Abstract
The IncN plasmids R46 and N3 each contain two copies of an insertion sequence which we denote IS46. This insertion sequence has single PstI and SalI restriction sites and is 0.81 kilobases long. All four copies of IS46 were capable of forming cointegrates, although the DNA between the insertion sequences, which in each case carries a tetracycline resistance gene, was not transposable in the form of a compound transposon. IS46-mediated cointegrates resolved in Rec+ but not in RecA- cells. Recombination between two copies of IS46, causing an inversion, accounts for the existence of two distinct forms of R46. IS46-mediated deletions were probably responsible for the formation of the plasmid pKM101 from R46. IS46 was not homologous to IS1 but did show homology with IS15.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T. F. Reflections on phage genetics. Annu Rev Genet. 1981;15:405–417. doi: 10.1146/annurev.ge.15.120181.002201. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Egner C., Hirschel B. J., Howard J., Johnsrud L., Jorgensen R. A., Tlsty T. D. Insertion, excision, and inversion of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):115–123. doi: 10.1101/sqb.1981.045.01.020. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Brown A. M., Willetts N. S. A physical and genetic map of the IncN plasmid R46. Plasmid. 1981 Mar;5(2):188–201. doi: 10.1016/0147-619x(81)90020-2. [DOI] [PubMed] [Google Scholar]
- Bräu B., Piepersberg W. Cointegrational transduction and mobilization of gentamicin resistance plasmid pWP14a is mediated by IS140. Mol Gen Genet. 1983;189(2):298–303. doi: 10.1007/BF00337820. [DOI] [PubMed] [Google Scholar]
- Chandler M., Clerget M., Galas D. J. The transposition frequency of IS1-flanked transposons is a function of their size. J Mol Biol. 1982 Jan 15;154(2):229–243. doi: 10.1016/0022-2836(82)90062-6. [DOI] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Phenotypic properties of R factors of Pseudomonas aeruginosa: R factors readily transferable between Pseudomonas and the Enterobacteriaceae. Genet Res. 1974 Jun;23(3):239–250. doi: 10.1017/s0016672300014890. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. Isolation of an IS1 flanked kanamycin resistance transposon from R1drd19. Mol Gen Genet. 1980;180(1):123–127. doi: 10.1007/BF00267360. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N., Hedges R. W. R factors from Proteus rettgeri. J Gen Microbiol. 