Abstract
The human hepatitis B virus (HBV) protein pX is a multifunctional regulatory protein that is known to affect both transcription and cell growth. Here we describe induction of apoptosis in NIH 3T3 polyclonal cell lines upon stimulation of pX expression from a dexamethasone inducible mouse mammary tumor virus (MMTV)-X expression vector. The effect of long-term pX expression on the cell survival of mouse fibroblasts was confirmed in colony generation assays. This effect is not shared either by the other HBV products and it is c-myc mediated, as shown by the use of a dominant negative deletion mutant of c-myc. pX also sensitize cells to programmed cell death after exposure to DNA damaging agents. Taking advantage of stable transfectants carrying the p53val135 temperature-sensitive allele, we directly demonstrate that induction of apoptosis by pX requires p53. In p53 null mouse embryo fibroblasts pX activates transcription and confers an evident growth advantage without loss of cell viability. Although pX protein was not detectable in the experimental conditions we used, our results indicate that its expression affects both cell growth and cell death control.
The hepatitis B virus (HBV) protein pX has been shown to function as a promiscous transactivator of many viral and cellular promoters, including those of genes involved in cell growth regulation—i.e., c-fos, c-jun, c-myc, and epidermal growth factor (EGF) receptor (1–4). Recent studies have demonstrated that pX is a dual function nuclear and cytoplasmic protein (5) able to activate specific transcriptional elements in the nucleus (5–8) and Ras dependent and independent signaling pathways in the cytoplasm (9–14), which are required for the activation of AP1 and NF-κB transcription factors. pX performs essential functions for viral infection (15, 16) and, similar to other viral regulatory proteins, possesses transforming properties, as assessed both in vivo [in the transgenic mouse model (17)] and in vitro [in immortalized mouse hepatocytes (18)]. Nevertheless, it is controversial whether pX transactivating activity itself is relevant for HBV-associated liver carcinogenesis or if other pX or HBV functions have to be considered. Aberrant induction of cell proliferation by viral oncoproteins (i.e., E1a, E7) is known to prompt cells to p53-dependent programmed cell death (19–22). Indeed, full and efficient transformation of rodent cells by E1a or E7 requires binding and inactivation of p53 by other viral products (E1b55Kd, E6) (19–22). Despite its ability to induce cell cycle progression (23), to bind p53 (24, 25), and to inactivate its transactivating function (24, 25), the relative potency of pX in the transformation of mouse fibroblasts in in vitro assays is unexpectedly low (26), and it is clearly enhanced by the presence of the entire HBV genome (26). Here we describe the effects of HBV X gene on cell viability, and characterize, using different experimental approaches, their dependency on wild-type p53 expression.
MATERIALS AND METHODS
Cell Culture.
REF-52 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. NIH 3T3, BALB/c3T3 cells and p53−/− mouse embryo fibroblasts (MEFs) were maintained in DMEM supplemented with 10% newborn calf serum. The BALB/ctsp53 clone, carrying the p53-val135 temperature sensitive (ts) allele and the parental BALB/c3T3 cells, are described elsewhere (27). MEFs from p53 null mice (p53−/− MEFs) have been described (28).
Plasmids.
The pX expression vector pMMTV-X or the pMMTV-CAT control plasmid, both carrying the hygromycin resistance gene, are derived from the Epstein–Barr virus-based episomal plasmid pHEBo (29). The E2F1 cDNA was cloned into the pcDNA1 (Invitrogen) as a BamHI–XbaI fragment to obtain pCMV-E2F1. pE1A(12S) express the E1A(12S) under the control of its own regulatory sequences.
Chloramphenicol Acetyltransferase (CAT) Assays.
For the transient expression assays p53−/− MEFs were plated at 6 × 105 per 6-cm dish and cultured in DMEM containing 1% serum for 24 hr before transfection. The cells were then transfected using the calcium phosphate coprecipitation technique as described (13). All experiments were repeated three to five times with at least two different preparations of DNA.
Nuclear Labeling Index.
REF-52 cells were transfected by the calcium phosphate coprecipitation method using 5 μg of pSV-CAT, pUC18 as a nonspecific carrier, and 1 μg of pSV-X or 500 ng of pCMV-E2F1 and pE1a(12S), where appropriate, for a total of 20 μg in 1 ml. The DNA precipitate (0.1 ml) was added to each 100 mm Lab-Tek cell culture chamber slide well (Nunc). Cells were exposed to the DNA precipitates for 18 hr, washed, and then placed in 0.1 fetal calf serum containing DMEM for 48 hr. 5-Bromodeoxyuridine (BrdUrd; Sigma), at a final concentration of 10 μM, was then added and the cells were further incubated for 12 hr before being processed as described (30). For stable transfections, early passages NIH 3T3 cells were transfected with 10 μg of either pMMTV-X or pMMTV-CAT by the calcium phosphate method. On the following day, cells were replated and selected in medium. After 2 weeks of selection in medium containing hygromycin at a concentration of 400 μg/ml, resistant clones were pooled and plated in 100 mm Lab-Tek cell culture chamber slides (Nunc) at 1 × 104 cells per well in 0.1% fetal calf serum containing DMEM. After 48 hr cells were exposed to BrdUrd with or without dexamethasone (Sigma) at a concentration of 10−6 M for 24 hr before being stained as for BrdUrd incorporation as described (30). Alternatively, serum-starved cells were examined for morphological changes after 24, 48, and 72 hr of dexamethasone stimulation to induce pX or control gene expression.
