Abstract
Because plants produce cytotoxic compounds to which they, themselves, are susceptible and are exposed to exogenous toxins (microbial products, allelochemicals, and agrochemicals), cell survival is contingent on mechanisms for detoxifying these agents. One detoxification mechanism is the glutathione S-transferase-catalyzed glutathionation of the toxin, or an activated derivative, and transport of the conjugate out of the cytosol. We show here that a transporter responsible for the removal of glutathione S-conjugates from the cytosol, a specific Mg2+-ATPase, is encoded by the AtMRP1 gene of Arabidopsis thaliana. The sequence of AtMRP1 and the transport capabilities of membranes prepared from yeast cells transformed with plasmid-borne AtMRP1 demonstrate that this gene encodes an ATP-binding cassette transporter competent in the transport of glutathione S-conjugates of xenobiotics and endogenous substances, including herbicides and anthocyanins.
Keywords: anthocyanins, herbicides, heterologous expression, vacuolar membrane, xenobiotic detoxification
The recent finding that intact vacuoles and vacuolar membrane vesicles isolated from vascular plants mediate MgATP-dependent accumulation of glutathione S-conjugates (GS-conjugates) in the absence of a transmembrane H+-electrochemical potential difference (1, 2) is seminal in two respects. Not only does it implicate MgATP as a direct energy source for organic solute transport across plant membranes but, because the compounds transported include glutathionated herbicides (1, 2), isoflavonoid phytoalexins (3), and possibly anthocyanins (4), a mechanism for the vacuolar sequestration of GS-conjugable xenobiotics and endogenous substances appears to have been discovered.
Despite its strategic value for manipulating and investigating toxin compartmentation in plants, neither the protein nor the gene encoding the plant vacuolar GS-conjugate pump have been identified. Significant therefore are three clues as to its possible identity. (i) The resemblance of plant vacuolar GS-conjugate transport to that mediated by the GS-conjugate transporting Mg2+-ATPase, GS-X pump (5), of mammalian cells. In both cases, transport is selective for GS-conjugates and oxidized glutathione (GSSG), but not reduced glutathione (GSH), directly energized by MgATP and potently inhibited by the phosphoryl transition state analog, vanadate (1, 2). (ii) The ability of the human multidrug resistance-associated protein gene (HmMRP1), an ATP-binding cassette transport protein (ABC transporter) gene isolated from drug-resistant small cell lung carcinoma cell lines (6), to confer GS-X pump activity, specifically MgATP-energized transport of leukotriene C4 and related GS-conjugates, on transfected cells (7, 8). (iii) The functional and structural equivalence of the Saccharomyces cerevisiae (yeast) cadmium factor (ScYCF1) gene product, identified according to its ability to confer cadmium resistance (9), to HmMRP1, its localization to the vacuolar membrane (10) and the capacity of HmMRP1 for alleviating the cadmium-hypersensitive phenotype and restoring GS-conjugate transport in membranes prepared from yeast strains deleted for the YCF1 gene (11, 12). Here we describe the isolation of a gene from Arabidopsis thaliana (AtMRP1), encoding a protein belonging to the same subclass of ABC transporters as HmMRP1 and ScYCF1, whose heterologous expression in yeast confers MgATP-energized GS-conjugate transport. In so doing, we provide a functional definition of an ABC transporter from vascular plants and identify an element involved in the removal of GS-conjugable compounds from the cytosol in a manner independent of the transmembrane H+ gradient.
MATERIALS AND METHODS
Isolation of AtMRP1.
On the basis of the functional resemblance between plant vacuolar GS-conjugate transport and that mediated by ScYCF1 and HmMRP1, and the 44.6% sequence identity (63.9% similarity) between ScYCF1 and HmMRP1, degenerate oligonucleotide primers corresponding to the second ATP-binding cassette of ScYCF1 and HmMRP1 (positions 1,300–1,321 and 1,474–1,494, respectively)—two of the most ScYCF1- and HmMRP1-specific sequences common to both—were used for the isolation of plant genes likely involved in GS-conjugate transport by PCR amplification of A. thaliana genomic DNA. The sequences of the two primers yielding a 0.6-kb HmMRP1- and ScYCF1-hybridizing amplification product by this approach were: 5′-GARAARGTIGGIATHGTIGGIMGIACIGGIGC-3′ (MRP2) and 5′-TCCATDATIGTRTTIARICKTGIGC-3′ (MRP4), where I = inosine, K = T or G, M = C or A, and R = A or G. MRP2 corresponds to amino acid residues 1,300–1,310 and 1,321–1,331 of ScYCF1 and HmMRP1, respectively; MRP4 corresponds to residues 1,466–1,474 and 1,486–1,494 (6, 9). After determining that the 0.6-kb PCR product exhibited greatest similarity to ScYCF1 and HmMRP1 plus the putative translation product of an unidentified 1.6-kb A. thaliana expressed sequence tag (EST) (ATTS1246) (13), a mixed probe consisting of this and the PCR product was used to screen approximately 3 × 105 plaques of a size-fractionated (3–6 kb) A. thaliana cDNA library constructed in λZAPII (14). The cDNA insert (3.5 kb) of the largest of the clones isolated from this screen was subcloned into pBluescript SK−, sequenced, and, of all the sequences in the GenBank/EMBL database release 90 (15), was found to align best with ScYCF1 and HmMRP1. On the assumption that the complete ORF of the gene corresponding to this cDNA was similar in size to those of ScYCF1 and HmMRP1, it was estimated to be missing a minimum of 1.5 kb of 5′ coding sequence. Hence, to isolate clones containing this missing sequence, 500 bp of the 5′ terminus of the 3.5-kb cDNA was used to screen two A. thaliana bacterial artificial chromosome (BAC) libraries, UCD (16) and TAMU (17). This procedure yielded eight genomic clones: U1L22, U8C12, U12A2, U23J22, U4I9, T9C22, T1B17, and T4K22. After establishing that BAC clone U1L22 encompassed the 3.5-kb cDNA, a 3-kb BglII fragment containing the 5′ sequence missing from the cDNA was used to rescreen approximately 2 × 106 plaques from the A. thaliana λZAPII cDNA library. Twenty-six independent positive clones were obtained, and the one containing the longest cDNA insert, 5.2 kb, was subcloned, sequenced, and designated AtMRP1.