1972 Oct;72(3):543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- Crisona N. J., Nowak J. A., Nagaishi H., Clark A. J. Transposon-mediated conjugational transmission of nonconjugative plasmids. J Bacteriol. 1980 May;142(2):701–713. doi: 10.1128/jb.142.2.701-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Compatibility groups among fi - R factors. Nature. 1971 Nov 26;234(5326):222–223. doi: 10.1038/234222a0. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., Willetts N. S. Plasmid co-integrates of prophage lambda and R factor R100. J Bacteriol. 1976 Apr;126(1):166–176. doi: 10.1128/jb.126.1.166-176.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd H. M., Bennett P. M. R46 encodes a site-specific recombination system interchangeable with the resolution function of TnA. Plasmid. 1983 May;9(3):247–261. doi: 10.1016/0147-619x(83)90003-3. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. On the molecular mechanisms of transposition. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4858–4862. doi: 10.1073/pnas.78.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Harshey R. M., Bukhari A. I. A mechanism of DNA transposition. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1090–1094. doi: 10.1073/pnas.78.2.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W. Genetics of Pseudomonas. Bacteriol Rev. 1969 Sep;33(3):419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Iida S., Hänni C., Echarti C., Arber W. Is the IS1-flanked r-determinant of the R plasmid NR1 a transposon? J Gen Microbiol. 1981 Oct;126(2):413–425. doi: 10.1099/00221287-126-2-413. [DOI] [PubMed] [Google Scholar]
- Iida S., Meyer J., Linder P., Goto N., Nakaya R., Reif H. J., Arber W. The kanamycin resistance transposon Tn2680 derived from the R plasmid Rts1 and carried by phage P1Km has flanking 0.8-kb-long direct repeats. Plasmid. 1982 Sep;8(2):187–198. doi: 10.1016/0147-619x(82)90056-7. [DOI] [PubMed] [Google Scholar]
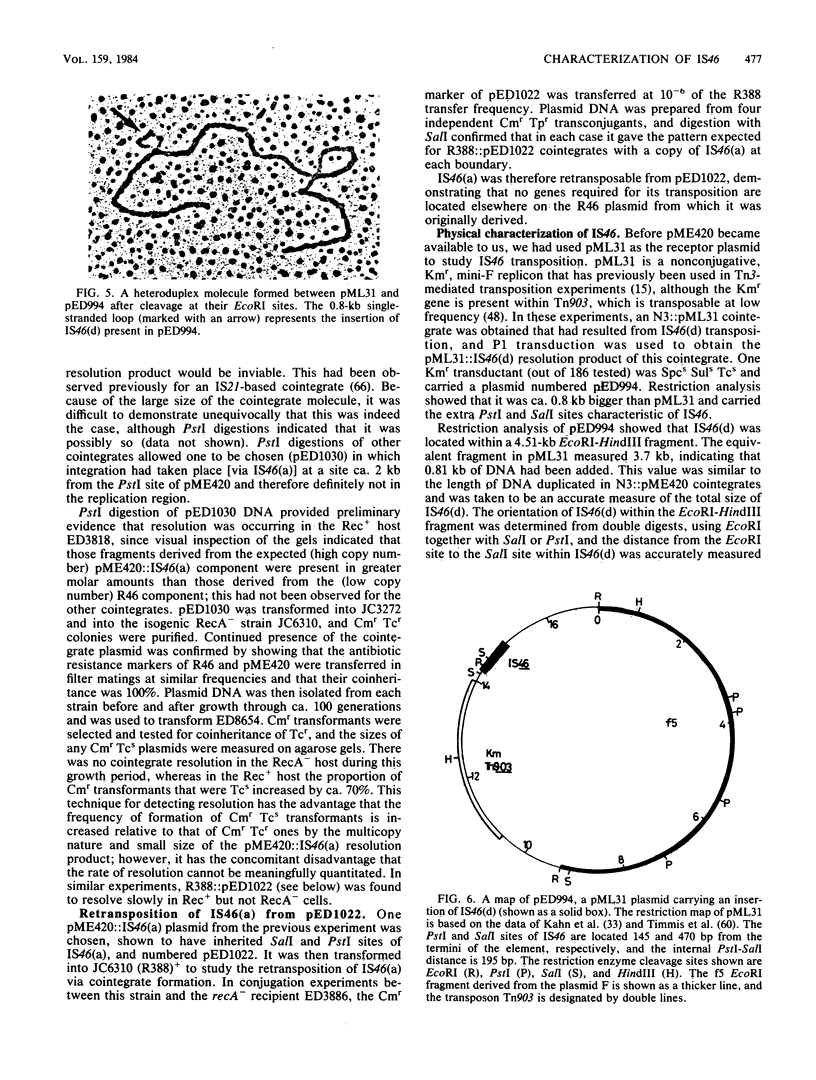
- Kahn M. L., Figurski D., Ito L., Helinski D. R. Essential regions for replication of a stringent and a relaxed plasmid in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):99–103. doi: 10.1101/sqb.1979.043.01.015. [DOI] [PubMed] [Google Scholar]