Colony Generation Assays.
Early passage NIH 3T3 cells were cotransfected with the indicated amounts of the different expression vectors and a fixed amount (200 ng) of the pAG60 selection plasmid. Twenty-four hours after cotransfection cells were split and selected in G418 (0.7 mg/ml) (Sigma) for 2–3 weeks. Resistant colonies were counted after fixing and staining plates in a solution containing crystal violet (Sigma) in 20% ethanol.
Western Blot Analysis.
Immunoblots were performed as described (30). The anti-p53 antibody pAb2 (Oncogene Science) has been used at the dilution of 1:200.
DNA Fragmentation Analysis.
Unexpanded pSV-X and pSV-CAT colonies, obtained as described above, were treated with 0.1 mg/ml of etoposide for 48 hr and then incubated for 5 min with bisbenzimide H 33258 at a concentration of 1 mg/ml (Boehringer Mannheim) for chromatin staining. Low molecular weight DNA was isolated from 106 adherent and nonadherent pSV-X or pSV-CAT pooled cells 48 hr after exposure to etoposide cells. DNA was resuspended in TE buffer, resolved on a 1.5% agarose gel, and visualized by ethidium bromide staining as described (31).
RESULTS AND DISCUSSION
pX Induces Both DNA Synthesis and Loss of Cell Viability in Serum-Starved Cells.
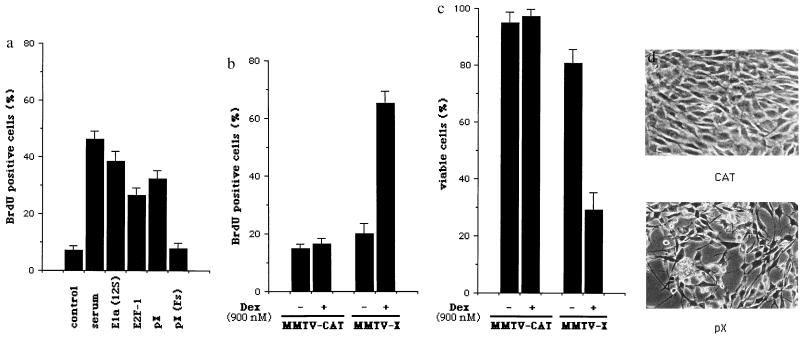
The ability of pX to induce cell cycle progression was assessed both in transiently transfected quiescent rat embryo fibroblasts (REF-52 cells) and in NIH 3T3-derived polyclonal cell lines in which pX expression is under the control of the dexamethasone inducible mouse mammary tumor virus (MMTV) promoter. The synthesis of DNA in REF-52 cells was measured by BrdUrd incorporation into cells that were transfected with various plasmids and then deprived of serum. Cotransfection of the CAT gene expressing plasmid pSV-CAT (1) allowed the identification of cells that were successfully transfected. The addition of serum to a culture of quiescent REF-52 cells induces DNA synthesis in about 45% of the CAT positive cells (Fig. 1a). Using this assay, we found that cotransfection of the pX expression vector pSV-X (1), but not of the pSV-X(FS) mutant (1), results in a large proportion of the CAT positive REF-52 cells becoming BrdUrd positive, with an efficiency similar to that of expression vectors for E2F1, a key regulator of cell proliferation (32), and E1a (Fig. 1a). However, from these experiments, it is not possible to determine whether pX maintains cells in a proliferative state by preventing cell cycle exit or whether it drives otherwise quiescent cells to enter S phase. To address this question, NIH 3T3 cells were transfected with the pX expression vector pMMTV-X, which also contains the gene for hygromycin selection, and resistant clones were pooled. In these polyclonal NIH 3T3 cell lines, made quiescent by serum deprivation, dexamethasone resulted in a sharp increase in the number of BrdUrd positive cells (Fig. 1b). Interestingly, while polyclonal cell lines transfected with the control pMMTV-CAT vector display the typical appearance of serum-starved NIH 3T3 cells, between 48 to 72 hr after dexamethasone stimulation up to 60% of the pMMTV-X cells die, as determined by trypan blue dye exclusion (Fig. 1c), are found to round up, display cytoplasmic blebs, and detach (Fig. 1d). These experiments demonstrate that pX is capable of inducing S phase in resting cells and that its overexpression can alter cell viability.