The genomic sequence of AtMRP1 was determined from BAC clone T1B17, whose exon sequences were found in the course of another series of investigations to match exactly the coding sequence of the cDNA. After digestion with HindIII the resulting 9.2- and 3.5-kb restriction fragments of T1B17 were subcloned into pBluescript SK− and sequenced. Double-stranded DNA sequencing was performed using a dye terminator cycle sequencing kit (Perkin–Elmer) by primer walking or by the generation of nested deletions with exonuclease III (Pharmacia). The positions and sizes of the introns of the genomic clone were deduced by comparison with the cDNA sequence of AtMRP1.
Northern Analyses.
Expression of AtMRP1 in A. thaliana was determined by Northern analysis. Total RNA was extracted from roots, stems leaves, and flowers of 5-week-old plants by the phenol/LiCl method (14) in homogenization buffer supplemented with aurintricarboxylic acid (25 μM). The RNA samples were resolved on 10% (vol/vol) formaldehyde-agarose gels, transferred to Hybond-N+ membrane filters, and baked at 80°C for 2 hr. The filters were prehybridized in 7% (wt/vol) SDS/1 mM EDTA/0.3 M sodium phosphate buffer (pH 7.2) at 65°C for 2 hr before hybridization overnight in the same buffer containing 32P-labeled, random-primed 3.5-kb SacI/HindIII restriction fragment corresponding to the coding sequence of AtMRP1. The filters were washed twice in 1× standard saline citrate/0.1% (wt/vol) SDS (5 min, room temperature) and once in 0.2× SSC/0.1% (wt/vol) SDS (15 min, 65°C). 32P-labeled bands were visualized using a Molecular Dynamics PhosphorImager.
Determination of Physical Map Position of AtMRP1.
The physical map position of the AtMRP1 gene was determined by screening two A. thaliana yeast artificial chromosome (YAC) libraries, CIC and YUP (18), with the full-length AtMRP1 cDNA clone. Thirteen positive YAC clones (CIC12A12, CIC8B11, CIC9B11, CIC11B11, CIC12B11, CIC11E10, CIC12E10, YUP16B3, YUP16C2, YUP15D4, YUP16H7, YUP11E12, and YUP3F11) were recovered of which seven (YUP15D4, YUP3F11, YUP11E12, YUP16H7, CIC12A12, CIC12B11, and CIC12E10) yielded PCR product of the length (400 bp) and sequence expected of AtMRP1 after amplification with oligonucleotide primers AtMRP11250S (5′-CACTGATTGCTGCTGTGTTG-3′) and AtMRP9A1 (5′-TCAGTGCTATAATTATACG-3′). From a search of the A. thaliana genome database all seven YAC clones were found to map to a region between SSLP markers nga392 and nga280 on chromosome 1.
Heterologous Expression of AtMRP1 in S. cerevisiae ycf1Δ mutants.
For constitutive expression of AtMRP1 in S. cerevisiae a derivative of the yeast-Escherichia coli shuttle vector pYES2 (Invitrogen) was constructed in which the 831-bp XbaI/NotI restriction fragment containing the 3-phosphoglycerate kinase (PGK) gene promoter from plasmid pFL61 (19) was inserted between the SpeI/NotI restriction sites of pYES2, so replacing the galactose-inducible yeast GAL1 promoter of pYES2 with the constitutive yeast PGK promoter to generate pYES3. AtMRP1 cDNA, from which the 5′ untranslated region had been removed by PCR, was inserted into the multiple cloning site located between the PGK promoter and cytochrome c1 gene (CYC1) termination sequences of pYES3. After confirming the fidelity of the constructs by sequencing, S. cerevisiae ycf1Δ strain DTY168 (MATα his6 leu2–3,-112, ura 3–52 ycf1::hisG) (9) was transformed with pYES3-AtMRP1 or empty vector lacking the AtMRP1 insert (pYES3) by the LiOAc/polyethylene glycol method (20) and selected for uracil prototrophy (21).
Measurement of GS-Conjugate Transport.
Cells were grown and vacuolar membrane-enriched vesicles were purified as described (21). Uptake of S-(2,4-dinitrophenyl)[3H]glutathione (DNP-GS), [3H]GSSG, [3H]cyanidin 3-glucoside-GS (C3G-GS), or [14C]metolachlor-GS was measured in 200-μl reaction volumes containing membrane vesicles (10–20 μg), 3 mM ATP, 3 mM MgSO4, 5 μM gramicidin D, 10 mM creatine phosphate, 16 units/ml creatine kinase, 50 mM KCl, 1 mg/ml BSA, 400 mM sorbitol, and 25 mM Tris⋅Mes (pH 8.0). Uptake was terminated by the addition of 1 ml of ice-cold wash medium (400 mM sorbitol/3 mM Tris⋅Mes, pH 8.0) and vacuum filtration of the suspension through prewetted Millipore cellulose nitrate filters (pore size 0.45 μm). The filters were rinsed twice with wash medium, and the radioactivity retained was determined by liquid scintillation counting. Nonenergized uptake was estimated by the same procedure except that ATP was omitted from the uptake media.
