Abstract
Gene activation and cellular differentiation induced by interleukin-6 (IL-6) and transcription factor Stat3 are suppressed by several factors, including ionomycin, granulocyte/macrophage-colony-stimulating factor, and phorbol 12-myristate 13-acetate (PMA), that block IL-6-induced Stat3 activation. These inhibitory agents activate mitogen activated protein kinases (MAPKs), and thus the role of MAPKs in the mechanism of inhibition of Stat3 activation was investigated. Inhibition of IL-6-induced Stat3 activation by PMA and ionomycin was rapid (within 5 min) and did not require new RNA or protein synthesis. Inhibition of Stat3 DNA-binding activity and tyrosine phosphorylation by PMA, ionomycin, and granulocyte/macrophage-colony-stimulating factor was reversed when activation of the extracellular signal-regulated kinase (ERK) group of MAPKs was blocked by using specific kinase inhibitors. Expression of constitutively active MEK1, the kinase that activates ERKs, or overexpression of ERK2, but not JNK1, inhibited Stat3 activation. Inhibition of Stat3 correlated with suppression of IL-6-induction of a signal transducer and activator of transcription (STAT)-dependent reporter gene. In contrast to IL-6, activation of Stat3 by interferon-α was not inhibited. MEKs and ERKs inhibited IL-6 activation of Stat3 harboring a mutation at serine-727, the major site for serine phosphorylation, similar to inhibition of wild-type Stat3, and inhibited Janus kinases Jak1 and Jak2 upstream of Stat3 in the Jak-STAT-signaling pathway. These results demonstrate an ERK-mediated mechanism for inhibiting IL-6-induced Jak-STAT signaling that is rapid and inducible, and thus differs from previously described mechanisms for downmodulation of the Jak-STAT pathway. This inhibitory pathway provides a molecular mechanism for the antagonism of Stat3-mediated IL-6 activity by factors that activate ERKs.
The Janus kinase-signal transducer and activator of transcription (Jak-STAT) signal transduction pathway is used by many cytokines and growth factors that regulate gene expression and cellular activation, proliferation, and differentiation (1, 2). The binding of these cytokines to their receptors activates Jak protein tyrosine kinases, followed by tyrosine phosphorylation and activation of latent cytoplasmic transcription factors termed STATs, which dimerize and translocate to the nucleus. Ligation of cytokine receptors typically results in a transient activation of Jaks and STATs, and STAT activation can be inhibited by antagonistic factors (2). This suggests that pathways that down-regulate Jak-STAT signaling exist, and, indeed, several inhibitory mechanisms have been described recently. Constitutive pathways for downmodulating STAT activity include dephosphorylation, proteolytic degradation, or association with inhibitory molecules (3–8). One inducible mechanism for inhibiting the Jak-STAT pathway is cytokine-mediated induction of expression of SOCS/JAB/SSI proteins, which interact with and inhibit Jaks (9–11). Many additional stimuli, including crosslinking of Fc or complement CR3 receptors (12, 13), antagonistic cytokines such as TGFβ, granulocyte/macrophage-colony-stimulating factor (GM-CSF), and angiotensin II (14–17), activation of calcium fluxes (17), or activation of protein kinase A or protein kinase C (17, 18, 19), inhibit Jak-STAT signaling by blocking signaling upstream of the activation of STATs. The mechanisms of inhibition of Jak-STAT signaling by these agents have not been resolved.
ERKs constitute one family of MAPKs that are downstream effector kinases in a signaling pathway activated by a large number of extracellular ligands (20–22). Interaction between ERK and Jak-STAT pathways can lead to synergistic activation of genes by interferon-α (IFNα) (23), but ERKs also can antagonize Jak-STAT signaling in several systems (24–30). The molecular basis for the interaction of ERK and Jak-STAT pathways has not yet been resolved. One hypothesis is that ERKs modify STATs directly by phosphorylating STATs on a conserved carboxy-terminal sequence, PXSP (corresponding to serine-727 in Stat1 and Stat3), that is the predominant site for serine phosphorylation and resembles a substrate for proline-directed kinases such as ERKs (23, 31). However, evidence from several laboratories suggests that kinases other than ERKs phosphorylate STATs on serine residues (31–40). Serine phosphorylation of STATs has varying effects on function and has been reported to potentiate transcriptional activity (39), potentiate tyrosine phosphorylation and DNA binding (34, 38), inhibit tyrosine phosphorylation (31), or have no effect on tyrosine phosphorylation or DNA binding (36, 37, 39). Thus, the kinases that phosphorylate STATs on serine-727 and the functional significance of serine phosphorylation will likely vary according to extracellular stimulus, cell type, and activation status.