Figure 1.
pX induces both DNA synthesis and loss of cell viability in serum-starved cells. (a) REF-52 cells were transfected with pSV-CAT together with pSV-X, pSV-X(Fs), pCMV-E2F1, and pE1A(12S). The percentage of the CAT positive REF-52 cells also positively displaying BrdUrd was calculated. The data plotted represent the average of three independent experiments. Error bars refer to standard error (SEM). (b) Early passage NIH 3T3 cells were transfected with either the pX expression vector pMMTV-X or the pMMTV-CAT control plasmid. Resistant clones were pooled, made quiescent by serum deprivation, and visualized for BrdUrd incorporation in the presence or absence of dexamethasone. pX transcripts were barely detectable in unstimulated cells and increased several fold after dexamethasone treatment, as assessed by Northern blot analysis (data not shown). Plotted data represent the average of three independent experiments ± standard error (SEM). (c) Serum-starved NIH 3T3 polyclonal populations, obtained as described above, were examined for cell loss by trypan blue dye exclusion and morphological changes at different times after induction of pX or CAT expression. A representative experiment at 72 hr after dexamethasone stimulation is shown (d).
Long-Term pX Expression Reduces Cell Clonogenic Survival in Mouse Fibroblasts Expressing Wild-Type p53.
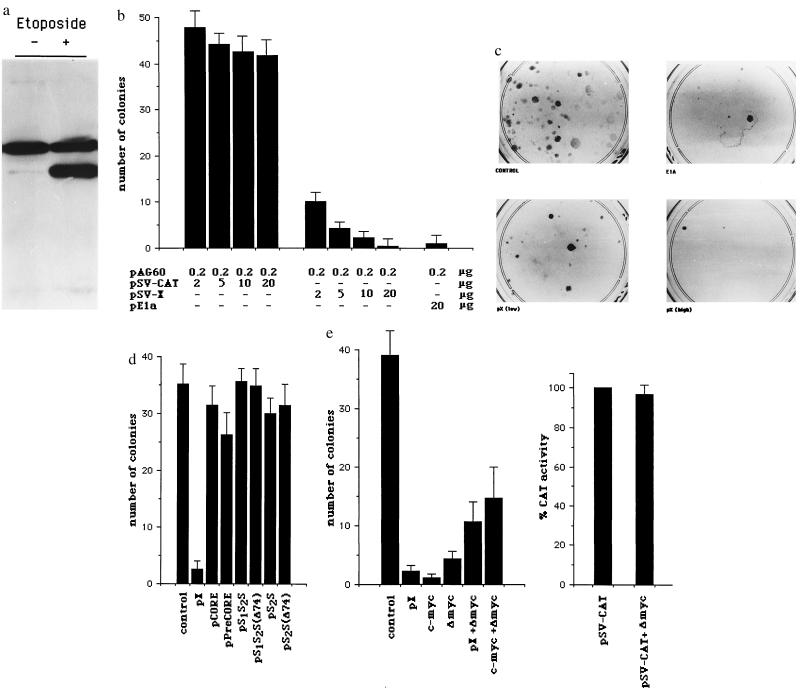
These observations prompted us to evaluate whether pX proliferative potential is also associated with induction of apoptosis, thus explaining its low oncogenic potential when expressed alone in vitro (26). Indeed, the aberrant induction of cell proliferation by viral and cellular oncoproteins, such as E1a and c-myc, is coupled to a cell suicide response that is mediated by the tumor suppressor protein p53 (19, 20, 22, 28, 33). Although E1a- and c-myc-associated apoptosis can be observed during normal propagation of cells, the level of cell death dramatically increases when cells are in conditions of growth limitation, as realized by mitogens deprivation or high cell density (20, 33). Clonogenic assays appear to be a very sensitive way to assess the phenomenon (34). Therefore, to determine the long-term effects of pX expression on cell survival, we performed colony generation assays either in mouse fibroblasts expressing wild-type p53 or in p53-null cells. Early passages NIH 3T3 cells were cotransfected with either the pX expression vector pSV-X or the pSV-CAT control plasmid and the plasmid pAG60, carrying the geneticin resistance gene. p53 expression in these cells was determined by indirect immunofluorescence using the anti-p53 monoclonal antibody pAb122 (Oncogene Science), which reacts with both the native and mutant conformers, and p53 status with pAb246 (Oncogene Science), which reacts only with the native protein. With both antibodies a weak and predominantly nuclear staining was obtained (data not shown), a pattern compatible with the presence of wild-type p53 protein. p53 wild-type status was further confirmed by its accumulation after treatment of cells with etoposide (Fig. 2a). The number of pX stable transfectants obtained is always significantly lower than in controls (Fig. 2 b and c). Titration of 2–20 μg of the pX expression vector demonstrates a tight correlation between the amount of transfected plasmid and the effect on cell survival (Fig. 2b). Inhibition of colony formation in this type of assay can be explained by either a block of cell cycling or induction of cell death. However, as shown in Fig. 1b, in the same cells we have evidence that pX induces efficiently cell cycle progression, thus ruling out the first possibility. The observed effect is specific to pX, in that none out of the other HBV products has similar effects on cell viability (Fig. 2d). Interestingly, the truncated preS/S transactivating proteins of HBV (38), which share with pX many target genes (35), do not affect the clonogenic potential in this system (Fig. 2d). Moreover, in REF-52 cells, in which E1a expression does not result in an evident loss of cell viability (ref. 20 and data not shown), pX expression does not affect colony formation (data not shown). Because pX up-regulates c-myc gene expression in several cell lines (ref. 3; P.C., unpublished observations) and c-myc functions as a transcriptional regulator controlling both cell proliferation and cell death (28, 33), we studied the effects of a dominant negative deletion mutant of c-myc (ΔMyc) (37) on pX ability to reduce colony formation in NIH 3T3 cells. The expression of ΔMyc per se results in a decreased number of colonies (Fig. 2e,), probably as a consequence of its negative effect on cell growth (37). ΔMyc expression in cotransfection experiments does not affect c-myc and pX expression (Fig. 2e) but greatly reduces the negative effects of both c-myc and pX on NIH 3T3 colony formation (Fig. 2e), strongly suggesting that pX modulation of cell survival in our system is indeed mediated by c-myc.
