Abstract
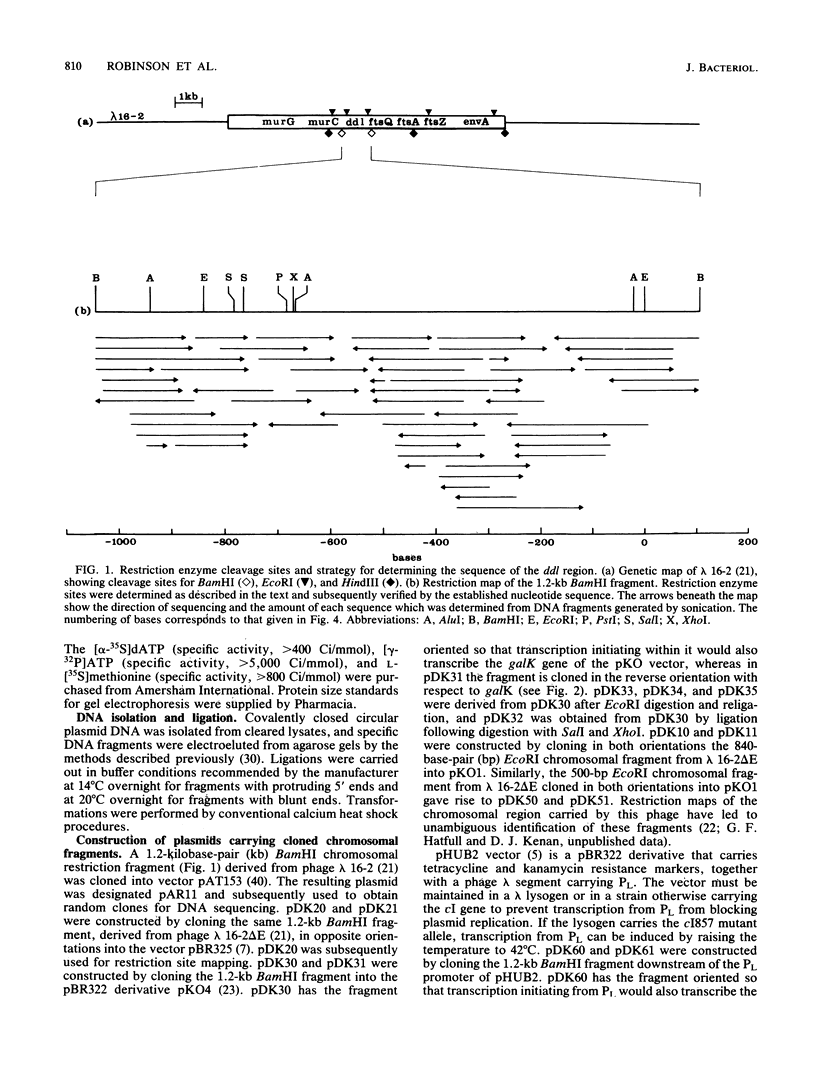
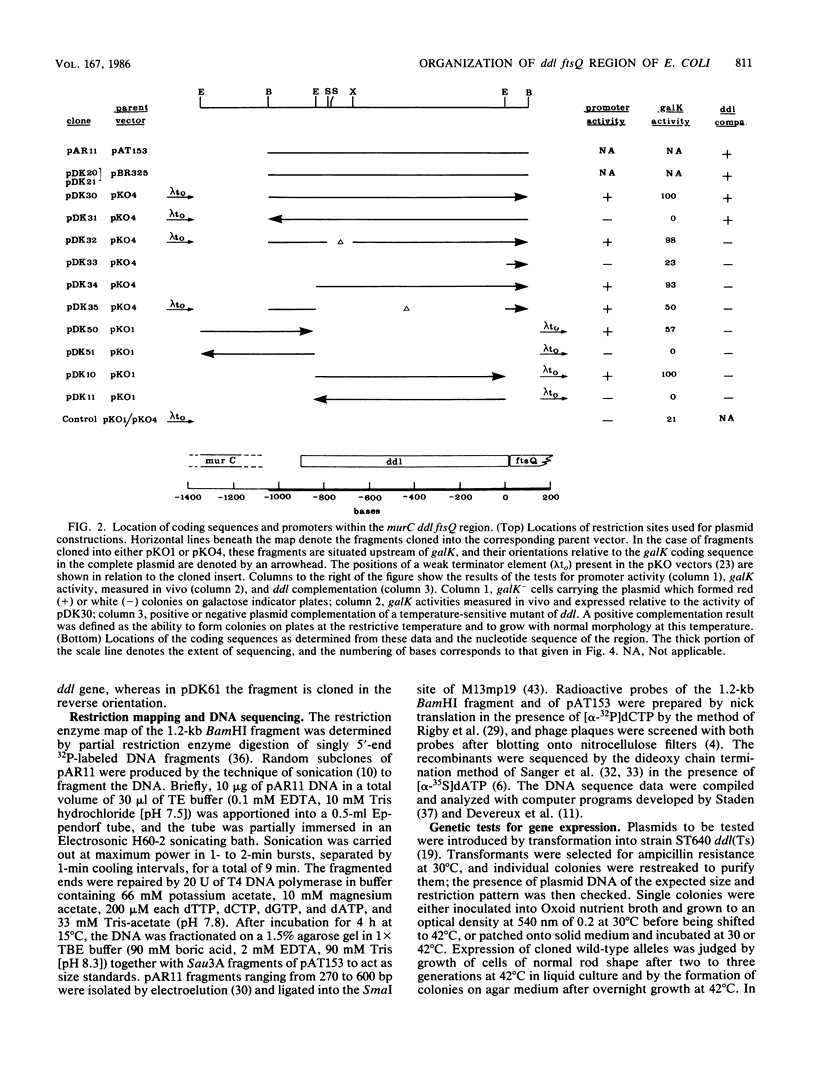
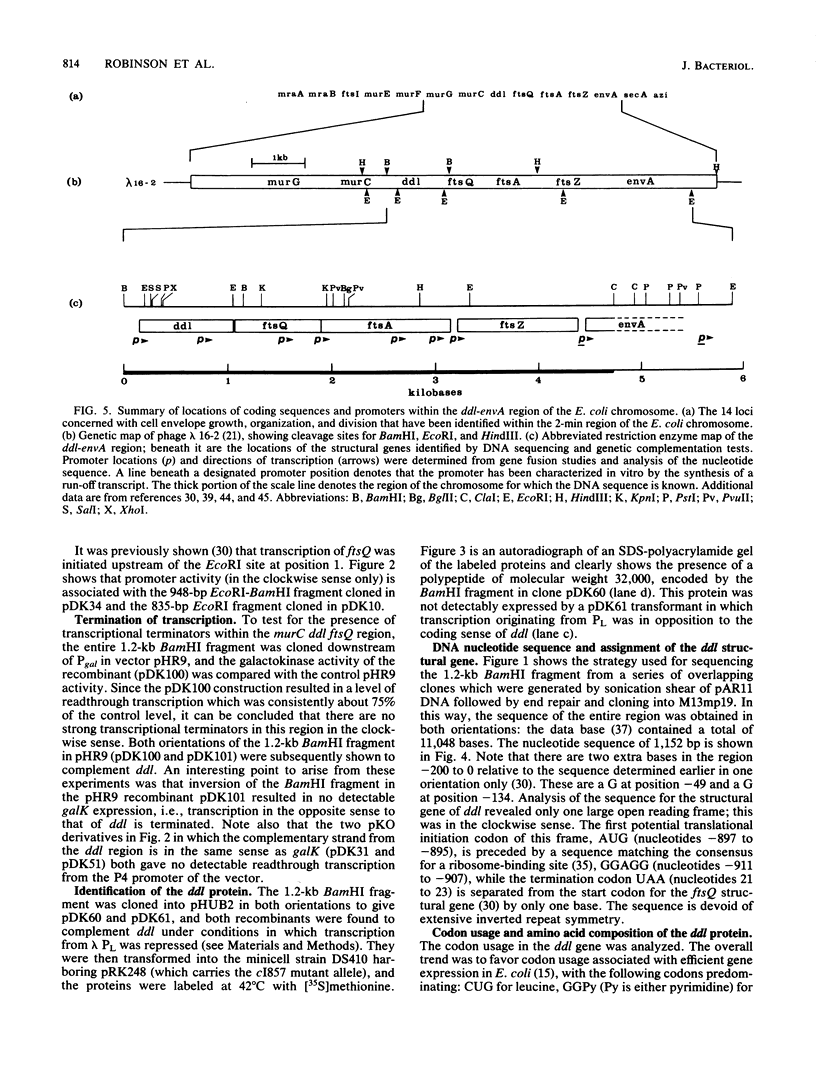
A 1.2-kilobase-pair BamHI fragment from a cell envelope-cell division gene cluster of Escherichia coli containing ddl and part of ftsQ was cloned and sequenced, and the sequence was interpreted with the aid of genetic complementation and promoter fusion data for the region. Both ddl and ftsQ were transcribed in the same direction (clockwise on the genetic map). ddl was shown to be capable of independent expression from a promoter of its own, and a promoter was identified within the ddl structural gene. The structural gene of ddl consisted of 918 nucleotides, encoding a 306-residue polypeptide of molecular weight 32,840; the synthesis of a protein of this molecular weight was shown to be directed from the 1.2-kilobase-pair BamHI fragment in minicells. Analysis of the DNA sequence further showed that the termination codon of ddl is separated from the initiation codon of ftsQ by one base, which suggests that these two genes may be translationally coupled when transcription is initiated upstream of ddl. This represents a second instance of potential translational coupling within this gene cluster and also indicates that the ddl and ftsQ transcriptional units must overlap (as has been reported earlier for ftsQ and ftsA and for ftsA and ftsZ).
