Abstract
We isolated 142 Hir- (host inhibition of replication) mutants of an Escherichia coli K-12 Mu cts Kil- lysogen that survived heat induction and the killing effect of Mu replicative transposition. All the 86 mutations induced by insertion of Tn5 or a kanamycin-resistant derivative of Tn10 and approximately one-third of the spontaneous mutations were found by P1 transduction to be linked to either zdh-201::Tn10 or Tn10-1230, indicating their location in or near himA or hip, respectively. For a representative group of these mutations, complementation by a plasmid carrying the himA+ gene or by a lambda hip+ transducing phage confirmed their identification as himA or hip mutations, respectively. Some of the remaining spontaneously occurring mutations were located in gyrA or gyrB, the genes encoding DNA gyrase. Mutations in gyrA were identified by P1 linkage to zei::Tn10 and a Nalr gyrA allele; those in gyrB were defined by linkage to tna::Tn10 and to a gyrB(Ts) allele. In strains carrying these gyrA or gyrB mutations, pBR322 plasmid DNA exhibited altered levels of supercoiling. The extent of growth of Mu cts differed in the various gyrase mutants tested. Phage production in one gyrA mutant was severely reduced, but it was only delayed and slightly reduced in other gyrA and gyrB mutants. In contrast, growth of a Kil- Mu was greatly reduced in all gyrase mutant hosts tested.
Full text
PDF






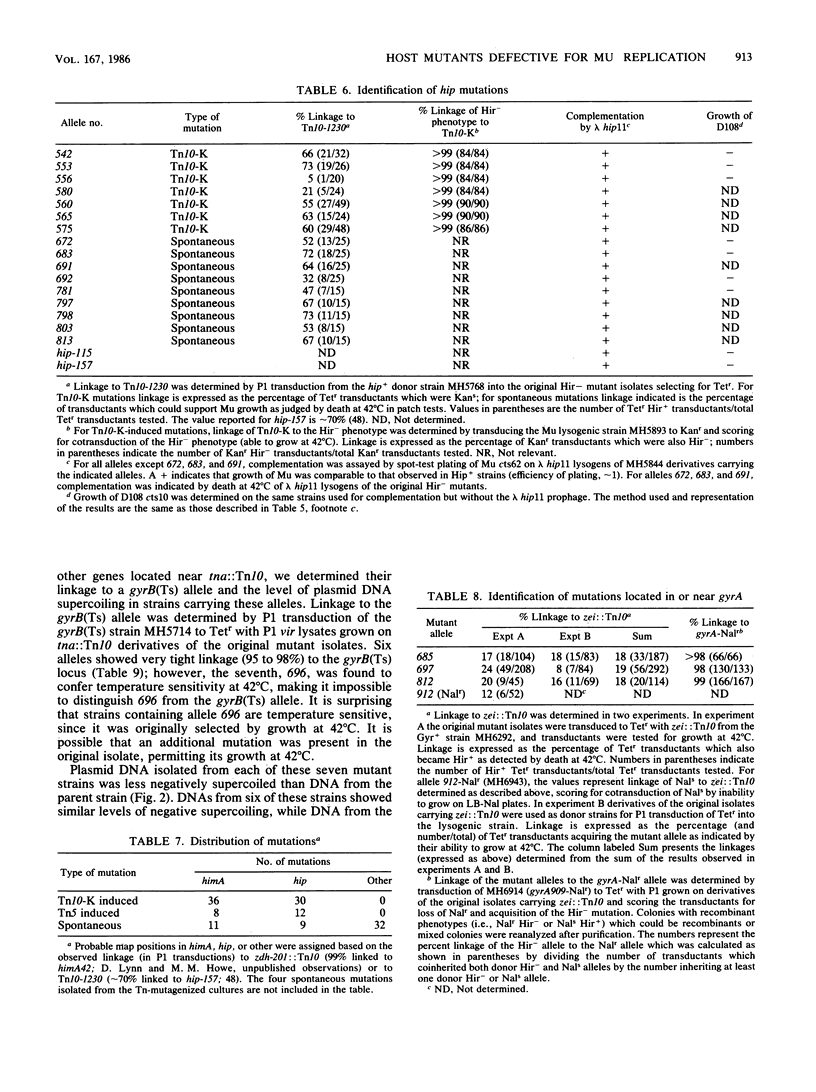
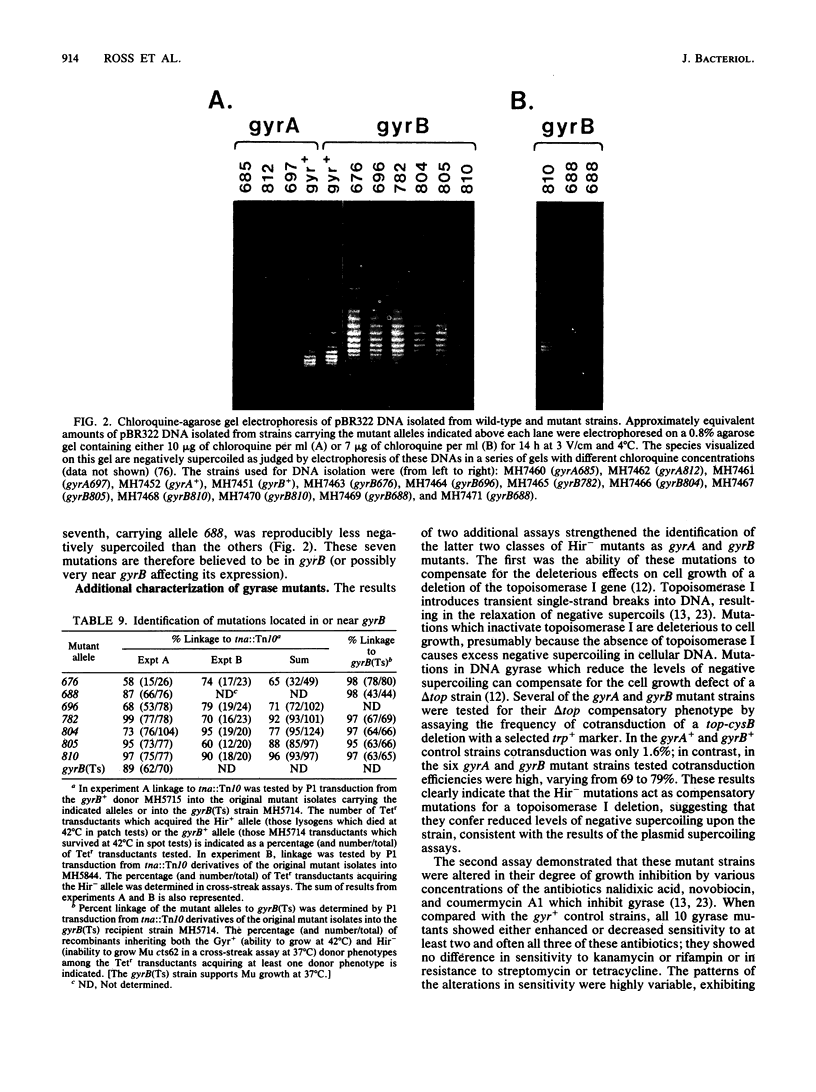





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B. Mu insertion duplicates a 5 base pair sequence at the host inserted site. Cell. 1979 Jan;16(1):123–129. doi: 10.1016/0092-8674(79)90193-4. [DOI] [PubMed] [Google Scholar]
- Alper M. D., Ames B. N. Positive selection of mutants with deletions of the gal-chl region of the Salmonella chromosome as a screening procedure for mutagens that cause deletions. J Bacteriol. 1975 Jan;121(1):259–266. doi: 10.1128/jb.121.1.259-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I. Bacteriophage mu as a transposition element. Annu Rev Genet. 1976;10:389–412. doi: 10.1146/annurev.ge.10.120176.002133. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Lupski J. R., Svec P., Godson G. N. Comparison of left-end DNA sequences of bacteriophages Mu and D108. Gene. 1985;33(2):235–239. doi: 10.1016/0378-1119(85)90098-8. [DOI] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985 Jul;41(3):867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Degnen G. E., Cox E. C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974 Feb;117(2):477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubow M. S., Bukhari A. I. Effects of prophage Mu induction on expression of adjacent host genes. Mol Biol Rep. 1980 Dec 31;6(4):229–234. doi: 10.1007/BF00777530. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A. Isolation of conditional defective mutants of temperate phage Mu-1 and deletion mapping of the Mu-1 prophage. Virology. 1973 Jul;54(1):117–124. doi: 10.1016/0042-6822(73)90121-9. [DOI] [PubMed] [Google Scholar]
