Abstract
Mutations in the presenilin 1 (PS1) and presenilin 2 (PS2) genes increase the production of the highly amyloidogenic 42-residue form of amyloid β-protein (Aβ42) in a variety of cell lines and transgenic mice. To elucidate the molecular mechanism of this effect, wild-type (wt) or mutant PS1 and PS2 genes were stably transfected into Chinese hamster ovary cells expressing endogenous or transfected β-amyloid precursor protein (APP). By immunoprecipitation/Western blot analysis, APP was consistently found to coimmunoprecipitate with PS1 or PS2 proteins. Several distinct PS1, PS2, or APP antibodies precipitated PS–APP complexes that were detectable by blotting with either APP or PS antibodies. Importantly, complex formation could be detected at endogenous protein levels in nontransfected cells. In various Chinese hamster ovary cell lines, the amounts of APP coprecipitated by PS antibodies were proportional to the expression levels of both APP and PS. APP–PS complexes also were recovered from human 293 and HS683 cells. Full maturation of APP was not required for the interaction; most APP molecules complexed with PS were solely N-glycosylated. Treatment of cells with brefeldin A or incubation at 20°C did not block complex formation, suggesting that the association between APP and PS occurs in part in the endoplasmic reticulum. Complex formation was detected for both wt and mutant PS and APP proteins. Deletion of the APP C-terminal domain did not abrogate complex formation, suggesting that the interaction does not occur in the cytoplasmic domains of the proteins. Our results demonstrate that wt and mutant PS1 and PS2 proteins form complexes with APP in living cells, strongly supporting the hypothesis that mutant PS interacts with APP in a way that enhances the intramembranous proteolysis of the latter by a γ-secretase cleaving at Aβ42.
It is now well established that Alzheimer disease (AD) is genetically heterogeneous. Causative mutations or polymorphisms have been identified in four different genes so far, and several additional genetic loci are anticipated (for review, see ref. 1). A central issue in AD research is whether these disparate genetic alterations operate through a common pathogenic mechanism to induce the clinical phenotype. The gene most frequently implicated in autosomal dominant AD to date is the presenilin 1 (PS1) gene (2). Families bearing mutations in this gene or in the highly homologous presenilin 2 (PS2) gene (3, 4) invariably develop an aggressive form of AD that becomes symptomatic in the forties or fifties and leads to death within one to two decades thereafter.
Recent studies in PS-transfected cell lines (5–7, 16), PS-transgenic mice (5, 7, 8), and in the plasma (9), skin fibroblast media (9), and brain tissue (10) of humans carrying PS mutations all demonstrate a selective and highly significant increase in the levels of the 42-residue form of the amyloid-β-protein (Aβ42) as a direct consequence of expressing mutant presenilins. Aβ42 is proteolytically released from the β-amyloid precursor protein (APP) and secreted by neural and nonneural cells of humans and many lower mammals throughout life (11). The mechanism by which mutations in PS1 and PS2 lead to a selective increase in Aβ42 production is unknown. Because both PS and APP are known to be expressed in the endoplasmic reticulum (ER) and Golgi apparatus (12–15), it is possible that a direct or indirect interaction of the two proteins in these compartments could underlie the effect of mutant PS on APP proteolytic processing. To address this hypothesis, we systematically searched for evidence of APP–PS complexes in both nonhuman [Chinese hamster ovary (CHO)] and human (293 and HS683) cells. Our experiments demonstrate that such complexes indeed exist under physiological conditions and that APP or PS proteins carrying familial AD-linked missense mutations still can form complexes.
MATERIALS AND METHODS
Generation of PS1- or PS2-Transfected CHO Cells.
Wild-type (wt) and mutant PS1 or PS2 cDNAs were subcloned into the cytomegalovirus-based expression vectors PCI-neo (PS1) (Promega) or pZeo (PS2) (Invitrogen) (6). The neo gene in the PS1 vector then was replaced with a transferrin receptor (TR) gene driven by the simian virus 40 promoter. Using lipofectin (Life Technologies), the wt PS1 or PS2 vectors were stably transfected into plain CHO cells (resulting in lines C-PS1 and C-PS2, respectively). Both wt and mutant PS1 or PS2 vectors were stably transfected into CHO cells stably expressing wt human APP751 [resulting in lines PS1Wt-1, PS1Wt-2, PS1ML-2 (M146L missense mutation), and PS1CY-1 (C410Y missense mutation), and lines PS2Wt, PS2VG (N141I missense mutation), and PS2It (M239V missense mutation)] (6). Cells were maintained in 200 μg/ml G418 (Life Technologies) plus 25 μg/ml puromycin (for PS1) or 250 μg/ml zeocin (Invitrogen) (for PS2). We also examained CHO lines singly transfected with wt APP751 or APP751 bearing the KM651/652NL or the V698F missense mutations.
Antibodies.