We have previously shown that inhibition of IL-6-induced immediate early gene activation, Stat3 activation, and Jak1 activity in monocytes by the calcium ionophore ionomycin was rapid (occurring within 5 min of addition of ionomycin) and independent of new RNA and protein synthesis (17). This result suggested the existence of a novel inhibitory pathway that acted by direct modification of Jak-STAT-signaling components. Because ligands that activate ERKs can antagonize STAT-mediated effects on gene activation and cell differentiation (12–14, 17, 19, 24–29, 41), we investigated whether the mechanism of inhibition of IL-6 activation of Stat3 involved ERKs. We present evidence demonstrating that rapid inhibition of IL-6 signaling and Stat3 activation by several factors is mediated by the ERK group of MAPKs and that inhibition occurs upstream of Stat3 activation.
MATERIALS AND METHODS
Tissue Culture.
MM6 human myeloid cells (42), 293T, CHO, and Hep-G2 cells were cultured in RPMI 1640, DME, or F12 (Ham) medium supplemented with 10% fetal bovine serum. When agents that were dissolved in dimethyl sulfoxide were used (phorbol 12-myristate 13-acetate (PMA), ionomycin, PD98059, U0126), dimethyl sulfoxide was added to control cells to keep concentrations of dimethyl sulfoxide (0.1 or 0.2%) equal in all wells. Cell extracts were prepared as described (43).
Electrophoretic Mobility Shift Assays (EMSAs).
Extracts corresponding to 3.3 × 105 cells (≈20 μg of protein) were incubated for 15 min at room temperature with 0.5 ng of 32P-labeled double-stranded hSIE oligonucleotide (17) in 15 μl of binding reaction containing 40 mM NaCl and 2 μg of poly-deoxyinosine-deoxycytidine (Pharmacia), as described (43), and complexes were resolved on nondenaturing 4.5% polyacrylamide gels.
Immunoblotting, Immunoprecipitation (IP), and Kinase Assays.
Extracts corresponding to 3.3 × 105 cells or 25% of an immunoprecipitate (see below) were fractionated on 7.5% SDS-polyacrylamide gels, transferred to poly(vinylidene difluoride) membranes, and incubated with specific Stat3 antiserum (44), phospho-specific (tyr-705) anti-Stat3 (New England Biolabs), ERK2 mAb (Transduction Laboratories, Lexington, KY) or FLAG antibody M2. Detection was by enhanced chemiluminescence (Amersham). For IPs, extracts corresponding to 10–50 × 106 cells were adjusted to 0.5 ml of volume in IP buffer (17), precleared by using protein G-agarose beads (Pierce), and incubated with 4 μg of affinity-purified ERK2, FLAG, Stat3, Jak1, or Jak2 antibodies. Immunoprecipitates were collected by using protein G-agarose beads and washed three times with IP buffer and one time with PBS. For kinase assays, 25% of the IPs were saved for immunoblot analysis and the remaining 75% were washed and resuspended in 50 μl of kinase buffer and Jak assays carried out as described (17), and ERK2 activity was assayed by incubation at room temperature for 30 min with 10 μCi of [γ-32P]ATP and 5 μg of myelin basic protein (MBP).
Transient Transfections.
CHO cells were transfected by using lipofectamine and 293T and Hep-G2 cells by using the calcium coprecipitation technique. The Stat3 expression plasmid (35, 39), the plasmids encoding ERK2, JNK1, β-galactosidase, and the constitutively active MEK1 (containing the S218E, S222D mutations, and an amino terminal deletion of residues 32–51) have been described (45, 46).
RESULTS
Inhibition of IL-6-Induced Stat3 DNA Binding by PMA, Ionomycin, and GM-CSF Is Reversed by MEK Inhibitors.