Measurement of Protein.
Protein was estimated by a modification of the Lowry method (22).
Chemicals.
DNP-[3H]GS (17.4 mCi/mmol) was synthesized enzymically from 1-chloro-2,4-dinitrobenzene and [3H]GSH and purified as described (2). [3H]GSSG was synthesized enzymically by incubation of 100 μM [3H]GSH (44 Ci/mmol) and 100 μM hydrogen peroxide with 10 units of glutathione peroxidase (type III from baker’s yeast) for 4 hr at 25°C in 10 mM phosphate buffer, pH 7.5. [14C]Metolachlor-GS was synthesized by general base catalysis by adding 100 μM GSH and 50 μM [14C]metolachlor (8.3 mCi/mmol) to 25 mM sodium borate buffer, pH 9.0, and incubating overnight at 55°C. C3G-[3H]GS was synthesized enzymically by incubating equimolar C3G (8.8 mM) and [3H]GSH (44 Ci/mmol) at 25°C overnight with the total glutathione S-transferase (GST) fraction from maize shoots. Total maize GSTs were extracted and affinity-chromatographed as described (23) except that 1 μM acivicin was added to the extraction buffer to minimize γ-glutamyl transpeptidase activity. All of the conjugates were purified by reverse-phase FPLC as described (2). All other reagents were of analytical grade and purchased from Fisher, Fluka, Research Organics, or Sigma.
RESULTS
Isolation of AtMRP1.
PCR amplification of A. thaliana genomic DNA using oligonucleotide primers corresponding to the second ATP-binding cassette of ScYCF1 and HmMRP1 (Fig. 1) yielded a 0.6-kb product that hybridized with the equivalent PCR products of ScYCF1 and HmMRP1. Sequence analysis disclosed that this 0.6-kb PCR product comprised a 76-bp intron and 510-bp exon, the latter of which shared 60.6% and 66.5% amino acid sequence identity with the corresponding regions of ScYCF1 and HmMRP1 and complete nucleotide sequence identity with the 5′ segment of a 1.6-kb A. thaliana EST, ATTS1246 (13). Given that their deduced amino acid sequences were as similar to ScYCF1 and HmMRP1 as ScYCF1 and HmMRP1 are to each other, the A. thaliana PCR product and EST were used as probes for the isolation of full-length clones.
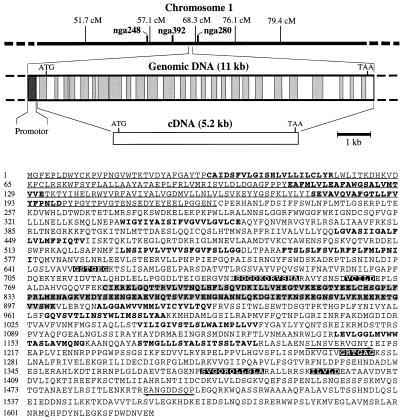
Figure 1.
Chromosomal location and organization of genomic clone of AtMRP1 and deduced amino acid sequence of polypeptide encoded by AtMRP1 cDNA. The relative positions and sizes of the introns and exons of the genomic clone, determined by comparison with the cDNA sequence, are indicated by shaded and open rectangles, respectively. The Walker A and B boxes (sequences GSTGEG and GRTGAG, and VCILD and ILVLD, respectively) and the ABC signature motifs (sequences SGGQKQRVSMA and SVGQRQLLSLA) of the deduced amino acid sequence of AtMRP1 are indicated by white text on a black background. The N-terminal extension and CFTR-like truncated regulatory (R) domain are indicated by underlined plain typeface and shaded bold typeface, respectively. Putative transmembrane spans, identified using the TopPred II program (Claros and von Heijne, Karolinska Institute, Stockholm, Sweden), are indicated by unshaded bold typeface. The positions and directions of the two PCR primers used to amplify A. thaliana genomic DNA during the initial steps of cloning AtMRP1 are indicated by arrows.
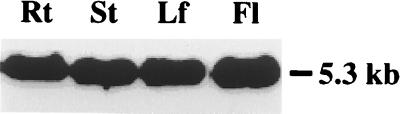
Full-length clones were isolated in three stages. A cDNA library was screened with a mixed probe consisting of the 0.6-kb PCR product and EST to isolate a partial (3.5 kb) clone exhibiting 42.5% and 43.5% amino acid identity to ScYCF1 and HmMRP1, respectively; 0.5 kb of the most 5′ portion of the 3.5-kb cDNA was used to screen two BAC libraries to identity a clone (U1L22) encompassing the sequence of the cDNA; and a 3-kb restriction fragment of clone U1L22, containing the 5′ sequence missing from the 3.5-kb cDNA, was used to rescreen the cDNA library. The longest cDNA insert (5.2 kb) recovered from the rescreen was designated AtMRP1. After verifying that this cDNA is derived from A. thaliana rather than a nonplant contaminant of the cDNA library, by showing that random-primed AtMRP1 hybridized at high stringency with a single 5.3-kb band on Northern blots of total RNA extracted from roots, stems, leaves, and flowers of 5-week-old A. thaliana plants (Fig. 2), its chromosomal location and complete sequence were determined.
Figure 2.