- Feiss M., Frackman S., Sippy J. Essential interaction between lambdoid phage 21 terminase and the Escherichia coli integrative host factor. J Mol Biol. 1985 May 25;183(2):239–246. doi: 10.1016/0022-2836(85)90216-5. [DOI] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Friden P., Voelkel K., Sternglanz R., Freundlich M. Reduced expression of the isoleucine and valine enzymes in integration host factor mutants of Escherichia coli. J Mol Biol. 1984 Feb 5;172(4):573–579. doi: 10.1016/s0022-2836(84)80024-8. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. J., Carver D., Gellert M. Synergistic effect of himA and gyrB mutations: evidence that him functions control expression of ilv and xyl genes. J Bacteriol. 1984 Feb;157(2):484–489. doi: 10.1128/jb.157.2.484-489.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I., Plantefaber L. C., Olson E. J., Carver D., O'Dea M. H., Gellert M. Mutations in the DNA gyrB gene that are temperature sensitive for lambda site-specific recombination, Mu growth, and plasmid maintenance. J Bacteriol. 1984 Feb;157(2):490–497. doi: 10.1128/jb.157.2.490-497.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. On the molecular mechanisms of transposition. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4858–4862. doi: 10.1073/pnas.78.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Ghelardini P., Liebart J. C., Marchelli C., Pedrini A. M., Paolozzi L. Escherichia coli K-12 gyrB gene product is involved in the lethal effect of the ligts2 mutant of bacteriophage Mu. J Bacteriol. 1984 Feb;157(2):665–668. doi: 10.1128/jb.157.2.665-668.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. S., Hull R. C., Curtiss R., 3rd Mutator bacteriophage D108 and its DNA: an electron microscopic characterization. J Virol. 1981 Jan;37(1):420–430. doi: 10.1128/jvi.37.1.420-430.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van Heuvel M., Moolenaar G. F., van de Putte P. Regulation of Mu transposition. II. The escherichia coli HimD protein positively controls two repressor promoters and the early promoter of bacteriophage Mu. Gene. 1984 Dec;32(3):419–426. doi: 10.1016/0378-1119(84)90017-9. [DOI] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Regulation of Mu transposition. I. Localization of the presumed recognition sites for HimD and Ner functions controlling bacteriophage Mu transcription. Gene. 1984 Oct;30(1-3):41–46. doi: 10.1016/0378-1119(84)90103-3. [DOI] [PubMed] [Google Scholar]
- Goosen T., Giphart-Gassler M., Van de Putte P. Bacteriophage Mu DNA replication is stimulated by non-essential early functions. Mol Gen Genet. 1982;186(1):135–139. doi: 10.1007/BF00422925. [DOI] [PubMed] [Google Scholar]