PS1 polyclonal antibodies X81, 311/2a (gift of B. Hyman, Massachusetts General Hospital, Boston), 4624, and 4627, and mAb 13A11 (gift of P. Seubert, Athena Neurosciences, San Francisco) were raised against residues 1–81 (X81), 17–33 (311/2a), 294–309 (13A11), 343–357 (4624), or 457–467 (4627) of PS1 (6). PS2 polyclonal antibodies 2972 (gift of C. Haass, University of Heidelberg, Germany) (6, 16), PS2L (gift of T. Iwatsubo, T. Saido, K. Maruyama, and T. Tomita, Tokyo) (16) and PS2n (gift of L. D’Adamio, National Institutes of Health) (17) were raised to PS2 residues 1–75 (2972), 316–339 (PS2L), or 341–377 (PS2n). Polyclonal antibody B5 is against APP444–592 (APP695 numbering) (18). mAbs 8E5 and 13G8 are against APP444–592 (8E5) or APP676–695 (13G8) (gifts of P. Seubert and D. Schenk, Athena Neurosciences, San Francisco). mAb 22C11 (against residues 60–100 of APP) was from Boehringer Mannheim. Antibody TR Ab-1 to human TR was from CalBiochem.
Immunoprecipitation and Western Blotting.
Cells were incubated in methionine-free, fetal bovine serum-free medium for 15 min, labeled with 100 μCi/ml [35S]methionine for 60 min, and lysed in a buffer containing 50 mM Tris at pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, and a protease inhibitor cocktail [5 μg/ml leupeptin/5 μg/ml aprotinin/2 μg/ml pepstatin A/0.25 mM phenylmethylsulfonyl fluoride (Sigma)]. Immunoprecipitates (6) were washed in 50 mM Tris, pH 7.6/500 mM NaCl/2 mM EDTA and the same protease inhibitors for 15 min at 4°C, followed by additional washes in 50 mM Tris, pH 7.6/150 mM NaCl/2 mM EDTA and the inhibitors. Coimmunoprecipitates were eluted with 2× sample buffer (40% glycerol/6% SDS/6% 2-mercaptoethanol), electrophoresed on 8–16% Tris-glycine gels (Novex), and transferred to poly(vinylidene difluoride). Western blotting using ECL detection was performed as per the manufacturer (Amersham).
RESULTS
Interaction of Presenilin Proteins with APP in Intact Cells.
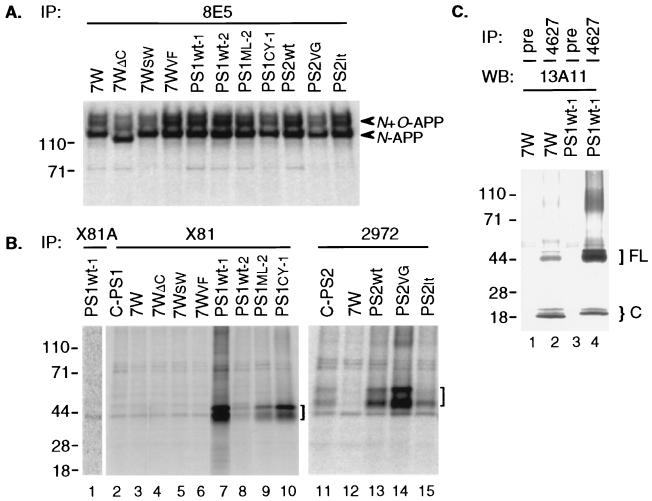
A variety of CHO cell lines stably expressing human APP and/or PS1 or PS2 proteins were established (6). Expression levels of APP and PS proteins were determined by metabolic labeling and immunoprecipitation of cell lysates. Cell lines solely transfected with APP included 7W (wt APP751) (19), 7WΔC [APP751 with deletion of almost the entire cytoplasmic tail (residue 710–751)] (20), 7WSW (APP751 with the “Swedish” KM651/652NL double-mutation) (21), and 7WVF (APP751 with the V698F mutation) (19). The cell line 7W was used as a parental line to generate PS1 or PS2 double-transfectants. Therefore, the amounts of APP expressed in these double-transfected lines (PS1Wt-1, PS1Wt-2, PS1ML-2, PS1CY-1, PS2Wt, PS2VG, and PS2It) were essentially the same (Fig. 1A). PS1 or PS2 expression levels were determined by precipitation of the same lysates shown in Fig. 1A with the PS1 antibody X81 or the PS2 antibody 2972 (Fig. 1B). Cell line C-PS1 transfected solely with wt PS1 expressed a low level of this protein, as did the double-transfectant PS1Wt-2 (Fig. 1B). Double-transfectants PS1Wt-1, PS1ML-2 (M146L missense mutation), and PS1CY-1 (C410Y missense mutation) expressed high levels of PS1 (Fig. 1B) (22). PS2 levels were moderate in the single-transfected line C-PS2 and the double-transfectant PS2It [M239V (Italian) missense mutation], and were high in the double-transfectants PS2Wt and PS2VG [N141I (Volga German) missense mutation] (Fig. 1B).
Figure 1.