Northern analysis of expression of AtMRP1 transcript in roots, stems, leaves, and flowers of A. thaliana. Total RNA (10 μg per lane) was extracted, separated, blotted, and hybridized with 32P-labeled, random-primed AtMRP1 as described in Materials and Methods. The 5.3-kb band indicated was the only 32P-labeled band detected.
Physical Map Position and Sequence of AtMRP1.
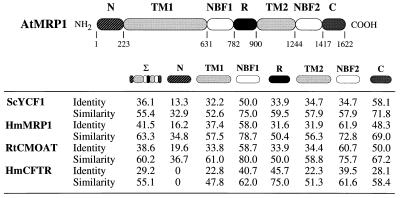
AtMRP1 mapped between SSLP markers nga392 and nga280 on chromosome 1 and encompassed a single ORF of 1,622 amino acid residues (Fig. 1). The 181-kDa polypeptide encoded exhibited at least 36% overall sequence identity (55% similarity) to ScYCF1, HmMRP1, and another established GS-X pump, the rat canalicular multispecific organic anion transporter, and 29% identity (55% similarity) to the human cystic fibrosis transmembrane conductance regulator (CFTR) (Fig. 3). AtMRP1 possessed two nucleotide-binding folds (NBF1 and NBF2), each containing the Walker A and B motifs and C domain, characteristic of ABC transporters (24), and 12 putative transmembrane spans, whose location was consistent with the topologies inferred for HmMRP1 (6) and ScYCF1 (9) (Figs. 1 and 3). In addition, two subclass-specific structures were evident: a putative “regulatory” (R) domain contiguous with NBF1, rich in charged amino acids (Figs. 1 and 3), common to the MRP and CFTR subclasses, but which is truncated in the former (117–161 vs. 256 amino acid residues) (Fig. 3); and a 192–223 amino acid residue N-terminal extension, absent from the CFTR subclass but present in all MRP-subclass members (Fig. 3).
Figure 3.
Identity and similarity analysis of putative domains of AtMRP1 against ScYCF1, HmMRP1, rat canalicular multispecific organic anion transporter (RtCMOAT), and human CFTR (HmCFTR). In addition to the overall sequences (Σ), the regions examined were the N-terminal extension (N), transmembrane segments 1 and 2 (TM1 and TM2, respectively), CFTR-like putative regulatory domain (R), NBF1 and NBF2, and C terminus (C). The compilation was derived from the results of GAP comparisons using the default parameters of the GCG package (version 8.0, University of Wisconsin, Madison, WI). The GenBank/EMBL accession numbers for the proteins shown were: AF008124, AtMRP1; L35237, ScYCF1; L05628, HmMRP1; L49379, RtCMOAT; M28668, HmCFTR.
A dendrogram derived from a paup analysis comparing AtMRP1 with other representative members of the ABC transporter superfamily clearly demonstrated that AtMRP1, MRP1, and canalicular multispecific organic anion transporter from various sources, ScYCF1, three other ABC transporters, for which GS-X pump activity has yet to be reported—the rabbit epithelial basolateral conductance regulator (RbEBCR), Leishmania P-glycoprotein-related molecule (LEPGP1), and yeast (S. cerevisiae) oligomycin resistance protein (ScYOR1)—belong to the same subclass, a subclass within the same cluster as CFTR but remote from the multidrug resistance proteins (MDRs or PGPs), major histocompatibility complex class II-linked peptide transporters (TAPs), Schizosaccharomyces pombe heavy metal tolerance protein (SpHMT1), and S. cerevisiae mating factor export protein (ScSTE6) (data not shown).
AtMRP1 Catalyzes GS-Conjugate Transport.
To determine if the structural similarity between AtMRP1, ScYCF1, and HmMRP1 signifies a functional equivalence, the capacity of heterologously expressed AtMRP1 for MgATP-energized, uncoupler-insensitive GS-conjugate transport was tested. For this purpose, S. cerevisiae ycf1Δ strain DTY168, from which 95% of the coding sequence of the YCF1 gene had been deleted (9) and high-affinity, MgATP-dependent, uncoupler-insensitive vacuolar GS-conjugate transport is abolished (10–12), was transformed with empty expression vector, pYES3, or vector containing the entire ORF of AtMRP1 (pYES3-AtMRP1) under the control of the constitutive yeast phosphoglycerate kinase gene (PGK) promoter. After growth on selective media, vacuolar membrane-enriched vesicles (21) were prepared from these and untransformed DTY168 cells and assayed for GS-conjugate transport. To facilitate direct comparisons of the transport activity of AtMRP1 with those of ScYCF1, HmMRP1, and the endogenous plant vacuolar GS-conjugate pump, the model compound, DNP-GS—the only compound whose transport characteristics have been determined in all three of these systems (1, 2, 7, 8, 10–12)—was used as transport substrate for the initial experiments.
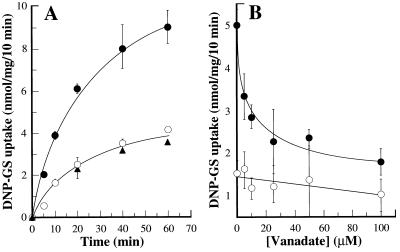
Vector alone exerted negligible effect on DNP-GS uptake, implying that the cDNA insert of pYES3-AtMRP1 was the factor responsible for the increases in transport measured. When assayed at an initial DNP-[3H]GS concentration of 61.3 μM, the rates of MgATP-dependent, uncoupler-insensitive uptake by vacuolar membrane-enriched vesicles purified from untransformed DTY168 cells (“DTY168 cells”) and pYES3-transformed DTY168 cells (“DTY168/pYES3 cells”) were indistinguishable and more than 3- to 4-fold lower than for the equivalent membrane fraction from pYES3-AtMRP1-transformed DTY168 cells (“DTY168/pYES3-AtMRP1 cells”) (Fig. 4A).