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman G., Nomura M. Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell. 1983 Oct;34(3):979–988. doi: 10.1016/0092-8674(83)90555-x. [DOI] [PubMed] [Google Scholar]
- Begg K. J., Hatfull G. F., Donachie W. D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol. 1980 Oct;144(1):435–437. doi: 10.1128/jb.144.1.435-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bernard H. U., Remaut E., Hershfield M. V., Das H. K., Helinski D. R., Yanofsky C., Franklin N. Construction of plasmid cloning vehicles that promote gene expression from the bacteriophage lambda pL promoter. Gene. 1979 Jan;5(1):59–76. doi: 10.1016/0378-1119(79)90092-1. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R., George J. Inducible sfi dependent division inhibition in Escherichia coli. Mol Gen Genet. 1980;177(4):629–636. doi: 10.1007/BF00272673. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Jones C. A., Holland I. B. Inactivation of essential division genes, ftsA, ftsZ, suppresses mutations at sfiB, a locus mediating division inhibition during the SOS response in E. coli. EMBO J. 1984 May;3(5):1181–1186. doi: 10.1002/j.1460-2075.1984.tb01948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg E. J., v Schijndel-van Dam A. Temperature-sensitive mutant of Escherichia coli K-12 with an impaired D-alanine:D-alanine ligase. J Bacteriol. 1973 Jan;113(1):96–104. doi: 10.1128/jb.113.1.96-104.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg E. J., v Schijndel-van Dam A. Temperature-sensitive mutants of Escherichia coli K-12 with low activities of the L-alanine adding enzyme and the D-alanyl-D-alanine adding enzyme. J Bacteriol. 1972 Apr;110(1):35–40. doi: 10.1128/jb.110.1.35-40.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg E. J., v Schijndel-van Dam A. Temperature-sensitive mutants of Escherichia coli K-12 with low activity of the diaminopimelic acid adding enzyme. J Bacteriol. 1972 Apr;110(1):41–46. doi: 10.1128/jb.110.1.41-46.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983 Jun;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wolf-Watz H., Donachie W. D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol. 1980 May;142(2):615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wu H. C. Determination of transcriptional units and gene products from the ftsA region of Escherichia coli. J Bacteriol. 1980 Sep;143(3):1281–1288. doi: 10.1128/jb.143.3.1281-1288.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirelman D., Yashouv-Gan Y., Schwarz U. Regulation of murein biosynthesis and septum formation in filamentous cells of Escherichia coli PAT 84. J Bacteriol. 1977 Mar;129(3):1593–1600. doi: 10.1128/jb.129.3.1593-1600.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Vaughn V., Phillips T. A., Bloch P. L. Gene-protein index of Escherichia coli K-12. Microbiol Rev. 1983 Jun;47(2):231–284. doi: 10.1128/mr.47.2.231-284.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Ma J. C., Howe K. M., Garner I., Hayward R. S. Evidence that rifampicin can stimulate readthrough of transcriptional terminators in Escherichia coli, including the attenuator of the rpoBC operon. Nucleic Acids Res. 1982 Nov 25;10(22):7409–7424. doi: 10.1093/nar/10.22.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S. Genetics of a chain-forming mutant of Escherichia coli. Transduction and dominance of the envA gene mediating increased penetration to some antibacterial agents. Genet Res. 1970 Aug;16(1):63–78. doi: 10.1017/s0016672300002287. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., McKenney K., Sobieski D. A., Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982 Oct;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. F., Donachie W. D. Overlapping functional units in a cell division gene cluster in Escherichia coli. J Bacteriol. 1984 Jun;158(3):1198–1201. doi: 10.1128/jb.158.3.1198-1201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. F., Donachie W. D. Transcriptional organization within an Escherichia coli cell division gene cluster: direction of transcription of the cell separation gene envA. J Bacteriol. 1984 Nov;160(2):724–732. doi: 10.1128/jb.160.2.724-732.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Wijsman H. J. A genetic map of several mutations affecting the mucopeptide layer of Escherichia coli. Genet Res. 1972 Aug;20(1):65–74. doi: 10.1017/s0016672300013598. [DOI] [PubMed] [Google Scholar]
- Wolf-Watz H., Normark S. Evidence for a role of N-acetylmuramyl-L-alanine amidase in septum separation in Escherichia coli. J Bacteriol. 1976 Nov;128(2):580–586. doi: 10.1128/jb.128.2.580-586.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Lutkenhaus J. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene. 1985;36(3):241–247. doi: 10.1016/0378-1119(85)90179-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]