Expression of PS and APP proteins in stably transfected CHO cells. Cells were metabolically labeled for 60 min and lysates immunoprecipitated (IP) with the indicated APP (A) or PS1 or PS2 (B) antibodies. (A) APP antibody 8E5 precipitated high levels of APP in all of these APP751-transfected lines. Note the slightly faster migration of both N and N+O-glycosylated APP in cell line 7WΔC, which expresses APP lacking the C-terminal cytoplasmic domain. The slightly lower amounts of APP seen in PS1CY-1 and PS2VG lanes represent gel loading variation and were not consistent. (B) IP with X81 (to PS1) or 2972 (to PS2) precipitated the characteristic PS1 doublet at 43–45 kDa or PS2 doublet at 50–55 kDa (brackets). Both of the PS1 bands have been radiosequenced in the PS1Wt-1 cell line, confirming their identities as SDS-stable conformers of full-length human PS1 (6). The band at ≈44 kDa is a background band that is detected even by X81 preabsorbed with its cognate antigen (lane 1). Endogenous full-length PS1 in 7W, 7WΔC, 7WSW, and 7WVF (lanes 3–6) is not clearly detectable by IP alone. (C) Endogenous hamster PS1 was detected in 7W cell lysate by IP with 4627 followed by Western blotting (WB) with 13A11 (lane 2). Pre, preimmune serum. Transfected human PS1 in line PS1Wt-1 (lane 4) includes full-length (FL) proteins at 43–45 kDa and C-terminal fragments (C) at 18–19 kDa.
To detect the steady state levels of endogenous and transfected presenilins and their endoproteolytic fragments (23) and to search for PS–APP complexes, we used a highly sensitive immunoprecipitation/Western blotting technique (22). The method is exemplified in Fig. 1C, in which the APP-transfected 7W cells and the PS1/APP double-transfected PS1Wt-1 cells were precipitated with PS1 polyclonal antibody 4627 followed by Western blotting with PS1 mAb 13A11 (22). In the 7W cells, endogenous full-length hamster PS1 and its characteristic C-terminal endoproteolytic fragments (CTF) were detected (Fig. 1C, lane 2). These bands were not precipitated by preimmune serum (Fig. 1C, lane 1). In the PS1Wt-1 cells, the amount of full-length PS1 (representing the transfected human form) was markedly increased, and the resultant ≈19-kDa human CTF replaced the endogenous ≈18-kDa hamster CTF (Fig. 1C, lane 4), as reported previously in cells overexpressing human PS (23).
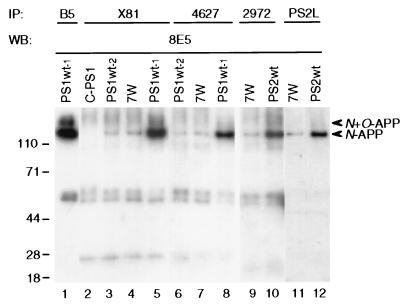
To determine whether APP and PS proteins interact in CHO cells, we performed immunoprecipitation/Western blotting using numerous distinct PS and APP antibodies. Confluent cells were lysed and precipitated with polyclonal antibody X81 against the N-terminal domain of PS1. The PS1 immunoprecipitates were separated on SDS/PAGE, and any coprecipitated APP was detected by blotting with mAb 8E5. In cells expressing transfected human APP and endogenous PS1 (line 7W), we found that APP coprecipitated with PS1 (Fig. 2, lane 4). Because antibody 8E5 is human APP specific, blotting with this antibody in line C-PS1 expressing endogenous hamster APP and transfected human PS1 showed no APP (Fig. 2, lane 2). APP also was coprecipitated by X81 in the PS1Wt-2 line that expresses transfected APP and a low level of transfected PS1 (but still higher than endogenous PS1) (Fig. 2, lane 3). A large amount of APP was coprecipitated by the PS1 antibody in the PS1Wt-1 line, which expresses high levels of both PS1 and APP (Fig. 2, lane 5). The amounts of APP–PS1 complexes detected in the various cell lines examined were consistently related to the levels of expression of the two proteins. Highly similar results were obtained when antibody 4627 (to the PS1 C-terminus) was used to immunoprecipitate, followed by blotting with APP antibody 8E5. The amount of precipitated APP again correlated with the expression levels of both PS1 and APP (Fig. 2, lanes 6–8). When the APP molecules coprecipitating with PS1 were electrophoretically compared with the total N- and N+O-glycosylated APP proteins precipitated by the APP polyclonal antibody B5 (Fig. 2, lane 1), the coprecipitating APP was almost entirely the N-glycosylated form.
Figure 2.
Complex formation between PS1 or PS2 and APP in CHO cells. CHO cells stably transfected with PS1 or PS2 were lysed and precipitated (IP) with the indicated PS1 or PS2 antibodies, followed by blotting (WB) with APP antibody 8E5. Lower arrowhead indicates the coprecipitating N-APP polypeptide [compare with the N- and N+O-glycosylated APP precipitated by APP antibody B5 (lane 1)]. IP with antibodies to either PS1 (X81; 4627) or PS2 (2972; PS2L) coprecipitates APP indistinguishably. Bands at 50 and 25 kDa are IgG heavy and light chains.