Figure 4.
DNP-GS uptake by vacuolar membrane-enriched vesicles purified from S. cerevisiae ycf1Δ strain DTY168 before and after transformation with empty pYES3 vector (pYES3) or vector containing the coding sequence of AtMRP1 (pYES3-AtMRP1). (A) Time course of MgATP-dependent, uncoupler-insensitive uptake by DTY168/pYES3-AtMRP1 (•), DTY168/pYES3 (○), and DTY168 membranes (▴). (B) Sensitivity of MgATP-dependent, uncoupler-insensitive uptake by DTY168/pYES3-AtMRP1 (•) and DTY168/pYES3 membranes (○) to inhibition by vanadate. Uptake was measured at a DNP-[3H]GS concentration of 61.3 μM throughout. MgATP-dependent uptake was enumerated as the increase in uptake consequent on the provision of 3 mM ATP. The data were fitted to a single negative exponential function by nonlinear least-squares analysis (25) to yield an I50 for inhibition by vanadate (exclusive of the uninhibitable, AtMRP1-independent component) of 8.3 ± 3.2 μM. Values shown are means ± SE (n = 3–6).
The increases in MgATP-dependent DNP-GS uptake exhibited by DTY168/pYES3-AtMRP1 membranes were exclusively attributable to vanadate-sensitive, uncoupler-insensitive transport. Protonophores (carbonylcyanide p-trifluoro-methoxyphenylhydrazone, gramicidin D) inhibited uptake to a similar extent as the V-ATPase inhibitor, bafilomycin A1, neither class of inhibitor markedly augmented the effects of the other, and the inhibitions measured were the same whether the membranes were from DTY168/pYES3 or DTY168/pYES3-AtMRP1 cells (Table 1). Uncoupler- and V-ATPase inhibitor-insensitive, MgATP-dependent uptake, on the other hand, comprised two components: (i) an AtMRP1-dependent component accounting for more than 60% of total MgATP-dependent uptake by DTY168/pYES3-AtMRP1 membranes that was absent from both DTY168 and DTY168/pYES3 membranes; and (ii) an AtMRP1-independent component accounting for less than 40% of total MgATP-dependent uptake by DTY168/pYES3-AtMRP1 membranes that was present at the same level in DTY168 and DTY168/pYES3 membranes. Of these two components, only the former was potently inhibited by vanadate. The rate of MgATP-dependent, uncoupler-insensitive DNP-[3H]GS uptake by DTY168/pYES3-AtMRP1 membrane vesicles decreased as a single negative exponential function of vanadate concentration to yield an I50 of 8.3 ± 3.2 μM (Fig. 4B) whereas uptake by DTY168/pYES3 (and DTY168) membranes was essentially insensitive to vanadate at concentrations of 25 μM and less (Fig. 4B).
Table 1.
Effect of inhibitors and of adenosine 5′-[β, γ-imino] triphosphate (AMP-PNP) on MgATP-dependent DNP-GS uptake by vacuolar membrane-enriched vesicles purified from pYES3-transformed and pYES3-AtMRP1-transformed DTY168 cells
| Treatment | MgATP-dependent DNP-[3H]GS uptake, nmol/mg per 10 min
|
|
|---|---|---|
| DTY168/pYES3 | DTY168/pYES3-AtMRP1 | |
| Control | 2.46 ± 0.08 (100.0) | 4.68 ± 0.31 (100.0) |
| MgAMP-PNP (−MgATP) | 0.03 ± 0.1 (1.2) | 0.0 (0.0) |
| Bafilomycin A1 | 1.73 ± 0.14 (70.3) | 4.32 ± 0.33 (92.3) |
| FCCP | 1.41 ± 0.12 (57.3) | 4.20 ± 0.30 (89.7) |
| Gramicidin D | 1.77 ± 0.09 (72.0) | 4.38 ± 0.20 (93.6) |
| Gramicidin D + bafilomycin A1 | 1.58 ± 0.23 (64.2) | 4.08 ± 0.34 (87.2) |
MgATP-dependent DNP-[3H]GS uptake was measured as described in the legend to Fig. 5 at a MgATP concentration of 3 mM unless otherwise indicated. MgAMP-PNP, bafilomycin A1, carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP), and gramicidin D were added at concentrations of 3 mM, 0.5 μM, 5 μM, and 5 μM, respectively. Values outside parentheses are means ± SE (n = 3−6); values inside parentheses are mean rates of uptake expressed as percentage of control.
Its absence from DTY168 and DTY168/pYES3 membranes and selective inhibition by micromolar concentrations of vanadate meant that AtMRP1-dependent transport could be enumerated in two ways. Either as the difference between the rates of MgATP-dependent, uncoupler-sensitive GS-conjugate uptake by DTY168/pYES3-AtMRP1 membranes by comparison with DTY168 or DTY168/pYES3 membranes or as the vanadate-sensitive component of MgATP-dependent, uncoupler-insensitive uptake by DTY168/pYES3-AtMRP1 membranes. Because the results were qualitatively and quantitatively similar whichever method was used, “AtMRP1-dependent” transport from hereon refers to uptake measured as the increment consequent on transformation of DTY168 cells with pYES3-AtMRP1 vs. pYES3.
AtMRP1-Mediated Transport Resembles Vacuolar GS-Conjugate Transport.