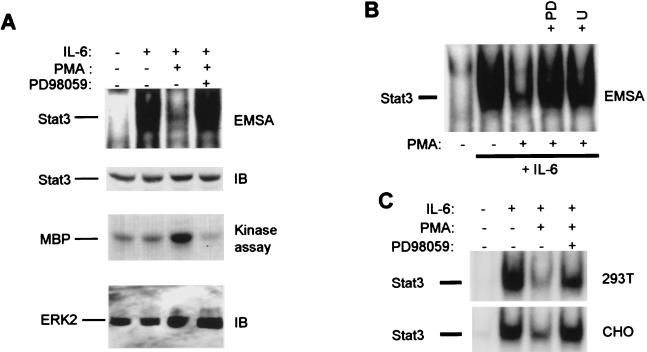
The potential role of ERKs in inhibition of IL-6-induced activation of Stat3 DNA binding was investigated by using PMA, which activates protein kinase C and the Raf-MEK-ERK-signaling pathway (22). Stimulation of MM6 myeloid cells with IL-6 resulted in the rapid induction of a DNA-protein complex (Fig. 1A, first panel, lane 2) that binds the hSIE oligonucleotide (17, 44), and was shown by competition and supershift experiments to consist predominantly of Stat3, consistent with our previous results (data not shown; see ref. 17). Addition of PMA 5 min before adding IL-6 resulted in strong suppression of Stat3 activation (lane 3). Preincubation with the MEK inhibitor PD98059 resulted in a near-complete reversal of PMA-mediated inhibition of Stat3 DNA-binding activity (lane 4). PD98059 appears to be a specific inhibitor of MEK1 and MEK2, the kinases directly upstream of ERK-1 and ERK-2, and, at the doses used, does not inhibit at least 22 other kinases tested (47, 48). Immunoblotting of the same extracts with Stat3 antibody demonstrated comparable levels of Stat3 protein in all lanes (Fig. 1A, second panel). PMA induced ERK2 activity, as expected (Fig. 1A, third panel, lane 3) and complete inhibition of ERK2 activity (lane 4) correlated with reversal of inhibition of Stat3 binding. Inhibition of Stat3 DNA binding by PMA also was reversed by compound U0126, a specific MEK inhibitor (data not shown) that is structurally unrelated to PD98059 (Fig. 1B); inhibition was not reversed by several different kinase inhibitors, including p38 kinase inhibitors (data not shown). MEK-dependent inhibition of Stat3 activation was not limited to myeloid cells but was observed in several cell lines including 293T and CHO cells (Fig. 1C). These results suggest that MEKs and likely ERKs play a role in the inhibition of Stat3 activation.
Figure 1.
Inhibition of IL-6 activation of Stat3 by PMA is reversed by MEK inhibitors. MM6 cells (A and B) or 293T and CHO cells (C) were incubated in complete medium with PMA for 5 min (A) or 15 min (B and C), followed by a 12-min stimulation with IL-6 (20 ng/ml). Cells were preincubated with PD98059 (20 μM) or U0126 (10 μM) for 45 min. Cell extracts were assayed for binding to the hSIE oligonucleotide by using EMSA as described (17). In A, the same extracts were analyzed by immunoblotting with specific Stat3 antiserum or were immunoprecipitated with ERK2 antibodies. Seventy-five percent of each immunoprecipitate was used in a kinase assay with MBP substrate, and 25% was subjected to immunoblotting with ERK2 antibodies. IB, immunoblot; PD, PD98059; U, U0126.
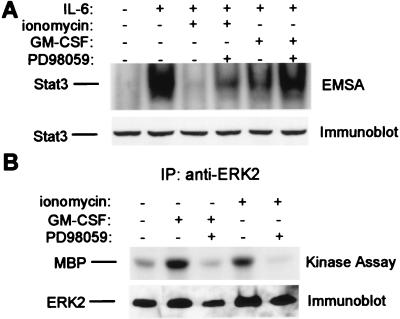
We previously demonstrated that ionomycin and GM-CSF inhibited IL-6-triggered Stat3 activation and expression of a Stat3 target gene (17). Treatment of MM6 cells with ionomycin and GM-CSF resulted in, respectively, near-complete and partial inhibition of IL-6-triggered Stat3 activation (Fig. 2A, lanes 3 and 5). Incubation with PD98059 resulted in a partial reversal of the ionomycin effect (lane 4) and a near-complete reversal of the GM-CSF effect (lane 6). Both GM-CSF and ionomycin induced ERK2 activity (Fig. 2B, lanes 2 and 4), and inhibition of kinase activity by PD98059 (lanes 3 and 5) correlated with reversal of inhibition of DNA binding. These results suggest that ERKs play a significant role in inhibition of Stat3 DNA binding by at least three different inhibitory factors. Inhibition of Stat3 DNA binding by a MEK-dependent pathway occurred as soon as 5 min after treatment with ionomycin or PMA, and did not require new RNA or protein synthesis (Fig. 1, ref. 17, and data not shown), suggesting that inhibition occurred via modification of preexisting IL-6-signaling components.