Figure 2.
Long-term pX expression reduces cell clonogenic survival in mouse fibroblasts expressing wild-type p53. (a) p53 immunoblotting in untreated and etoposide-treated NIH 3T3 cells using pAb240, which detects both mutant and wild-type p53 in Western blot analysis. The upper band, present in both untreated and etoposide-treated cells, is unspecific. The lower band, whose intensity greatly increases in etoposide-treated cells, is p53. (b) Early passage NIH 3T3 cells were cotransfected with increasing amounts (2–20 μg, as indicated) of either pSV-X or control pSV-CAT expression vectors, pSV-0 as a nonspecific carrier, for a total of 20 μg/ml of DNA, and a fixed amount (200 ng) of the pAG60 geneticin selection plasmid. Resistant colonies were counted after 2–3 weeks. For each experimental condition plotted data represent the average number of colonies obtained in five independent experiments ± standard error (SEM). In c, stained colonies from a representative experiment are shown. (d) Colony generation assays in NIH 3T3 cells using expression vectors for the HBV core, precore/core, large (preS1S2S) and middle (preS2S) envelope proteins (35, 36) and for the truncated envelope proteins with transactivating properties preS1S2S(D74) and preS2S(D74) (35). (e) c-myc mediates pX effects on cell clonogenic survival in NIH 3T3 cells. Cells were cotransfected with pSV-X (10 μg), pCMV-Myc (10 μg), and pCMV-ΔMyc (10 μg), alone or in combination and 200 ng of pAG60 (Left). pCMV-ΔMyc encodes for a c-myc protein carrying a large deletion (amino acids 70–178) in the N-terminal transactivation domain and acts as a dominant negative mutant of wild-type c-myc (37). Colony formation was scored and represented as in a. Cotransfection of pCMV-ΔMyc (2 μg) does not influence the simian virus 40 (SV40) promoter-driven CAT expression (pSV-CAT, 2 μg) (Right).
pX Clones Are Sensitized to p53-Mediated Apoptosis.
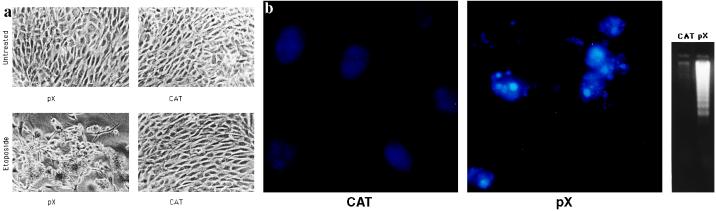
Oncogene overexpression increases cell susceptibility to programmed cell death induced by a variety of exogenous stimuli, including several anti-cancer drugs, which share the ability to increase p53 levels in the cell (39) and tumor necrosis factor α (40). To probe the role of p53 in pX induced loss of cell viability we treated pX clones with low dosages (0.1 mg/ml) of the topoisomerase II inhibitor etoposide. Colony regression (Fig. 3a) was observed in 69% of NIH 3T3 pX clones as compared with 9% in the control SV-CAT clones (Table 1). During apoptosis, loss of membrane integrity is typically preceded by chromatin condensation and internucleosomal cleavage of genomic DNA. Colony regression of pX clones is indeed due to apoptosis, as clearly demonstrated by both the fragmentation of Hoechst 33258 stained nuclei (Fig. 3 b Center) and by DNA ladder formation (Fig. 3 b Right). Since at the concentration used etoposide requires p53 for the execution of the death program in oncogene prompted cells (20), our results are consistent with the hypothesis that pX sensitizes cells to p53-mediated programmed cell death and alters the cell response to DNA damage from growth arrest to apoptosis. To evaluate more directly the role of p53 status in pX induced cell death we made use of a well-characterized 3T3-derived clone carrying the p53-val135 temperature sensitive (ts) allele (BALB/ctsp53) (27). As expected, DNA synthesis, assessed by BrdUrd incorporation, dramatically decreased after shifting the cells at the permissive temperature (32°C), due to the expression of the p53-val135 protein in a native active conformation (data not shown). At the nonpermissive temperature (38°C) the p53-val135 protein indeed behave as a dominant interfering mutant, and inhibition of DNA synthesis was completely reversed (ref. 27 and data not shown). BALB/c3T3-val135 cells were cotransfected with either pSV-X or pSV-CAT and the pSV-Hygro expression