- Groenen M. A., Timmers E., van de Putte P. DNA sequences at the ends of the genome of bacteriophage Mu essential for transposition. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2087–2091. doi: 10.1073/pnas.82.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Involvement of the invertible G segment in bacteriophage mu tail fiber biosynthesis. Virology. 1984 Apr 30;134(2):296–317. doi: 10.1016/0042-6822(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Morphogenetic structures present in lysates of amber mutants of bacteriophage Mu. Virology. 1985 Jun;143(2):485–504. doi: 10.1016/0042-6822(85)90388-5. [DOI] [PubMed] [Google Scholar]
- Harshey R. M., Bukhari A. I. A mechanism of DNA transposition. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1090–1094. doi: 10.1073/pnas.78.2.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R. M., Getzoff E. D., Baldwin D. L., Miller J. L., Chaconas G. Primary structure of phage mu transposase: homology to mu repressor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7676–7680. doi: 10.1073/pnas.82.22.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Manlapaz-Ramos P., Gandhi R. T., Olivera B. M. Bacteriophage Mu: a transposing replicon. Cell. 1983 Jun;33(2):623–628. doi: 10.1016/0092-8674(83)90443-9. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Moncecchi D., Manlapaz-Ramos P., Olivera B. M. Bacteriophage Mu DNA replication in vitro. J Biol Chem. 1983 Apr 10;258(7):4293–4297. [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hopkins J. D., Clements M., Syvanen M. New class of mutations in Escherichia coli (uup) that affect precise excision of insertion elements and bacteriophage Mu growth. J Bacteriol. 1983 Jan;153(1):384–389. doi: 10.1128/jb.153.1.384-389.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Schumm J. W., Taylor A. L. The S and U genes of bacteriophage mu are located in the invertible G segment of mu DNA. Virology. 1979 Jan 15;92(1):108–124. doi: 10.1016/0042-6822(79)90218-6. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Knight D. M., Das A., Miller H. I., Echols H. Control of phage lambda development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA and himD genes. Cell. 1982 Dec;31(3 Pt 2):565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- Hull R. A., Gill G. S., Curtiss R., 3rd Genetic characterization of Mu-like bacteriophage D108. J Virol. 1978 Sep;27(3):513–518. doi: 10.1128/jvi.27.3.513-518.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina T. S., Nechaeva E. V., Romanova YuM, Smirnov G. B. Isolation and mapping of Escherichia coli K12 mutants defective in Tn9 transposition. Mol Gen Genet. 1981;181(3):384–389. doi: 10.1007/BF00425616. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. DNA gyrase is a host factor required for transposition of Tn5. Cell. 1982 Aug;30(1):9–18. doi: 10.1016/0092-8674(82)90006-x. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- KONDO E., MITSUHASHI S. DRUG RESISTANCE OF ENTERIC BACTERIA. IV. ACTIVE TRANSDUCING BACTERIOPHAGE P1 CM PRODUCED BY THE COMBINATION OF R FACTOR WITH BACTERIOPHAGE P1. J Bacteriol. 1964 Nov;88:1266–1276. doi: 10.1128/jb.88.5.1266-1276.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R., Kamp D. Nucleotide sequences of the attachment sites of bacteriophage Mu DNA. Nature. 1979 Jul 19;280(5719):247–250. doi: 10.1038/280247a0. [DOI] [PubMed] [Google Scholar]