Next, we searched for evidence of complex formation between PS2 and APP. When cell lysates were precipitated with the polyclonal PS2 N-terminal antibody 2972, followed by Western blotting with APP antibody 8E5, APP was again found to coprecipitate (Fig. 2, lanes 9–10). Cell line PS2Wt, which expressed high levels of both PS2 and APP, showed the most coprecipitating APP, but it also was detected in the 7W cells. When the PS2 “loop” antibodies, PS2L (Fig. 2, lanes 11–12) and PS2n (not shown), were used for precipitation, highly similar results were obtained: APP was observed in both 7W and PS2Wt cells, with a higher level in the latter. Thus, APP and PS2 were consistently coprecipitated by both N-terminal and “loop” region PS2 antibodies.
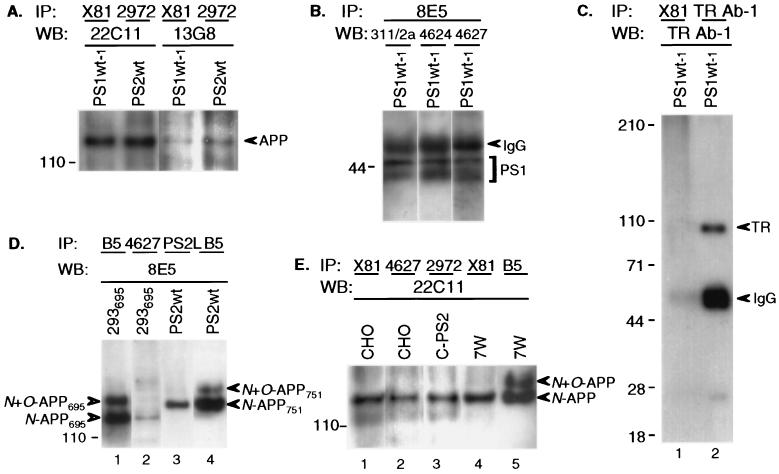
To confirm that the APP–PS interaction was specific, we used several antibodies to different regions of APP to detect the coprecipitates in the CHO transfectants. Using either 22C11 or 13G8 for blotting, we again detected N-glycosylated APP that had been precipitated with PS1 or PS2 antibodies (Fig. 3A). We also used the antibodies in reverse order: lysates were precipitated with APP antibody 8E5 and any PS1 then detected by blotting with N-terminal antibody 311/2a, “loop” antibody 4624 or C-terminal antibody 4627. As expected, all three antibodies identified PS1 that had precipitated with APP (Fig. 3B). To address the possibility that simply overexpressing PS leads to nonspecific interaction with APP, we performed coprecipitations of PS1 and human TR in cell line PS1Wt-1, which coexpressed PS1 and TR. Precipitation with either X81 (Fig. 3C, lane 1) or 4627 (not shown) and blotting with TR antibody revealed no TR, whereas precipitation of the same lysate with TR antibody and blotting with that antibody revealed a clear TR band.
Figure 3.
APP–PS complex formation demonstrated under various conditions. (A) Coprecipitating APP is detectable in PS1Wt-1 or PS2Wt CHO cells using different APP antibodies (22C11; 13G8) for WB. (B) Reverse order of IP/WB: APP antibody (8E5) for IP and various PS1 antibodies (311/2a, 4624, 4627) for WB in PS1Wt-1 cells. IgG bands represent crossreaction of goat anti-rabbit secondary antibody used for WB with primary mouse mAb (8E5) used for IP. (C) IP of PS1Wt-1 CHO cells stably cotransfected with APP, PS1, and human TR using PS1 antibody X81 reveals no coprecipitating TR upon WB with anti-TR (lane 1), whereas both IP and WB with anti-TR reveals TR. Note the expected IgG crossreaction of the anti-rabbit IgG secondary WB antibody with the rabbit primary antibody used for IP. (D) Lane 2, 4627 (PS1) coprecipitates N-glycosylated APP695 (blotted by 8E5) in 293 cells stably transfected with APP695. Lane 3, PS2L (PS2) coprecipitates N-glycosylated APP751 in PS2Wt CHO cells cotransfected with PS2 and APP751. Lanes 1 and 4: IP with B5 as control. (E) Lanes 1 and 2, coprecipitating endogenous APP is detected by 22C11 in nontransfected CHO cells precipitated with X81 or 4627 PS1 antibodies. Lane 3, endogenous APP interacts with transfected PS2 in line C-PS2. Lane 4, transfected APP interacts with endogenous PS1 in line 7W. Lane 5, total APP in 7W cells.
We next searched for APP–PS1 complex formation in another cell type, human 293 cells, in which APP processing has been extensively characterized heretofore. N-glycosylated APP was precipitated with PS1 by antibody 4627 in 293 cells stably expressing APP695 (Fig. 3D, lane 2). Electrophoretic comparison to the N-glycosylated APP751 species coprecipitated in PS-transfected CHO cells (Fig. 3D, lane 3) showed the expected faster migration of the 695-residue isoform, supporting the authenticity of the APP molecules precipitated by PS antibodies.
Detection of Endogenous APP–PS Complex Formation.