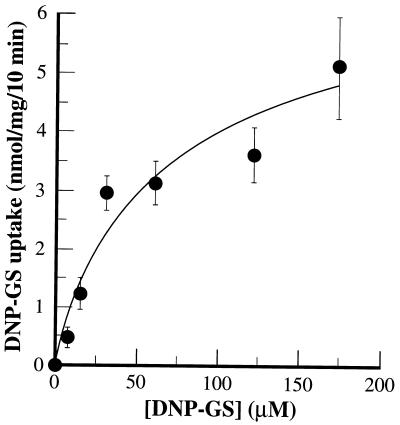
The most rigorously characterized plant GS-conjugate pump is that associated with the vacuolar membrane fraction from mung bean (Vigna radiata) hypocotyls (2). Direct comparisons between the activity of this transporter and heterologously expressed AtMRP1 revealed a basic functional equivalence. Not only did the I50 for inhibition of MgATP-dependent, uncoupler-insensitive DNP-[3H]GS uptake into DTY168/pYES3-AtMRP1 membrane vesicles by vanadate (Fig. 4B) coincide with that for uptake by mung bean vacuolar membrane vesicles (7.5 ± 3.9 μM) (2) but the two processes also exhibited similar concentration dependencies and GS-conjugate preferences. AtMRP1-dependent uptake conformed to Michaelis–Menten kinetics with respect to DNP-GS concentration to yield a Km (48.7 ± 15.4 μM) (Fig. 5) within the range reported for the equivalent transport function in vacuolar membranes from mung bean (81.3 ± 41.8 μM) (2). The rates of AtMRP1-dependent uptake of DNP-GS, GSSG, and glutathionated derivatives of the herbicide metolachlor (metolachlor-GS) and the anthocyanin C3G-GS fell in the same rank order as for uptake by mung bean vacuolar membrane vesicles. These were C3G-GS > metolachlor-GS > DNP-GS ≥ GSSG (uptake ratio = 2.7:1.5:1.0:0.7) for AtMRP1-dependent uptake by yeast vacuolar membrane-enriched vesicles (Table 2) and C3G-GS (26.43 ± 9.56 nmol/mg per 10 min) > metolachlor-GS (18.45 ± 5.00 nmol/mg per 10 min) > DNP-GS (4.83 ± 0.59 nmol/mg per 10 min) ≥ GSSG (3.85 ± 0.43 nmol/mg per 10 min) (uptake ratio = 5.5:3.9:1.0:0.8) for MgATP-dependent, uncoupler-insensitive uptake by mung bean vacuolar membrane vesicles. In neither membrane preparation was MgATP-dependent, uncoupler-insensitive uptake of the unconjugated precursors of these compounds, DNP, GSH, metolachlor, and C3G, detectable (data not shown).
Figure 5.
Concentration dependence of AtMRP1-dependent DNP-GS uptake. AtMRP1-dependent DNP-[3H]GS uptake rates were calculated by subtracting the radioactivity taken up by vacuolar membrane-enriched membrane vesicles prepared from pYES3-transformed DTY168 cells from that taken up by the equivalent membrane fraction from pYES3-AtMRP1-transformed DTY168 cells. The data were fitted to a single Michaelis–Menten function by nonlinear least-squares analysis (25) to yield a Km and Vmax of 49.7 ± 15.4 μM and 6.0 ± 1.7 nmol/mg per 10 min, respectively. Values shown are means ± SE (n = 3–6).
Table 2.
AtMRP1-dependent uptake of different GS-conjugates
| GS-conjugate | AtMRP1-dependent uptake, nmol/mg per 10 min |
|---|---|
| DNP-GS | 2.91 ± 0.42 |
| GSSG | 2.07 ± 0.71 |
| Metolachlor-GS | 4.44 ± 0.82 |
| C3G-GS | 7.85 ± 0.75 |
MgATP-dependent, uncoupler-insensitive uptake by DTY168/pYES3-AtMRP1 and DTY168/pYES3 membrane vesicles was measured and AtMRP1-dependent uptake was calculated as described in the legend to Fig. 5. All of the GS-conjugates were present at a concentration of 61.3 μM. Values shown are means ± SE (n = 3−6).
The overall equivalence of the kinetics of uptake and uptake ratios for different glutathionated derivatives by the membrane preparations from mung bean and pYES3-AtMRP1-transformed yeast, together with the pronounced osmotic dependence of uptake by both preparations (data not shown), indicate that genuine transport into the intravesicular space, rather than ATP-dependent adsorption to membranes containing an abundance of endogenous or heterologous protein, is what was measured in these experiments.
AtMRP1, like the endogenous vacuolar GS-conjugate pump of plants (Z.-S.L. and P.A.R., unpublished work) but unlike the YCF1-encoded function of S. cerevisiae (12), was not competent in the GSH-dependent transport of Cd2+. Transformation with pYES2-AtMRP1 neither suppressed the Cd2+ hypersensitive phenotype of DTY168 cells nor conferred on their membranes the capacity for MgATP-energized, GSH-dependent Cd2+ transport (data not shown).