Figure 2.
Inhibition of Stat3 by ionomycin and GM-CSF is reversed by the MEK inhibitor PD98059. MM6 cells were treated with ionomycin (1 μg/ml) or GM-CSF (100 ng/ml), followed by a 12-min stimulation with IL-6 (20 ng/ml). (A) Cell extracts were assayed for binding to the hSIE oligonucleotide, and the same extracts were analyzed by immunoblotting with specific Stat3 antiserum. (B) Cell extracts were immunoprecipitated with ERK2 antibodies. Seventy-five percent of each immunoprecipitate was used in a kinase assay, and 25% was analyzed by immunoblotting.
Inhibition of Stat3 Tyrosine Phosphorylation by the MEK-Dependent Pathway.
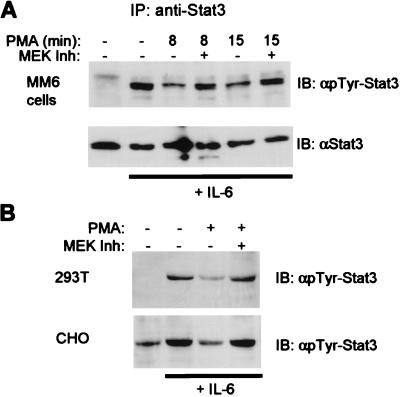
Tyrosine phosphorylation of Stat3 at a conserved tyrosine residue 705 is necessary for dimerization and DNA binding. The role of ERKs in the regulation of Stat3 tyrosine phosphorylation was examined by using IP and immunoblotting with phosphotyrosine-specific antibodies (Fig. 3). Preincubation with PMA for short periods (8 or 15 min) resulted in suppression of IL-6-induced Stat3 tyrosine phosphorylation levels (Fig. 3A, lanes 3 and 5), and this suppression was reversed by the MEK inhibitor PD98059 (lanes 4 and 6). Similar results were obtained when ionomycin and GM-CSF were used to inhibit IL-6, and compound U0126 gave similar results to PD98059 (data not shown). Inhibition of tyrosine phosphorylation of Stat3 and reversal of inhibition by PD98059 also were observed when 293T and CHO cells were used (Fig. 3B). These data indicate that the MEK-ERK pathway regulates Stat3 at the level of tyrosine phosphorylation.
Figure 3.
Inhibition of tyrosine phosphorylation of Stat3 by PMA. MM6 cells (A) or CHO and 293T cells (B) were treated with PMA followed by a 12-min stimulation with IL-6. (A) Cell extracts were immunoprecipitated with Stat3 antibody and analyzed by using immunoblotting. (B) Cell extracts were fractionated by SDS/PAGE and analyzed by immunoblotting with antibodies specific for Stat3 phosphorylated on tyrosine-705.
Inhibition of Stat3 Activation by Expression of Constitutively Active MEK1 or Overexpression of ERK2.