To confirm unequivocally the physiological occurrence of APP–PS complexes at endogenous protein levels, we used X81 or 4627 to precipitate a large amount of lysate from nontransfected CHO cells expressing only endogenous APP and PS and then blotted with 22C11, which recognizes human and hamster APP. APP–PS1 coprecipitation was found in nontransfected cells, representing endogenous complex formation (Fig. 3E, lanes 1–2). Using the same 22C11 antibody, endogenous hamster APP precipitated with transfected human PS2 in line C-PS2 (Fig. 3E, lane 3). The endogenous coprecipitating species from CHO cells was N-glycosylated, as in the 7W cells (Fig. 3E, lanes 4 and 5). These results demonstrate clearly that APP–PS interactions occur at endogenous levels of each protein in vivo. Endogenous APP also coprecipitated with endogenous PS1 in nontransfected 293 and human glioma HS683 cells (data not shown). These results suggest a generalized occurrence of APP–PS complexes in mammalian cells.
The APP Cytoplasmic Domain Is Dispensable for PS Association.
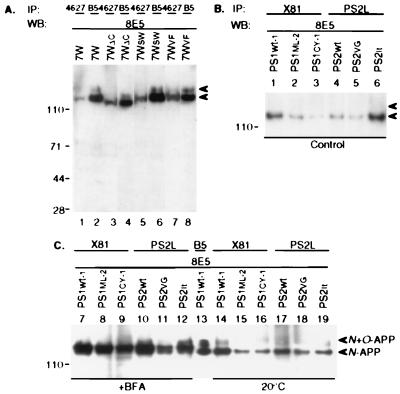
Based on topological studies of PS1, the N-, loop, and C-terminal domains of the protein all are oriented toward the cytoplasmic space (24, 25), in which the C-terminal domain of APP is found. To identify whether this region of APP is responsible for the interaction with PS, we analyzed PS1-APP complex formation in a stable CHO cell line (7WΔC) expressing APP751 truncated at residue 709 and thus lacking almost the entire cytoplasmic domain (710–751). 7WΔC cells were lysed, precipitated with 4627 (to PS1 C-terminus), and blotted with 8E5. We observed a coprecipitating APP species that: (i) migrated just below the full-length N-glycosylated APP precipitated by 4627 from the wt 7W cells (Fig. 4A, lane 3), and (ii) comigrated precisely with the truncated N-glycosylated APP precipitated by APP antibody B5 from the same 7WΔC lysate (Fig. 4A, lane 4). Thus, the cytoplasmic domain of APP is dispensable for the PS1 interaction.
Figure 4.
APP–PS complex formation in various cell lines and conditions. (A) IP by 4627 (PS1) or B5 (APP) reveals precipitated APP in CHO single transfectants expressing either wt (7W), C-terminal truncated (7WΔC), Swedish mutant (7WSW), or V698F mutant (7WVF) APP. Note reduced molecular weight of coprecipitated APP in 7WΔC (lanes 3 and 4), as expected. (B) IP by X81 (PS1) or PS2L (PS2) and WB by 8E5 (APP) reveals coprecipitating N-APP (lower arrowhead) in both wt and mutant PS1 and PS2 transfectants. (C) Persistent complex formation in various CHO lines after treatment with BFA (10 μg/ml, l hr) or 20°C temperature block (2.5 hr). The BFA-induced intermediate-sized APP species (27) coprecipitates with PS1 or PS2.
APP–PS Complex Formation in Cells Expressing Familial AD-Linked Missense Mutations.
To determine whether familial AD-causing mutations in APP, PS1, or PS2 affect the interaction between the two proteins, six stable CHO cell lines were used for immunoprecipitation/Western blot analyses: 7WSW and 7WVF expressing solely the KM651/652NL (Swedish) or V698F missense mutations in APP751; PS1ML-2 and PS1CY-1 expressing wt APP plus either the M146L or C410Y PS1 mutation; and PS2VG and PS2It expressing wt APP and either the N141I (Volga German) or M239V (Italian) PS2 mutation. When these various lines were lysed and precipitated with either X81, 4627, or PS2L, the characteristic N-glycosylated APP band was detected in all six lines (Fig. 4 A, lanes 5 and 7, and B, lanes 1–6). Therefore, both wt and mutant APP, PS1, and PS2 proteins can form complexes. No consistent qualitative or quantitative differences have been detected using the current methods.
Subcellular Localization of APP–PS Complex Formation.
Because PS1 and PS2 proteins have been localized principally to the ER, we expected the formation of APP–PS complexes to occur in this compartment. To examine this possibility, the wt and mutant PS–APP double-transfectants were treated with brefeldin A (BFA) for 1 hr followed by immunoprecipitation/Western blotting. BFA causes a collapse of the Golgi network, resulting in retention of proteins in the ER (26). It therefore changes APP glycosylation, resulting in one broad band of intermediate molecular weight migrating between the usual N- and N+O-glycosylated APP species on SDS gels (27). We found that BFA treatment did not noticeably affect the ability of APP to interact with PS1 or PS2 proteins. The altered form of APP was readily detected in both PS1 and PS2 immunoprecipitates (Fig. 4C, lanes 7–12). We also examined the effects of a 20°C temperature block, which results in protein retention principally in the trans-Golgi network (27, 28). Incubation of cells at 20°C for 2.5 hr followed by lysis and coprecipitation yielded a result consistent with the previous findings (Fig. 4C, lanes 14–19).
