Abstract
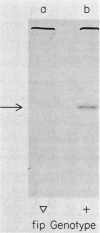
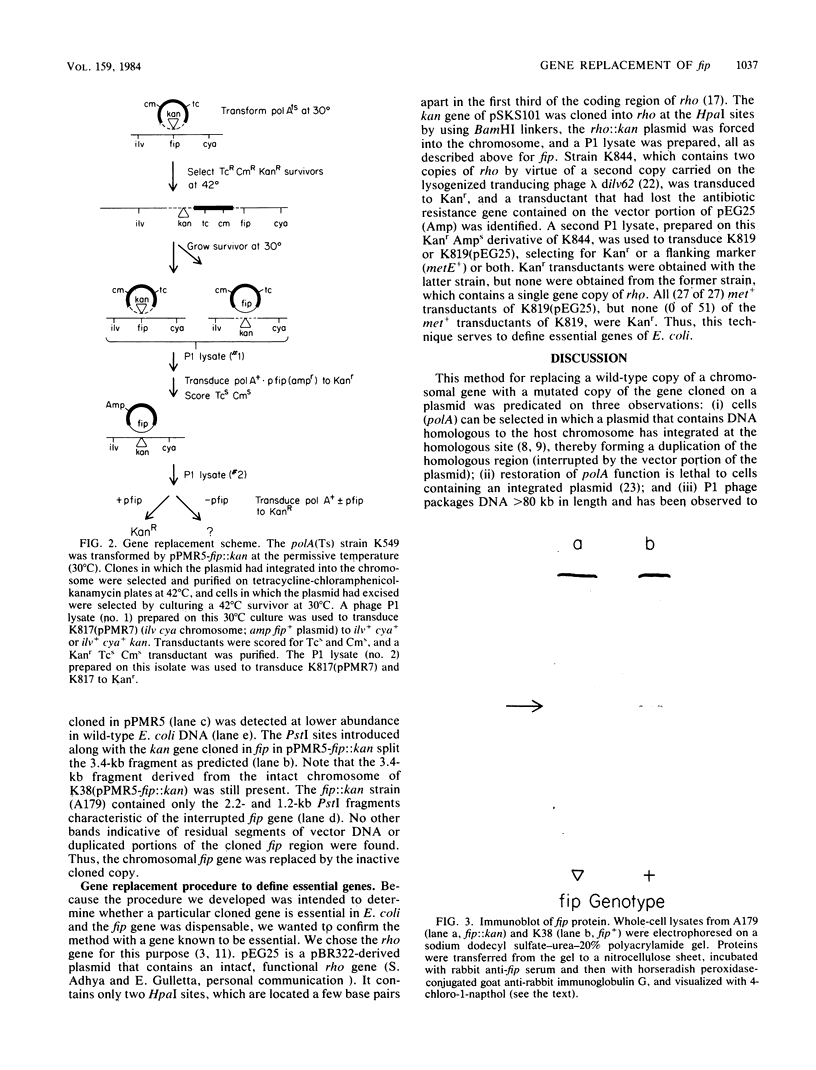
To determine whether the fip gene of Escherichia coli, which is required for filamentous phage assembly, is required for cell viability, we replaced the chromosomal copy of the gene with an inactive copy introduced on a plasmid. We found that the fip gene is dispensable. The method we devised, which should be generally useful, was also tested with an inactivated rho gene. As expected, the rho gene is essential.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F. Host mutant (tabD)-induced inhibition of bacteriophage T4 late transcription. II. Genetic characterization of mutants. J Mol Biol. 1975 Aug 25;96(4):601–624. doi: 10.1016/0022-2836(75)90141-2. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hohn B. Identification of a host protein necessary for bacteriophage morphogenesis (the groE gene product). Proc Natl Acad Sci U S A. 1978 Jan;75(1):131–135. doi: 10.1073/pnas.75.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C., Tilly K., Drahos D., Hendrix R. The B66.0 protein of Escherichia coli is the product of the dnaK+ gene. J Bacteriol. 1982 Mar;149(3):1175–1177. doi: 10.1128/jb.149.3.1175-1177.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener A., Hill C. W. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980 Oct;144(1):312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N. I., Koshland D. E., Jr Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid A., Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Inoko H., Shigesada K., Imai M. Isolation and characterization of conditional-lethal rho mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1162–1166. doi: 10.1073/pnas.74.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- LOEB T., ZINDER N. D. A bacteriophage containing RNA. Proc Natl Acad Sci U S A. 1961 Mar 15;47:282–289. doi: 10.1073/pnas.47.3.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- O'Connor K. A., Zusman D. R. Coliphage P1-mediated transduction of cloned DNA from Escherichia coli to Myxococcus xanthus: use for complementation and recombinational analyses. J Bacteriol. 1983 Jul;155(1):317–329. doi: 10.1128/jb.155.1.317-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham J. L., Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983 Jun 11;11(11):3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. A bacterial gene, fip, required for filamentous bacteriophage fl assembly. J Bacteriol. 1983 Jun;154(3):1064–1076. doi: 10.1128/jb.154.3.1064-1076.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Characterization of the cloned fip gene and its product. J Bacteriol. 1984 Feb;157(2):526–532. doi: 10.1128/jb.157.2.526-532.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Tessman I., Fassler J. S., Bennett D. C. Relative map location of the rep and rho genes of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1637–1640. doi: 10.1128/jb.151.3.1637-1640.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan M., Favre R., Gallay E., Caro L. Genetical and structural analysis of a group of lambda ilv and lambda rho transducing phages. Mol Gen Genet. 1981;182(3):462–470. doi: 10.1007/BF00293936. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Tomizawa J. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet. 1980;178(3):525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]