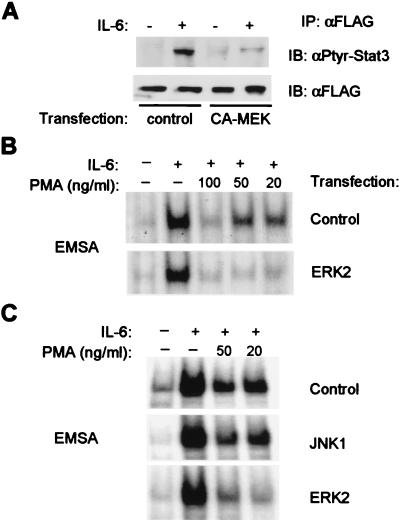
Transient transfection experiments were performed to confirm a role for MEKs and ERKs in regulation of IL-6 activation of Stat3. CHO cells were cotransfected with either a control expression vector or a vector encoding constitutively active MEK1 and a plasmid encoding wild-type Stat3 containing a carboxy-terminal FLAG tag (35, 39). Expression of constitutively active MEK1 dramatically suppressed tyrosine phosphorylation of Stat3 in response to IL-6 (Fig. 4A, lanes 2 and 4); suppression was not observed when catalytically inactive MEK1 was used (data not shown). Reblotting of the same filter with antibodies against the FLAG epitope confirmed equivalence of IP (Fig. 4A Lower). The role in inhibition of the ERKs downstream of MEKs was investigated by using overexpression of ERK2 in 293T cells. In these experiments, transfection efficiencies of >85% of cells were achieved, and thus the effect of ERK2 overexpression on activation of DNA binding of endogenous Stat3 could be examined. Activation of Stat3 in cells transfected with control expression vector was inhibited almost completely by 100 ng/ml PMA, and inhibition was partial when 50 ng/ml or 20 ng/ml PMA was used (Fig. 4B Upper). In contrast, Stat3 was strongly inhibited by all three doses of PMA in cells that overexpressed ERK2 (Fig. 4B Lower). Thus, ERK2 overexpression potentiated the inhibition of Stat3 activation, and overexpression of JNK1 did not (Fig. 4C). Similar results were obtained in four independent transfection experiments. These results, taken together with the results by using pharmacological inhibitors of MEK, strongly support a role for the MEK-ERK pathway in inhibition of Stat3 activation by IL-6.
Figure 4.
Inhibition of Stat3 by constitutively active MEK1 or overexpression of ERK2. (A) CHO cells were cotransfected with 3 μg of FLAG-Stat3 plasmid and 12 μg of control vector or vector encoding constitutively active MEK1 (CA-MEK). FLAG-Stat3 protein was immunoprecipitated by using FLAG antibodies, and immunoprecipitates were analyzed by using immunoblotting. (B and C) 293T cells were transfected with 15 μg of expression plasmids and 2.5 μg of a β-galactosidase-encoding plasmid that was used to monitor transfection efficiency (>85% of cells). Each transfected dish was split into replicate tissue culture dishes, and after 24 hr, cells were treated with PMA and IL-6 (20 ng/ml) and analyzed by using EMSA. Control, expression vector not containing an insert; ERK2 and JNK1, expression vectors containing an insert encoding, respectively, ERK2 or JNK1.
Inhibition of STAT-Dependent Reporter Gene Activity.
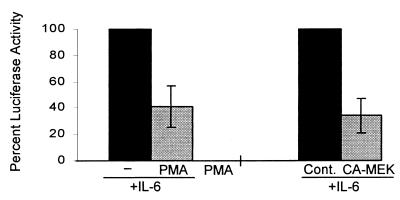
The functional consequences of activation of the MEK-ERK pathway on Stat3 regulation of transcription were investigated by using transient transfections with a reporter construct, 3 X Ly6E γ-activated sequence-luciferase, that contains three Stat3-binding sites upstream of the thymidine kinase promoter and has been shown previously to be dependent on STATs for cytokine-activated transcription (39). Hep-G2 cells were used because they showed significant levels of IL-6-inducible reporter gene activity (20- to 40-fold increase over baseline), and inhibition of IL-6 activation of Stat3 by the MEK-ERK pathway was verified in these cells (data not shown). IL-6-induced luciferase activity was inhibited by 61% when cells were treated with PMA (Fig. 5). Incubation of cells with MEK inhibitors nonspecifically suppressed reporter gene activity, precluding determination of the role of the MEK-ERK pathway by using this approach. Instead, a role for the MEK-ERK pathway in suppressing IL-6-activated, Stat3-dependent transcription was confirmed by using expression of constitutively active MEK1 (Fig. 5).
Figure 5.