DISCUSSION
Evidence from numerous laboratories has established that Aβ42 is the first amyloid peptide to be deposited in the brain in the course of AD-type neurodegeneration and that diffuse plaques composed of Aβ42 appear years or decades before mature (neuritic) plaques, microgliosis, astrocytosis, neurofibrillary tangles, and the other cytopathological features of AD (see e.g., refs. 29–32). A primary role for Aβ42 deposition is further supported by studies of a transgenic mouse model of AD, in which cerebral levels of Aβ42 rise sharply in the brain before the development of diffuse and neuritic plaques that closely resemble those of AD (33, 34). These findings are consistent with biochemical studies that demonstrate that Aβ42 aggregates into amyloid fibrils far more rapidly than Aβ40 (35). Moreover, familial AD-linked PS mutations consistently increase cellular Aβ42 production (5–10, 16). In view of these observations, elucidating the molecular mechanism whereby PS proteins regulate Aβ42 production is important for understanding the pathogenesis of AD in general.
Because PS and APP show partial subcellular colocalization, we addressed the hypothesis that these proteins interact to form stable complexes in cells. Here, we demonstrate a highly reproducible interaction between each of the presenilins and the N-glycosylated form of APP, the specificity of which is supported by several lines of evidence: (i) Numerous antibodies recognizing different epitopes in PS1, PS2, or APP all could precipitate the PS–APP complexes. (ii) PS and APP antibodies could be used for immunoprecipitation/Western blotting in either order with the same positive results. (iii) The amount of PS–APP complexes varied directly with expression levels in stable transfectants with widely varying PS and APP levels. (iv) Coexpression of a control transmembrane protein did not lead to association with PS1, suggesting that the APP–PS interaction is not simply due to overexpression. (v) Nontransfected cells showed PS–APP complex formation at endogenous protein levels. (vi) Treatment of cells with BFA changed the glycosylation of APP, as previously reported (27), and this modified form was specifically immunoprecipitated with PS1. (vii) Stable expression of different-sized APP polypeptides (APP695, APP751, and APPΔC) always led to specific coprecipitation of the appropriately sized species with PS. (viii) PS1 and PS2 each underwent highly similar complex formation with APP. (viv) The findings were confirmed in three distinct cell types. We conclude that PS and APP interact in intact living cells to form stable complexes.
None of four different missense mutations in PS1 or PS2 noticeably affected complex formation with APP, nor did the APP mutations KM651/652NL or V698F noticeably alter complex formation with PS1. However, the consistent augmentation of the intramembranous 42-γ-secretase cleavage of APP caused by PS mutations (5–10, 16) indicates that the interactions of mutant and wt PS with APP must differ at the molecular level. We now are conducting experiments aimed at detecting a subtle biochemical difference in the interactions. Of particular significance in this regard is our finding that a truncated APP protein lacking the C-terminal domain can coprecipitate with PS1, indicating that the cytoplasmic domain of APP is dispensable for the interaction. Further experiments examining whether forms of APP lacking the transmembrane domain or various segments of the ectodomain still bind to PS1 are underway. Based on the available data, we speculate that one or more of the 6–8 transmembrane regions of PS1 (24, 25) could interact with the single APP transmembrane domain. Such an interaction would be particularly significant in view of the localization of the γ-secretase cleavage site to the transmembrane domain of APP. We hypothesize that some APP and PS molecules form stable complexes and that any subtle alteration of this interaction brought about by PS mutations may enhance the access of a γ-secretase specifically cleaving APP at residue 42 of Aβ. This model could explain why missense mutations scattered widely in PS1 transmembranous and cytoplasmic domains all cause the same phenotype, an increase of Aβ42 production. Any conformational change caused by a PS1 mutation could lead to a subtle change in the topology of PS–APP complexes, with subsequent increased exposure of APP to a γ-secretase cleaving specifically at Aβ42. Evidence that pharmacologically distinct 40- and 42-specific γ-secretase activities actually exist in cells recently has been presented (36, 37).
Our results suggest that only a minor fraction of total cellular APP molecules may complex with PS. When a polyclonal APP antibody (e.g., B5) is used to immunoprecipitate a cell lysate, subsequent blotting with an APP antibody (e.g., 8E5) reveals substantially more APP than when the same lysate is precipitated with a polyclonal PS1 antibody (e.g., X81) (Fig. 4A). However, the fact that two different antibodies must be used to perform these precipitations precludes a firm quantitative estimate of the relative amounts of PS and APP that participate in the complexes. Whereas our experiments clearly demonstrate an interaction between APP and full-length PS, the question of whether PS endoproteolytic fragments also participate in APP complex formation will require further study. Furthermore, our results leave open the question of whether one or more other proteins are present in the complexes that contain PS and APP and whether the PS–APP interaction is direct or indirect.