vectors and selected in hygromycin containing cell culture medium at the nonpermissive temperature. When pSV-X or pSV-CAT unexpanded colonies were shifted to the permissive temperature (32.5°C) and exposed to low dosages of etoposide, 33% of the pX-expressing p53-val135-treated colonies regressed (Table 1) and displayed both morphological features of apoptosis and internucleosomal DNA cleavage (data not shown). No morphological changes and DNA degradation was observed upon etoposide treatment neither in pSV-CAT clones under any conditions or in pSV-X clones at the non permissive temperature (Table 1 and data not shown). The role of p53 in the phenomenon was further confirmed by the observation that no difference was observed in colony formation in both p53-deficient fibroblasts from p53−/− knock out mice (28) and HeLa cells, where p53 is functionally inactive, transfected with either pX or control vectors (Fig. 4a and data not shown). Early passage p53−/− MEFs were used in all the experiments to rule out the possibility that differences in the apoptotic behavior could be due to genetic alterations selected for by cell cultures conditions. Moreover, pX expressing p53−/− clones are insensitive to etoposide-induced cell death (Table 1).
Figure 3.
pX clones are sensitized to p53-mediated apoptosis. (a) Colony regression after exposure to low dosages of the anticancer agent etoposide. pSV-X (2 μg) and pSV-CAT (2 μg) were introduced into NIH 3T3 cells by calcium phosphate coprecipitation and resistant colonies were isolated by selection in G418 as described. Unexpanded pSV-X or pSV-CAT colonies were marked, inspected by microscopy, and photographed 72 hr after treatment with etoposide (Sigma) at the concentration of 0.1 mg/ml. (b) Analysis of chromatin structure (Left and Center) and internucleosomal cleavage of genomic DNA (Right) following treatment with etoposide. Low molecular weight DNA was extracted from equal numbers of cells from either pX or CAT-pooled unexpanded colonies.
Table 1.
Colony regression following etoposide treatment
| Cell | Exogenous gene | p53 phenotype/ genotype* | Colony viability
|
||
|---|---|---|---|---|---|
| Regressing | Resistant | % regressing | |||
| NIH 3T3 | HBV-X† | wt | 9 | 4 | 69 |
| CAT | wt | 5 | 50 | 9 | |
| REF-52 | HBV-X† | wt | 35 | 0 | 0 |
| CAT | wt | 40 | 0 | 0 | |
| BALB/c3T3 | HBV-X† | wt | 6 | 2 | 75 |
| CAT | wt | 3 | 40 | 9 | |
| BALB/c3T3 | p53-val135 | ||||
| 38°C | HBV-X† | mt | 2 | 32 | 6 |
| CAT | mt | 0 | 40 | 0 | |
| 32°C | HBV-X† | wt | 12 | 25 | 33 |
| CAT | wt | 1 | 30 | 4 | |
| MEFs | HBV-X† | −/− | 0 | 45 | 0 |
| CAT | −/− | 0 | 53 | 0 | |
| HeLa | HBV-X† | −/+ | 0 | 70 | 0 |
| CAT | −/+ | 0 | 67 | 0 | |
pSV-X (2 μg) and pSV-CAT (2 μg) were cotransfected together with either pAG60 or pSV-Hygro into early passages NIH 3T3 cells, REF-52 cells, BALB/c3T3 and the BALB/ctsp53 clone, early passages p53−/− MEFs and HeLa cells. Colonies were selected in G418 or hygromycin. Approximately 3 weeks after transfection the position of colonies was marked and 0.1 mg/ml of etoposide was added. Coded plates were inspected blindly by an independent observer and scored for significant morphological regression and cell death at 24, 48 and 72 hr after initiation of treatment. Colonies were considered regressing when at least >80% of cells displayed morphological alterations. Figures represent the average of three independent experiments. wt, wild type; mt, mutant.
p53 expression and status have been assessed by indirect immunofluorescence using the anti p53 antibodies pAb122 and pAb246 (NIH 3T3 and BALB/c3T3 cells) and pAb421 and pAb240 (REF-52 cells) and by immunoblot after etoposide and actinomicyn D treatment. See also Soddu et al. (27) and Lowe et al. (20).
HBV-X expression was assessed in at least five randomly selected clones by cDNA–PCR using primers derived from the X region (41).