Inhibition of an IL-6-inducible, STAT-dependent reporter gene. Hep-G2 cells were cotransfected with 3 μg of 3 X Ly6E GAS-luciferase (39), 1 μg of a β-galactosidase-encoding plasmid, and 11 μg of a control empty vector, or, as indicated, of the same vector containing a CA-MEK cDNA. Cells were split onto replicate plates and after an additional 24 hr, treated for 15 min with PMA followed by a 6-hr stimulation with IL-6. The mean luciferase activity (normalized according to β-galactosidase activity) relative to IL-6-induced activity (set at 100%) is shown. IL-6 induced luciferase activity 20- to 40-fold in these experiments. The results for PMA are based on five independent experiments and for CA-MEK on three independent experiments.
Stat3 Is Not the Target of Inhibition, Which Likely Occurs Upstream of Stat3 Tyrosine Phosphorylation in the IL-6-Signaling Pathway.
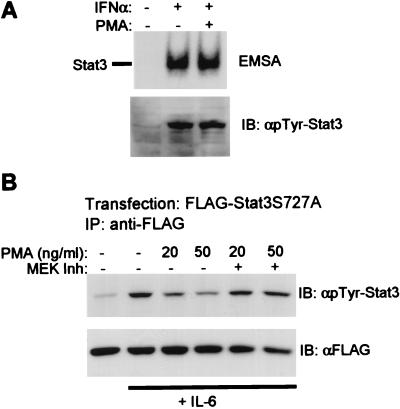
ERKs have been reported to phosphorylate Stat3 on serine-727, the major serine phosphorylation site. If the MEK-ERK pathway inhibits Stat3 by directly phosphorylating Stat3, one would predict that activation of Stat3 DNA-binding activity and tyrosine phosphorylation should be inhibited by ERKs, regardless of which cytokine is used to activate Stat3. This prediction was tested by activating Stat3 by using IFNα, which activates Stat3 (binds the hSIE oligonucleotide, Fig. 6A) in addition to activating ISGF3 (binds ISRE sequence; not detected by using hSIE) (1). In contrast to previous results when IL-6 was used (Figs. 1 and 3), pretreatment with PMA did not inhibit either DNA binding or tyrosine phosphorylation of Stat3 that was activated by using IFNα (Fig. 6A). Activation of Stat3 by IL-6 in the same experiment was inhibited by >80% (data not shown). Similar results were obtained when ionomycin or GM-CSF were used instead of PMA. These results suggest that the target for inhibition is not Stat3 but rather a molecule that is a component of the IL-6-, but not IFNα, signaling pathway. Inhibition was not mediated by modulation of levels of cell surface expression the IL-6 receptor (data not shown).
Figure 6.
(A) IFNα activation of Stat3 is not inhibited by PMA. MM6 cells were incubated with PMA for 15 min, followed by a 12-min stimulation with IFNα (8 ng/ml). Cell extracts were assayed for binding to the hSIE oligonucleotide by using EMSA. (B) Inhibition of Stat3S727A mutant. CHO cells were transfected with plasmids encoding Stat3S727A containing a FLAG epitope tag, stimulated with PMA and IL-6 and anti-FLAG immunoprecipitates analyzed by immunoblotting.
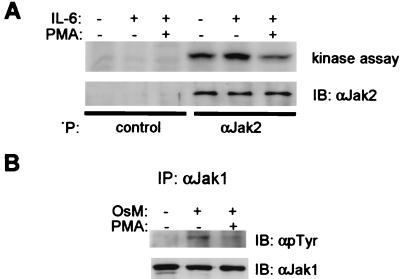
The role of serine-727 phosphorylation in regulating tyrosine phosphorylation of Stat3 in our system was addressed directly by using the Stat3S727A mutant that abolishes serine phosphorylation at this site; this is the same mutation studied in ref. 31. Tyrosine phosphorylation of Stat3S727A was inhibited by PMA, and inhibition was reversed when MEK was inhibited (Fig. 6B); inhibition of tyrosine phosphorylation was comparable to that of wild-type Stat3 in the same experiment (data not shown). These results indicate that inhibition of Stat3 by the ERK pathway was independent of phosphorylation on serine-727. To determine whether signaling events upstream of Stat3 tyrosine phosphorylation were inhibited, the activity of the Jak1 and Jak2 kinases that are activated by IL-6 (49) was analyzed. Modest but reproducible activation of Jak2 was detected and was inhibited by PMA pretreatment (Fig. 7A). Stronger activation over background of Jak1 was detected when oncostatin M, a cytokine similar to IL-6 that also signals through gp130, was used, and was inhibited by PMA (Fig. 7B), in parallel to inhibition of Stat3 tyrosine phosphorylation (data not shown). These results indicate that inhibition by the MEK-ERK pathway occurs, at least in part, upstream of Stat3 tyrosine phosphorylation.