- Kikuchi A., Flamm E., Weisberg R. A. An Escherichia coli mutant unable to support site-specific recombination of bacteriophage lambda. J Mol Biol. 1985 May 25;183(2):129–140. doi: 10.1016/0022-2836(85)90207-4. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979 Nov;140(2):424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. Behavior of bacteriophage Mu DNA upon infecton of Escherichia coli cells. J Mol Biol. 1979 Sep 25;133(3):339–357. doi: 10.1016/0022-2836(79)90397-8. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llyina T. S., Romanova YuM, Smirnov G. B. The effect of tnm mutations of Escherichia coli K12 on transposition of various movable genetic elements. Mol Gen Genet. 1981;183(2):376–379. doi: 10.1007/BF00270643. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marrs C. F., Howe M. M. AvaII and BglI restriction maps of bacteriophage Mu. Virology. 1983 Apr 30;126(2):563–575. doi: 10.1016/s0042-6822(83)80013-0. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Friedman D. I. An E. coli gene product required for lambda site-specific recombination. Cell. 1980 Jul;20(3):711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kikuchi A., Nash H. A., Weisberg R. A., Friedman D. I. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kirk M., Echols H. SOS induction and autoregulation of the himA gene for site-specific recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6754–6758. doi: 10.1073/pnas.78.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. I., Nash H. A. Direct role of the himA gene product in phage lambda integration. Nature. 1981 Apr 9;290(5806):523–526. doi: 10.1038/290523a0. [DOI] [PubMed] [Google Scholar]
- Miller H. I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- Miller J. L., Anderson S. K., Fujita D. J., Chaconas G., Baldwin D. L., Harshey R. M. The nucleotide sequence of the B gene of bacteriophage Mu. Nucleic Acids Res. 1984 Nov 26;12(22):8627–8638. doi: 10.1093/nar/12.22.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983 Dec;35(3 Pt 2):785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- Mozola M. A., Friedman D. I. A phi 80 function inhibitory for growth of lambdoid phage in him mutants of Escherichia coli deficient in integration host factor. I. Genetic analysis of the Rha phenotype. Virology. 1985 Jan 30;140(2):313–327. doi: 10.1016/0042-6822(85)90368-x. [DOI] [PubMed] [Google Scholar]