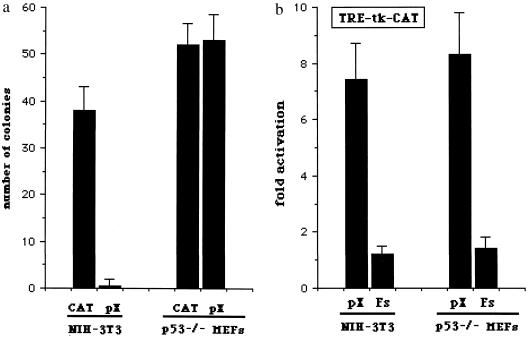
Figure 4.
p53 status dissects pX properties to induce apoptosis, activate transcription, and alter cell growth. (a) Effect of pX on colony formation in p53-null cells. Early passage p53-deficient MEFs from p53−/− knockout mice were cotransfected with 20 μg of either pSV-X or control pSV-CAT expression vectors, with pSV-0 as a nonspecific carrier, for a total of 20 μg/ml of DNA and 200 ng of pAG60. Colony generation assays were performed, and results are from five independent experiments. (b) Activation of AP1-dependent transcription by pX in serum-starved p53-null cells. p53−/− MEFs were costransfected with 1 μg of the CAT reporter vector pTRE-tk-CAT together with 2 μg of pSV-X, pSV-X(Fs) where appropriate, and pUC18 as nonspecific carrier DNA for a total of 20 μg/ml. The inability of pX to activate transcription from a minimal thymidine kinase (tk) promoter was confirmed using the reporter plasmid pBL2-CATdel.
p53 Status Dissects pX Properties to Induce Apoptosis, Activate Transcription, and Alter Cell Growth.
Interestingly, while the ability of pX to inhibit colony formation is clearly influenced by p53 status (Fig. 4a), in p53−/− cells pX is still able to activate AP1-dependent transcription (Fig. 4b) and to induce cell growth. In fact, pX expressing p53−/− grow faster as compared with control cells in both normal or low serum conditions (Table 2). Thus, pX activates transcription and induces cell cycle progression irrespective of p53 status. In cells expressing wild-type p53 these activities of pX may result in cell death, while in cells lacking functional p53, pX cannot affect cell viability and confers an evident growth advantage.
Table 2.
Effects of pX on cell growth rates of p53 null cells
| Clone | Number of cells (×103) ± SEM
|
|||||
|---|---|---|---|---|---|---|
| FCS 0.5%
|
FCS 10%
|
|||||
| 24 hr | 48 hr | 72 hr | 24 hr | 48 hr | 72 hr | |
| CAT1 | 220 ± 14 | 250 ± 16 | 210 ± 21 | 320 ± 35 | 645 ± 28 | 1,360 ± 78 |
| CAT2 | 240 ± 18 | 280 ± 25 | 300 ± 33 | 380 ± 43 | 755 ± 53 | 1,635 ± 96 |
| CAT3 | 205 ± 16 | 220 ± 14 | 240 ± 41 | 440 ± 54 | 980 ± 58 | 2,380 ± 126 |
| X1 | 370 ± 31 | 745 ± 68 | 1,340 ± 103 | 650 ± 75 | 2,110 ± 145 | 5,435 ± 378 |
| X2 | 410 ± 46 | 845 ± 65 | 1,570 ± 162 | 715 ± 49 | 2,475 ± 179 | 7,560 ± 387 |
| X3 | 435 ± 31 | 1,020 ± 87 | 1,770 ± 113 | 780 ± 83 | 2,780 ± 148 | 9,255 ± 520 |
Early pasage p53 deficient MEFs from p53−/− knockout mice were cotransfected with 20 μg of either pSV-X or control pSV-CAT expression vectors. pSV-0 as a nonspecific carrier, for a total of 20 μg/ml of DNA and 200 ng of pAG60. Colonies were selected in G418. Growth rates of pSV-X and pSV-CAT clones were determined in either low serum [fetal calf serum (FCS) 0.5%] or enriched medium conditions (FCS 10%). Cells were plated at 2 × 105 cells per 100-mm dish. Figures represent the mean ± SEM from three independent experiments.
CONCLUSIONS
Our results, obtained using different experimental approaches, strongly support the notion of pX as a multifunctional regulatory protein that affects transcription, cell growth, and cell viability. It is important to note that we did not demonstrate pX expression at the protein level and, therefore the “pX” and “pX expression” terminology does not imply the visualization of the protein in the cells being studied. The apparent dissociation between the detection of pX protein and the demonstration of pX effects is not new and it is matter of discussion (for both a review and discussion, see ref. 42). Our observations—i.e., the ability of pX to affect cell viability in a dose dependent manner—may indeed help to explain this paradoxical phenomenon.