After our studies were completed, Weidemann et al. (38) reported the detection of complexes between APP and PS2, but not PS1, in transiently transfected COS cells. Their finding about PS2 concurs with our results, although the high level of expression of the proteins in the COS transient system used to demonstrate the interaction contrasts to our detection of APP–PS complexes in stable transfectants expressing low levels of the proteins and in untransfected cells at endogenous protein levels. Dewji and Singer (39) described an in vitro interaction between cells expressing APP and other cells expressing PS, but their data does not unequivocally implicate PS directly in the intercellular binding; moreover, there is currently no evidence that PS is expressed at the cell surface in most cell types.
Based on the data in Fig. 4, the cellular localization of PS–APP complexes may be principally in the ER. BFA treatment did not affect complex formation, and the appropriate altered form of APP was coprecipitated with PS, indicating that the interaction occurs at least in part in the ER. APP–PS complex formation occurring even at 20°C further supports this event happening in an early vesicular compartment. The existence of a small amount of N+O-glycosylated APP that sometimes coprecipitated with PS1 in the PS1Wt-1 cells (Fig. 2) suggests that the early Golgi compartment also may be a site for complex formation. We recently have detected Aβ42 in both ER and Golgi fractions prepared from the same CHO cell lines used here (unpublished work), and thus APP–PS interactions in these compartments could well play a role in Aβ42 generation.
In summary, our findings provide a central clue for understanding the mechanism by which mutant PS proteins selectively alter APP processing. We conclude that PS and APP form complexes in vivo, predominantly in the ER, and that the transmembrane regions may be implicated in this process. These data establish a model that may explain the Aβ42 phenotype of cells expressing mutant PS1 and PS2, with attendant implications for the cerebral accumulation of Aβ42 plaques in Alzheimer disease in general.
Acknowledgments
We thank Drs. T. Iwatsubo, T. Saido, K. Maruyama, and T. Tomita for antibody PS2L; C. Haass for antibody 2972; P. Seubert and D. Schenk for antibodies 13A11, 8E5, and 13G8; B. Hyman for antibody 311/2a; L. D’Adamio for antibody PS2n; and Dr. M. Citron for helpful comments. This work was supported by National Institutes of Health Grants AG05134 and AG12749 (to D.J.S.) and AG12376 and NS01812 (to E.H.K.), and the Foundation for Neurologic Diseases.
ABBREVIATIONS
- Aβ
amyloid β-protein
- CHO
Chinese hamster ovary
- PS
presenilin
- AD
Alzheimer disease
- APP
β-amyloid precursor protein
- ER
endoplasmic reticulum
- BFA
brefeldin A
- TR
transferrin receptor
- wt
wild type
References
- 1.Selkoe D J. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 2.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, et al. Nature (London) 1995;375:754–760. [Google Scholar]
- 3.Rogaev E I, Sherrington R, Rogaeva E A, Levesque G, Ikeda M, et al. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 4.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell H, Yu C, Jondro P D, Schmidt S D, Wang K, Crowley A C, Fu Y-H, Guenette S Y, Galas D, Nemens E, Wijsman E M, Bird T D, Schellenberg G D, Tanzi R E. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 5.Borchelt D, Thinakaran G, Eckman C, Lee M, Davenport F, Ratovitsky T, Prada C, Kim G, Seekins S, Yager D, Slunt H, Wang R, Seeger M, Levey A, Gandy S, Copeland N, Jenkins N, Price D, Younkin S, Sisodia S. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 6.Xia W, Zhang J, Kholodenko D, Citron M, Podlisny M B, Teplow D B, Haass C, Seubert P, Koo E H, Selkoe D J. J Biol Chem. 1997;272:7977–7982. doi: 10.1074/jbc.272.12.7977. [DOI] [PubMed] [Google Scholar]
- 7.Citron M, Westaway D, Xia W, Carlson G, Diehl T, et al. Nat Med. 1997;3:67–72. doi: 10.1038/nm0197-67. [DOI] [PubMed] [Google Scholar]
- 8.Duff K, Eckman C, Zehr C, Yu X, Prada C, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, Morgan D, Gordon M, Holcomb L, Refolo L, Zenk B, Hardy J, Younkin S. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 9.Scheuner D, Eckman C, Jensen M, Song X, Citron M, et al. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 10.Lemere C A, Lopera F, Kosik K S, Lendon C L, Ossa J, Saido T C, Yamaguchi H, Ruiz A, Martinez A, Madrigal L, Hincapie L, Arango L, J C, Anthony D C, Koo E H, Goate A M, Selkoe D J, Arango V J C. Nat Med. 1996;2:1146–1150. doi: 10.1038/nm1096-1146. [DOI] [PubMed] [Google Scholar]