DISCUSSION
The results demonstrating that heterologous expression of AtMRP1 alone is sufficient for the reconstitution of vanadate-inhibitable, MgATP-dependent, uncoupler-insensitive GS-conjugate transport in yeast membranes suggest that a plant GS-conjugate pump has been cloned in its entirety. As such, they provide the first combined molecular and functional identification of an ABC transporter from vascular plants. Several sequence-related ESTs and one MDR-like gene (ATPGP1) have been isolated from A. thaliana (26) and a number of candidate ABC transporter activities, other than vacuolar GS-conjugate uptake, have been reported for plants (27, 28) but no transport function had been unambiguously assigned to a plant ABC transporter gene. The structural characteristics of AtMRP1, its mode of energization, and its membership of the CFTR-related MRP-subclass of ABC transporters defines an MgATP-energized organic solute transporter, antedating the evolutionary separation of animals and plants, which, contrary to the prevailing model for energy-dependent transport in plants (29), is not driven by a transmembrane H+-electrochemical potential difference. The acute sensitivity of AtMRP1 to vanadate, a specific inhibitor of aspartyl- or glutamyl-phosphate formation, its insensitivity to uncouplers, and the inability of the nonhydrolyzable ATP analog, adenosine 5′-[β,γ-imino]triphosphate to support uptake (Table 1) exclude chemiosmotic coupling and establish that AtMRP1-mediated transport is contingent on hydrolysis of the γ-phosphate of ATP and formation of a phosphoenzyme intermediate.
There is always a possibility, in view of the moderate rates of GS-conjugate transport catalyzed by AtMRP1 after its expression in yeast, that glutathionated compounds are not the sole substrates, or even the principal substrates for this transporter in vivo. However, the capacity of AtMRP1 for transport of the glutathionated chloroacetanilide herbicide, metolachlor, and by implication GS-conjugates of other herbicides, such as atrazine and simetryn, for which glutathionation in vivo (30) and transport by the endogenous vacuolar GS-conjugate pump in vitro (1) are also demonstrable, is consistent with the molecular identification of a transporter capable of removing these and related compounds from the cytosol. Given that some GS-conjugates are themselves toxic and most will end-product inhibit GSTs if their cytosolic concentrations increase sufficiently (29), transport functions such as that of AtMRP1 are critical if the detoxification of herbicides and other xenobiotics is to continue unimpeded. Analogously, its ability to transport C3G-GS at rates severalfold higher than DNP-GS, its functional resemblance to the endogenous vacuolar GS-conjugate pump of plants, and the results of analyses of the maize (Zea mays) gene Bronze-2 (Bz2) implicate AtMRP1 in plant cell pigmentation. Although it has been known for some time that the bronze coloration of bz2 mutants is due to the accumulation of C3G derivatives in the cytosol—in wild-type plants anthocyanins are accumulated in the vacuole as purple or red derivatives but in bz2 mutants they are restricted to the cytosol where they undergo oxidation to brown pigments—the biochemical basis of this lesion was not understood. Incisive, therefore, are experiments showing that Bz2 encodes a GST capable of conjugating C3G with GSH (4). In view of the efficacy of C3G-GS, but not C3G, as a substrate for transport by heterologously expressed AtMRP1 and the endogenous vacuolar GS-conjugate pump, the probable basis of the bz2 phenotype follows automatically. Being defective in the glutathionation of anthocyanins, bz2 mutants are unable to pump these pigments from the cytosol into the vacuole; a conclusion supported by the ability of vanadate treatment to phenocopy the bz2 mutation in wild-type protoplasts (4) and the exquisite sensitivity of AtMRP1-dependent transport to this compound.
Against this background it is instructive to note that the final steps in anthocyanin biosynthesis and the initial steps in xenobiotic detoxification require the same types of enzymes: cytochrome P450s, glucosyltransferases, and GSTs (31). When account is taken of this, the facility of cytochrome P450s for conferring the requisite electrophilicity on otherwise unreactive compounds for subsequent conjugation with GSH, the high capacity of the vacuolar GS-conjugate pump for the high affinity transport of glutathionated medicarpin, an isoflavonoid phytoalexin (3), and the amenability of GSSG, a product of peroxide detoxification and protein thiol reduction to transport by AtMRP1, the spectrum of processes that likely converge and depend on this and related transporters is extended beyond herbicide detoxification and cell pigmentation to include the alleviation of oxidative damage and storage of pathogen-elicited compounds. Thus, the identification of AtMRP1 not only provides fresh insights into the molecular basis of energy-dependent organic solute transport in plants but also offers the prospect of manipulating and investigating many fundamental processes that have hitherto been neglected at the transport level.
In the general context of GS-conjugate pumps, not only in plants but also other organisms, the findings reported here indicate a dichotomy. While AtMRP1 is evidently a structural homolog of yeast YCF1 and mammalian MRP1, its functional capabilities are not exactly equivalent. Unlike ScYCF1, which is active in the transport of both organic GS-conjugates (10, 11) and heavy metals, such as Cd2+, after their complexation with GSH (12), and HmMRP1, which is inferred to have a similarly broad substrate range (11), AtMRP1 appears capable of transporting only organic GS-conjugates: AtMRP1 neither confers resistance to nor mediates the GSH-dependent transport of Cd2+. As such, the properties of AtMRP1 highlight a basic functional bifurcation within the MRP1/YCF1 subclass of ABC transporters. Given our facility for expressing AtMRP1 in a functionally active state in yeast, the structural basis of these differences, and the important question of which domains of YCF1 and/or MRP1 confer the capacity for Cd2+ transport, may now be addressed by the construction and biochemical characterization of AtMRP1-YCF1 and/or AtMRP1-MRP1 protein chimeras.