- Konarska-Kozlowska M., Iyer V. N. Physical and genetic organization of the IncN-group plasmid pCU1. Gene. 1981 Aug;14(3):195–204. doi: 10.1016/0378-1119(81)90115-3. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A., Courvalin P. IS15, a new insertion sequence widely spread in R plasmids of gram-negative bacteria. Mol Gen Genet. 1983;189(1):102–112. doi: 10.1007/BF00326061. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A., Gerbaud G., Courvalin P. Translocation of sequences encoding antibiotic resistance from the chromosome to a receptor plasmid in Salmonella ordonez. Mol Gen Genet. 1981;182(3):390–408. doi: 10.1007/BF00293927. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A., Witchitz J., Courvalin P. Modular evolution of disseminated Inc 7-M plasmids encoding gentamicin resistance. Plasmid. 1982 Nov;8(3):215–231. doi: 10.1016/0147-619x(82)90060-9. [DOI] [PubMed] [Google Scholar]
- Langer P. J., Shanabruch W. G., Walker G. C. Functional organization of plasmid pKM101. J Bacteriol. 1981 Mar;145(3):1310–1316. doi: 10.1128/jb.145.3.1310-1316.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
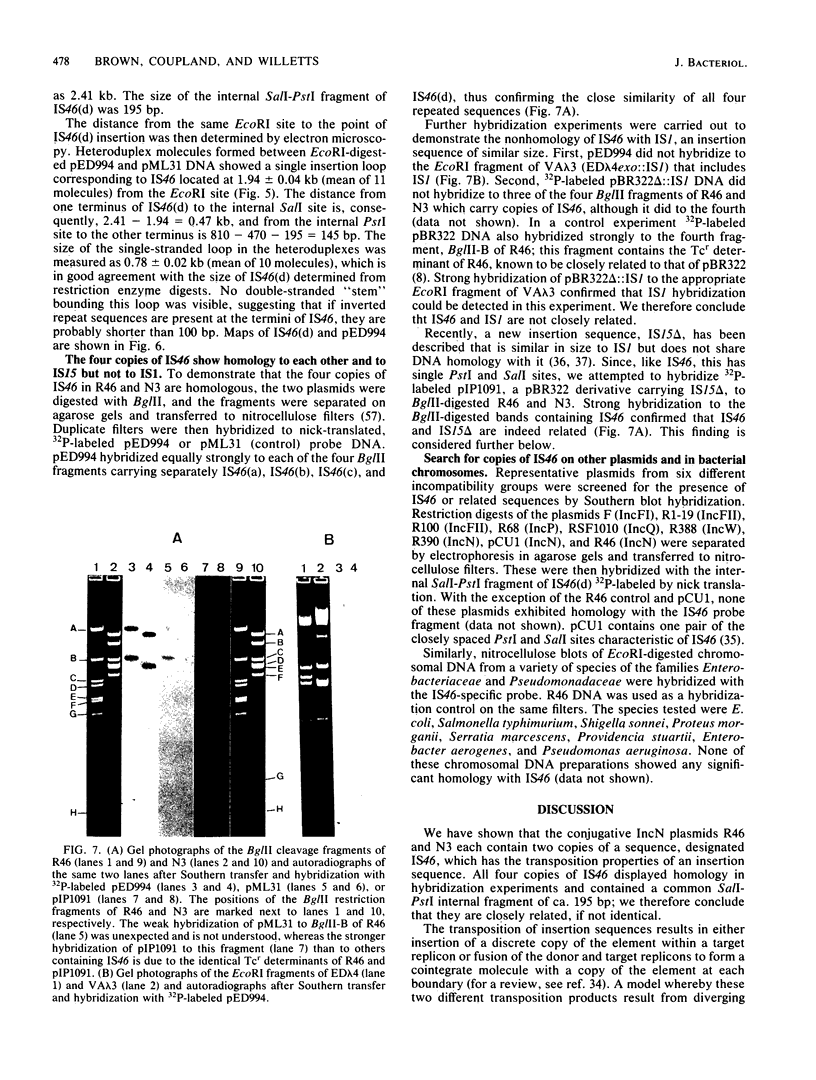
- Langer P. J., Walker G. C. Restriction endonuclease cleavage map of pKM101: relationship to parental plasmid R46. Mol Gen Genet. 1981;182(2):268–272. doi: 10.1007/BF00269669. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J., Spingarn N. E., Kobori J., Ames B. N. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci U S A. 1975 Mar;72(3):979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez B., Tachibana C., Levy S. B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980 Mar;3(2):99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- Meynell E., Datta N. Mutant drug resistant factors of high transmissibility. Nature. 1967 May 27;214(5091):885–887. doi: 10.1038/214885a0. [DOI] [PubMed] [Google Scholar]
- Mortelmans K. E., Stocker B. A. Segregation of the mutator property of plasmid R46 from its ultraviolet-protecting property. Mol Gen Genet. 1979 Jan 2;167(3):317–327. doi: 10.1007/BF00267425. [DOI] [PubMed] [Google Scholar]
- Nomura N., Yamagishi H., Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene. Gene. 1978 Feb;3(1):39–51. doi: 10.1016/0378-1119(78)90006-9. [DOI] [PubMed] [Google Scholar]
- Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature. 1981 Feb 12;289(5798):609–612. doi: 10.1038/289609a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K. L., Walker G. C. Identification of plasmid (pKM101)-coded proteins involved in mutagenesis and UV resistance. Nature. 1982 Nov 18;300(5889):278–281. doi: 10.1038/300278a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saedler H., Heiss B. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol Gen Genet. 1973 May 9;122(3):267–277. doi: 10.1007/BF00278602. [DOI] [PubMed] [Google Scholar]
- Smith D. I., Blattner F. R., Davies J. The isolation and partial characterization of a new restriction endonuclease from Providencia stuartii. Nucleic Acids Res. 1976 Feb;3(2):343–353. doi: 10.1093/nar/3.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M., Heffron F., McCarthy B. J. The E. coli gene encoding heat stable toxin is a bacterial transposon flanked by inverted repeats of IS1. Nature. 1979 Feb 8;277(5696):453–456. doi: 10.1038/277453a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Timmis K. N., Cabello F., Cohen S. N. Cloning and characterization of EcoRI and HindIII restriction endonuclease-generated fragments of antibiotic resistance plasmids R6-5 and R6. Mol Gen Genet. 1978 Jun 14;162(2):121–137. doi: 10.1007/BF00267869. [DOI] [PubMed] [Google Scholar]
- WATANABE T., NISHIDA H., OGATA C., ARAI T., SATO S. EPISOME-MEDIATED TRANSFER OF DRUG RESISTANCE IN ENTEROBACTERIACEAE. VII. TWO TYPES OF NATURALLY OCCURRING R FACTORS. J Bacteriol. 1964 Sep;88:716–726. doi: 10.1128/jb.88.3.716-726.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S., Crowther C., Holloway B. W. The insertion sequence IS21 of R68.45 and the molecular basis for mobilization of the bacterial chromosome. Plasmid. 1981 Jul;6(1):30–52. doi: 10.1016/0147-619x(81)90052-4. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Finnegan D. J. Characteristics of E. coli K12 strains carrying both an F prime and an R factor. Genet Res. 1970 Aug;16(1):113–122. doi: 10.1017/s0016672300002329. [DOI] [PubMed] [Google Scholar]
- Willetts N. S. Recombination and the Escherichia coli K-12 sex factor F. J Bacteriol. 1975 Jan;121(1):36–43. doi: 10.1128/jb.121.1.36-43.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Maule J. Investigations of the F conjugation gene traI:traI mutants and lambdatraI transducing phages. Mol Gen Genet. 1979 Feb 1;169(3):325–336. doi: 10.1007/BF00382278. [DOI] [PubMed] [Google Scholar]
- Willetts N. The transcriptional control of fertility in F-like plasmids. J Mol Biol. 1977 May 5;112(1):141–148. doi: 10.1016/s0022-2836(77)80161-7. [DOI] [PubMed] [Google Scholar]