- 11.Selkoe D J. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 12.Kovacs D M, Fausett H J, Page K J, Kim T-W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, Human B T, Tanzi R E, Wasco W. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 13.Cook D G, Sung J C, Golde T E, Felsenstein K M, Wojczyk B S, Tanzi R E, Trojanowski J Q, Lee V M-Y, Doms R W. Proc Natl Acad Sci USA. 1996;93:9223–9228. doi: 10.1073/pnas.93.17.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter J, Capell A, Grunberg J, Pesold B, Schindzielorz A, Prior R, Podlisny M, Fraser P, St. George Hyslop P, Selkoe D, Haass C. Mol Med. 1996;2:673–691. [PMC free article] [PubMed] [Google Scholar]
- 15.De Strooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, St. George-Hyslop P, Van Leuven F. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 16.Tomita T, Maruyama K, Saido T C, Kume H, Shinozaki K, Tokuhiro S, Capell A, Walter J, Gruenberg H, Haass C, Iwatsubo T, Obata K. Proc Natl Acad Sci USA. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vito P, Wolozin B, Ganjei J K, Iwasaki K, Lacana E, D’Adamio L. J Biol Chem. 1996;271:31025–31028. doi: 10.1074/jbc.271.49.31025. [DOI] [PubMed] [Google Scholar]
- 18.Oltersdorf T, Ward P J, Henriksson T, Beattie E C, Neve R, Lieberburg I, Fritz L C. J Biol Chem. 1990;265:4492–4497. [PubMed] [Google Scholar]
- 19.Koo E H, Squazzo S. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 20.Perez R, Koo E. In: Processing of the β-Amyloid Precursor Protein: Effects of C-Terminal Mutations on Amyloid Production. Iqbal K, Winblad B, Nishimura T, Takeda M, Wisniewski H M, editors. London: J. Wiley & Sons; 1997. pp. 407–416. [Google Scholar]
- 21.Perez R G, Squazzo S L, Koo E H. J Biol Chem. 1996;271:9100–9107. doi: 10.1074/jbc.271.15.9100. [DOI] [PubMed] [Google Scholar]
- 22.Podlisny M B, Citron M, Amarante P, Sherrington R, Xia W, Zhang J, Diehl T, Levesque G, Fraser P, Haass C, Koo E H, Seubert P, St. George-Hyslop P, Teplow D B, Selkoe D J. Neurobiol Dis. 1997;3:325–337. doi: 10.1006/nbdi.1997.0129. [DOI] [PubMed] [Google Scholar]
- 23.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Rotovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey A I, Gandy S E, Jenkins N A, Copeland N G, Price D L, Sisodia S S. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 24.Doan A, Thinakaran G, Borchelt D, Slunt H, Ratovitsky T, Podlisny M, Selkoe D, Seeger M, Gandy S, Price D, Sisodia S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann, S., Chiesa, R. & Harris, D. A. (1997) J. Biol. Chem., in press. [DOI] [PubMed]
- 26.Kelly R B. Cell. 1990;61:5–7. doi: 10.1016/0092-8674(90)90206-t. [DOI] [PubMed] [Google Scholar]
- 27.Haass C, Lemere C A, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe D J. Nat Med. 1995;1:1291–1296. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- 28.Matlin K S, Simons K. Cell. 1983;34:233–243. doi: 10.1016/0092-8674(83)90154-x. [DOI] [PubMed] [Google Scholar]
- 29.Mann D M A. Neurobiol Aging. 1989;10:397–399. doi: 10.1016/0197-4580(89)90073-0. [DOI] [PubMed] [Google Scholar]
- 30.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Neuron. 1995;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 31.Iwatsubo T, Mann D M, Odaka A, Suzuki N, Ihara Y. Ann Neurol. 1995;37:294–299. doi: 10.1002/ana.410370305. [DOI] [PubMed] [Google Scholar]
- 32.Lemere C A, Blustzjan J K, Yamaguchi H, Wisniewski T, Saido T C, Selkoe D J. Neurobiol Dis. 1996;3:16–32. doi: 10.1006/nbdi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 33.Johnson-Wood K, Lee M, Motter R, Hu K, Gordon G, Barbour R, Khan K, Gordon M, Tan H, Games D, Lieberburg I, Schenk D, Seubert P, McConlogue L. Proc Natl Acad Sci USA. 1997;94:1550–1555. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masliah E, Sisk A, Mallory M, Mucke L, Schenk D, Games D. J Neurosci. 1996;16:5795–5811. doi: 10.1523/JNEUROSCI.16-18-05795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarrett J T, Berger E P, Lansbury P T., Jr Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 36.Citron M, Diehl T, Gordon G, Biere A, Seubert P, Selkoe D. Proc Natl Acad Sci USA. 1996;93:13170–13175. doi: 10.1073/pnas.93.23.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klafki H W, Abramowski D, Swoboda R, Paganeti P A, Staufenbiel M. J Biol Chem. 1996;271:28655–28659. doi: 10.1074/jbc.271.45.28655. [DOI] [PubMed] [Google Scholar]
- 38.Weidemann A, Paliga K, Durrwang U, Czech C, Evin G, Masters C L, Beyreuther K. Nat Med. 1997;3:328–332. doi: 10.1038/nm0397-328. [DOI] [PubMed] [Google Scholar]
- 39.Dewji N N, Singer S J. Proc Natl Acad Sci USA. 1996;93:12575–12580. doi: 10.1073/pnas.93.22.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]