Acknowledgments
We thank Joe Ecker and his colleagues Ken Dewar, Christopher Kim, Paul Shinn, and Yaping Li, for the provision of BAC and YAC library filters; Dennis Thiele (University of Michigan, Ann Arbor, MI) for yeast strain DTY168; Herman Hofte (Laboratoire de Biologie Cellulaire, Versailles, France) for A. thaliana EST ATTS1246; the Arabidopsis Biological Research Center for the cDNA library used in these studies; Junsheng Zhan for technical assistance; Les Klimczak for sharing his computing expertise; Rui-Guang Zhen, Warren Davies. Jose Alonso, Said Nourizadeh, and Ji-Dong Feng for technical advice; and Virginia Walbot and Mark Alfenito (Stanford University, CA) for stimulating discussions of anthocyanin transport. This work was supported by grants from the United States Department of Agriculture (NRICGP 9303007), Department of Energy (DE-FG02–91ER20055), and University Research Foundation awarded to P.A.R.
ABBREVIATIONS
- ABC transporter
ATP-binding cassette transport protein
- GST
glutathione S-transferase
- C3G
cyanidin 3-glucoside
- DNP-GS
S-(2,4-dinitrophenyl)glutathione
- GS-conjugate
glutathione S-conjugate
- GSH
glutathione
- GSSG
oxidized glutathione
- MRP1
multidrug resistance-associated protein
- HmMRP1
human MRP1
- YCF1
yeast cadmium factor protein
- ScYCF1
Saccharomyces cerevisiae (yeast) cadmium factor
- EST
expressed sequence tag
- BAC
bacterial artificial chromosome
- CFTR
cystic fibrosis transmembrane conductance regulator
- NBF
nucleotide-binding fold
Footnotes
References
- 1.Martinoia E, Grill E, Tommasini R, Kreuz K, Amrheim N. Nature (London) 1993;364:247–249. [Google Scholar]
- 2.Li Z-S, Zhao Y, Rea P A. Plant Physiol. 1995;107:1257–1268. doi: 10.1104/pp.107.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, Z.-S., Alfenito, M. R., Rea, P. A., Walbot, V. & Dixon, R. A. (1997) Phytochemistry, in press. [DOI] [PubMed]
- 4.Marrs K, Alfenito M R, Lloyd A M, Walbot V. Nature (London) 1995;375:397–400. doi: 10.1038/375397a0. [DOI] [PubMed] [Google Scholar]
- 5.Ishikawa T. Trends Biochem Sci. 1992;17:463–468. doi: 10.1016/0968-0004(92)90489-v. [DOI] [PubMed] [Google Scholar]
- 6.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, Deeley R G. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 7.Leier I, Jedlitschky G, Buchholz U, Cole S P C, Deeley R G, Keppler D. J Biol Chem. 1994;269:27807–27810. [PubMed] [Google Scholar]
- 8.Muller M, Meijer C, Zaman G J R, Borst P, Scheper R J, Mulder N H, de Vries E G E, Jansen P L M. Proc Natl Acad Sci USA. 1994;91:13033–13037. doi: 10.1073/pnas.91.26.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 10.Li Z-S, Szczypka M, Lu Y-P, Thiele D J, Rea P A. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 11.Tommasini R, Evers R, Vogt E, Mornett C, Zaman G J R, Schinkel A H, Borst P, Martinoia E. Proc Natl Acad Sci USA. 1996;93:6743–6748. doi: 10.1073/pnas.93.13.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z-S, Lu Y-P, Zhen R-G, Szczypka M, Thiele D J, Rea P A. Proc Natl Acad Sci USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofte H, Desprez T, Amselem J, Chiapello H, Caboche M, et al. Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- 14.Kieber J J, Rothenberg M, Roman G, Feldman K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang G L, Warren R, Innes R, Osborne B, Baker B, Ronald P C. Plant Mol Biol Rep. 1996;14:107–114. [Google Scholar]
- 17.Choi S, Creelmann R A, Wing R A. Weeds World. 1995;2:17–20. [Google Scholar]
- 18.Creusot F, Fouilloux E, Dron M, Lafleuriel J, Pickard G, Billault A, Lepaslier D, Cohen D, Chaboute M E, Durr A, Fleck J, Gigot C, Camilleri C, Bellini C, Caboche M, Bouchez D. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- 19.Minet M, Dufour M E, Lacroute F. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- 20.Giest R D, Schiestl R H. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 21.Kim E J, Zhen R-G, Rea P A. J Biol Chem. 1995;270:2630–2635. doi: 10.1074/jbc.270.6.2630. [DOI] [PubMed] [Google Scholar]
- 22.Peterson G L. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 23.Holt D C, Lay V J, Clarke E D, Dinsmore A, Jepson I, Bright S W J, Greenland A J. Planta. 1995;196:295–302. doi: 10.1007/BF00201388. [DOI] [PubMed] [Google Scholar]
- 24.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 25.Marquardt D W. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- 26.Dudler R, Hertig C. J Biol Chem. 1992;267:5882–5888. [PubMed] [Google Scholar]
- 27.Hinder B, Schellenberg M, Rodon S, Ginsburg S, Vogt E, Martinoia E, Matile P, Hortensteiner S. J Biol Chem. 1996;271:27233–27236. doi: 10.1074/jbc.271.44.27233. [DOI] [PubMed] [Google Scholar]
- 28.Klein M, Weissenbock G, Dufaud A, Gaillard C, Kreuz K, Martinoia E. J Biol Chem. 1996;271:29666–29671. doi: 10.1074/jbc.271.47.29666. [DOI] [PubMed] [Google Scholar]
- 29.Bush D R. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:513–542. [Google Scholar]
- 30.Kreuz K. Proceedings of Brighton Crop Protection Conference: Weeds. UK: Brighton; 1993. pp. 1249–1258. [Google Scholar]
- 31.Marrs K A. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]