- O'Day K., Schultz D., Ericsen W., Rawluk L., Howe M. Correction and refinement of the genetic map of bacteriophage Mu. Virology. 1979 Mar;93(2):320–328. doi: 10.1016/0042-6822(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Orr E., Fairweather N. F., Holland I. B., Pritchard R. H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177(1):103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- Peacock S., Weissbach H., Nash H. A. In vitro regulation of phage lambda cII gene expression by Escherichia coli integration host factor. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6009–6013. doi: 10.1073/pnas.81.19.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess H., Kamp D., Kahmann R., Bräuer B., Delius H. Nucleotide sequence of the immunity region of bacteriophage Mu. Mol Gen Genet. 1982;186(3):315–321. doi: 10.1007/BF00729448. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Resibois A., Pato M., Higgins P., Toussaint A. Replication of bacteriophage mu and its mini-mu derivatives. Adv Exp Med Biol. 1984;179:69–76. doi: 10.1007/978-1-4684-8730-5_7. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Yin J. C., Yong-di Z., Johnson R. C., Reznikoff W. S. Genetic organization of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):99–105. doi: 10.1101/sqb.1981.045.01.018. [DOI] [PubMed] [Google Scholar]
- Schaus N. A., Wright A. Inhibition of Escherichia coli exonuclease V by bacteriophage Mu. Virology. 1980 Apr 15;102(1):214–217. doi: 10.1016/0042-6822(80)90083-5. [DOI] [PubMed] [Google Scholar]
- Schumm J. W., Moore D. D., Blattner F. R., Howe M. M. Correlation of the genetic and physical maps in the central region of the bacteriophage Mu genome. Virology. 1980 Aug;105(1):185–195. doi: 10.1016/0042-6822(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. DNA supercoiling: another level for regulating gene expression. Cell. 1981 Jun;24(3):599–600. doi: 10.1016/0092-8674(81)90085-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M., Desmet L., Allet B. The products of gene A of the related phages Mu and D108 differ in their specificities. Mol Gen Genet. 1983;190(1):70–79. doi: 10.1007/BF00330326. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Faelen M. The dependence of temperate phage Mu-1 upon replication functions of E. coli K12. Mol Gen Genet. 1974;131(3):209–214. doi: 10.1007/BF00267960. [DOI] [PubMed] [Google Scholar]
- Waggoner B. T., Marrs C. F., Howe M. M., Pato M. L. Multiple factors and processes involved in host cell killing by bacteriophage Mu: characterization and mapping. Virology. 1984 Jul 15;136(1):168–185. doi: 10.1016/0042-6822(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Waggoner B., Pato M., Toussaint A., Faelen M. Replication of mini-Mu prophage DNA. Virology. 1981 Aug;113(1):379–387. doi: 10.1016/0042-6822(81)90163-x. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- Yoshida R. K., Miller J. L., Miller H. I., Friedman D. I., Howe M. M. Isolation and mapping of Mu nu mutants which grow in him mutants of E. coli. Virology. 1982 Jul 15;120(1):269–272. doi: 10.1016/0042-6822(82)90027-7. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Westmaas G. C., Wijffelman C. Transfection with Mu-DNA. Virology. 1977 Aug;81(1):152–159. doi: 10.1016/0042-6822(77)90067-8. [DOI] [PubMed] [Google Scholar]