Kohno H, Sato S, Kitajima M, Hirohashi S. Cancer Res. 1996;56:2752–2757. [PubMed] [Google Scholar]
- 26.Hembrough T, Li L, Gonias S. FASEB J. 1996;10:A1417. (abstr.). [Google Scholar]
- 27.Erb K, Ditzel H, Rasmussen J W, Borup-Christensen P, Jensenius J C. Hum Antibodies Hybridomas. 1991;2:215–221. [PubMed] [Google Scholar]
- 28.Riopel C L, Butt I, Omary M B. Cell Motil Cytoskeleton. 1993;26:77–87. doi: 10.1002/cm.970260108. [DOI] [PubMed] [Google Scholar]
- 29.Asch H L, Mayhew E, Lazo R O, Asch B B. Mol Biol Int. 1993;29:1161–1169. [PubMed] [Google Scholar]
- 30.Chan R, Rossitto P V, Edwards B F, Cardiff R D. Cancer Res. 1986;46:6353–6359. [PubMed] [Google Scholar]
- 31.Bjorklund B, Bjorklund V. Cancer Detect Prev. 1983;6:41–50. [PubMed] [Google Scholar]
- 32.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 33.Altmannsberger M, Osborn M, Schauer A, Weber K. Lab Invest. 1981;45:427–434. [PubMed] [Google Scholar]