The molecular mechanisms underlying the different activities of pX are only partially defined. Indeed, pX has been shown to have pleiotropic and potentially conflicting effects on a variety of molecular targets, and both nuclear and peripheral sites of action of pX have been described. Several groups reported that pX modulates the intracellular signaling apparatus including the protein kinase C (9) and the Ras/Raf (11–13) signaling pathways. Moreover, pX has been shown to physically interact in vivo and/or in vitro with DNA-binding proteins involved in trascriptional regulation (6–8), with a putative DNA repair protein (43), with cellular proteases (44) and components of the proteasome complex (45), and, finally, with p53 (24, 25). Because pX binding to p53 is known to inhibit its ability to bind DNA and to transactivate target genes (refs. 24 and 25, and S.P., unpublished observations), the requirement of functional p53 for pX associated apoptosis is unexpected. However, recent evidences suggest that, in contrast to p53-induced growth arrest, which is mainly dependent from p53 transactivating function, two p53-dependent apoptotic pathways might exist, one requiring activation of specific target genes and the other independent from sequence-specific transactivation (46–49). Although it has been reported that pX attenuates apoptosis induced by direct injection of p53 expressing plasmids in human primary fibroblasts (50), differences in the relative ratio between pX and p53 in the two experimental conditions might be relevant to explain these apparently conflicting findings. Wang et al. (50) overexpressed both p53 and pX by microinjecton in primary human fibroblasts. The effect of p53 overexpression in their system appears to be a rapid induction of apoptosis rather than a growth arrest, as one might expect in untransformed cells bearing wild-type p53 alleles. Indeed, this apoptotic response is more commonly observed upon wild-type p53 overexpression in transformed cells, such as HeLa and SAOS-2, which lack functional endogenous p53 (47, 51). The effect of pX on p53-induced apoptosis in their experimental conditions correlate with the ability of pX to interfere with p53 transcriptional activation (as shown by a reduction of p53 induction of the downstream target p21). Our results indicate that, in a wild-type p53 context the chronic expression of pX, via its ability to perturb cell growth regulation, prompts cells to p53-mediated apoptosis and support a model in which pX does not block all p53 functions involved in the induction of apoptotis. This would be in agreement with the existence of multiple p53 apoptotic pathways (46–49) and pX, unique among viral transforming proteins, is able to dissect the p53 functions involved in transcription activation/growth arrest and cell death induction or execution. According to our findings, in the presence of wild-type p53 pX would induce both cell cycle progression and programmed cell death. In the presence of p53 gene mutations or conformational inactivation of p53 protein, pX would exert its effects on cell proliferation undisturbed. These observations and the increase of hepatocyte apoptosis described in pX transgenic mice (M. A. Buendia, personal communication), support our conclusions and suggest that p53 status play a role in determining the outcome of pX expression in HBV chronically infected hepatocytes.
Acknowledgments
We thank S. Soddu for the BALB/c Ts p53 clone, M. Perricaudet for pE1a(12S), A. Giordano for pCMV-E2F1, and D. Eick for p53−/− MEFs and the c-myc expression vectors. This project was supported by grants from Fondazione A. Cesalpino, Associazione Italiana per la Ricerca sul Cancro; P. F. Applicazioni Cliniche della Ricerche Oncologica, Consiglio Nazionale delle Ricerche, Italy; Progetto Epatiti Virali, ISS; and European Community Biomed 1. P.C. is supported by a fellowship from the Fondazione Cenci Bolognetti–Istituto Pasteur.
ABBREVIATIONS
- HBV
hepatitis B virus
- MMTV
mouse mammary tumor virus
- MEFs
mouse embryo fibroblasts
- CAT
chloramphenicol acetyltransferase
- SV40
simian virus 40
References
- 1.Avantaggiati M L, Natoli G, Balsano C, Chirillo P, Artini M, De Marzio E, Collepardo D, Levrero M. Oncogene. 1993;8:1567–1574. [PubMed] [Google Scholar]
- 2.Twu J S, Lai M Y, Chen D S, Robinson W S. Virology. 1993;192:346–350. doi: 10.1006/viro.1993.1041. [DOI] [PubMed] [Google Scholar]
- 3.Balsano C, Avantaggiati M L, Natoli G, De Marzio E, Will H, Perricaudet M, Levrero M. Biochem Biophys Res Commun. 1991;176:985–992. doi: 10.1016/0006-291x(91)90379-l. [DOI] [PubMed] [Google Scholar]
- 4.Menzo S, Clementi M, Alfani E, Bagnarelli P, Iacovacci S, Manzin A, Dandri M, Natoli G, Levrero M, Carloni G. Virology. 1993;196:878–882. doi: 10.1006/viro.1993.1550. [DOI] [PubMed] [Google Scholar]
- 5.Doria M, Klein N, Lucito R, Schneider R J. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maguire H F, Hoeffler J P, Siddiqui A. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 7.Cheong J H, Yi M K, Lin Y, Murakami S. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams J S, Andrisani O M. Proc Natl Acad Sci USA. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kekulè A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Nature (London) 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 10.Natoli G, Avantaggiati M L, Chirillo P, Costanzo A, Artini M, Balsano C, Levrero M. Mol Cell Biol. 1994;14:989–998. doi: 10.1128/mcb.14.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross J, Wen P, Rutter W J. Proc Natl Acad Sci USA. 1994;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benn J, Schneider R J. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 14.Chirillo P, Falco M, Puri P L, Artini M, Balsano C, Levrero M, Natoli G. J Virol. 1996;70:254–261. doi: 10.1128/jvi.70.1.641-646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoulim F, Saputelli J, Seeger C. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennat B C, Cote P J, Gerin J L, Purcell R H, Miller R H. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim C M, Koike K, Saito I, Miyamura T, Jay G. Nature (London) 1991;353:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 18.Hohne M, Schaefer S, Seifer M, Feitelson M A, Paul D, Gerlich W H. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debbas M, White E. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 20.Lowe S W, Ruley H E, Jacks T, Housman D E. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 21.Pan H, Griep A E. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 22.White E. Nature (London) 1994;371:21–22. doi: 10.1038/371021a0. [DOI] [PubMed] [Google Scholar]
- 23.Koike K, Moriya K, Yotsuyanagi H, Iino S, Kurikawa K. J Clin Invest. 1994;94:44–49. doi: 10.1172/JCI117343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J-R, Harris C C. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish J A. J Virol. 1995;69:1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifer M, Hohne M, Shaefer S, Gerlich W H. J Hepatol. 1991;13:61–65. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- 27.Soddu S, Blandino G, Scardigli R, Martinelli R, Rizzo M G, Crescenzi M, Sacchi A. Mol Cell Biol. 1996;16:487–496. doi: 10.1128/mcb.16.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermeking H, Eick D. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 29.Sugden B, Marsch K, Yates J. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puri P L, Balsano C, Burgio V L, Chirillo P, Natoli G, Ricci L, Mattei E, Graessmann A, Levrero M. Oncogene. 1997;14:1171–1184. doi: 10.1038/sj.onc.1200941. [DOI] [PubMed] [Google Scholar]
- 31.Natoli G, Ianni A, Costanzo A, De Petrillo G, Ilari I, Chirillo P, Balsano C, Levrero M. Oncogene. 1995;11:1157–1164. [PubMed] [Google Scholar]
- 32.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 33.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D D. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 34.Canman C E, Gilmer T M, Coutts S P, Kastan M B. Genes Dev. 1995;9:600–611. doi: 10.1101/gad.9.5.600. [DOI] [PubMed] [Google Scholar]
- 35.Natoli G, Avantaggiati M L, Balsano C, De Marzio E, Collepardo D, Elfassi E, Levrero M. Virology. 1992;187:663–670. doi: 10.1016/0042-6822(92)90469-6. [DOI] [PubMed] [Google Scholar]
- 36.JeanJean O, Levrero M, Will H, Perricaudet M, Rossignol J M. Virology. 1989;170:99–106. doi: 10.1016/0042-6822(89)90356-5. [DOI] [PubMed] [Google Scholar]
- 37.Hermeking H, Wolf D A, Kohlhuber F, Dickmanns A, Billaud M, Fanning E, Eick D. Proc Natl Acad Sci USA. 1994;91:10412–10416. doi: 10.1073/pnas.91.22.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kekulè A S, Lauer U, Meyer M, Caselmann W H, Hofschneider P H, Koshy R. Nature (London) 1990;343:457–461. doi: 10.1038/343457a0. [DOI] [PubMed] [Google Scholar]
- 39.Chen M-J, Holskin B, Strickler J, Gorniak J, Clark M A, Johnson P J, Mitcho M, Shalloway D. Nature (London) 1987;330:581–583. doi: 10.1038/330581a0. [DOI] [PubMed] [Google Scholar]
- 40.Fritsche M, Haessler C, Brandner G. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 41.Bertoletti A, Costanzo A, Chisari F V, Levrero M, Artini M, Sette A, Penna A, Giuberti T, Fiaccadori F, Ferrari C. J Exp Med. 1994;180:933–943. doi: 10.1084/jem.180.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen B Y. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 43.Lee T H, Elledge S J, Butel J. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takada S, Kido H, Fukutomi A, Takeshi M, Koike K. Oncogene. 1994;9:341–348. [PubMed] [Google Scholar]
- 45.Fisher M, Runkel L, Schaller H. Virus Genes. 1995;10:99–102. doi: 10.1007/BF01724303. [DOI] [PubMed] [Google Scholar]
- 46.Caelles C, Helmberg A, Karin M. Nature (London) 1994;370:220–2123. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 47.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 48.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 49.Bates S, Vousden K H. Curr Opin Genet Dev. 1996;6:12–18. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 50.Wang X W, Gibson M K, Vermeulen W, Yeh H, Forrester K, Sturzbecher H W, Hoeijmakers J H J, Harris C C. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 51.Yonish-Rouach E, Borde J, Gotteland M, Mishal Z, Viron A, May E. Cell Death Differ. 1994;1:39–47. [PubMed] [Google Scholar]