Figure 7.
Inhibition of Jaks by PMA. (A) MM6 cells were treated with PMA followed by a 12-min stimulation with IL-6. Seventy-five percent of immunoprecipitates were analyzed for Jak activity (17) and 25% analyzed by using immunoblotting. (B) CHO cells were treated with PMA followed by stimulation with 100 ng/ml of oncostatin M, and immunoprecipitates were analyzed by immunoblotting.
DISCUSSION
The results presented herein describe a pathway for inhibiting IL-6 activation of Stat3 that acts rapidly (within 5 min) and is mediated by the MEK-ERK cascade, likely by post-translational modification of preexisting IL-6-signaling components. Signaling by IL-6 is preferentially inhibited relative to signaling by IFNα, and the mechanism involves inhibition of tyrosine phosphorylation of Stat3 by blocking signaling upstream of this event. This inhibitory pathway differs from previously described mechanisms for inhibition of Jak-STAT signaling because it is inducible rather than basal (3–8) and because inhibition occurs rapidly without the time lag that is necessitated by a requirement for new protein synthesis (9–11). The MEK-dependent inhibitory pathway also could work together with other inhibitory mechanisms that are induced at later timepoints (9–11) to block cytokine signaling over an extended time frame.
Several lines of evidence support a role for MEKs and ERKs in inhibition of IL-6 activation of Stat3. Inhibition of Stat3 activation by several agents correlated with activation of ERK activity, and specific inhibition of MEK and ERK activity by using two structurally unrelated compounds, PD98059 and U0126, resulted in a reversal of inhibition of IL-6 signaling. PD98059 appears to be a specific inhibitor of MEKs (47, 48); similarly, compound U0126 has been tested against at least 15 kinases and appears specific for MEKs (P.A.S., unpublished data). Most convincingly, expression of constitutively active MEK1 blocked tyrosine phosphorylation of Stat3, and overexpression of ERK2 potentiated inhibition of Stat3 DNA binding. Potentiation of Stat3 inhibition after overexpression of ERK2 suggests that ERKs or other kinases downstream of ERK in the MEK-ERK cascade mediate inhibition.
The MEK-dependent inhibition of IL-6-, but not IFNα-induced Stat3 tyrosine phosphorylation, provides insight into molecular mechanisms whereby MAPK and Jak-STAT pathways can interact in either a synergistic or antagonistic fashion. ERKs have been reported to potentiate signaling and gene activation by IFNα (23). In this case, ERKs would not block IFNα signaling (Fig. 6) and instead could potentiate transcriptional activation by STATs through serine phosphorylation of serine-727 in the transcription activation domain (39). In contrast, ERKs antagonize or inhibit the action of IL-6 or related cytokines that signal through gp130 in several systems (17, 25, 26, 28). Inhibition of Stat3 tyrosine phosphorylation through activation of the MEK-ERK pathway identifies a plausible molecular mechanism that may underly differentiation of monocytic cells into dendritic-like cells (17, 26) and nerve growth factor inhibition of ciliary neurotrophic factor/Stat3-mediated differentiation of cerebral cortical cells into astrocytes, or of IL-6/Stat3-mediated regulation of neurite outgrowth in PC12 cells (25, 28).
The relative selectivity of inhibition of IL-6 relative to IFNα suggests that IL-6R subunits or associated molecules may be targets of inhibition. Pilot experiments with fusion receptors (50) suggest that regulation of inhibition is complex, and multiple receptor sequences may play a role in mediating inhibition (T.K.S. and L.B.I, unpublished data). Many cytokines, including those that signal by using gp130, simultaneously activate signal transduction pathways leading to activation of ERKs and Stat3 (49, 51), although the relative intensity of ERK activation varies according to cell type (ref. 52 and L.B.I., unpublished data). Our data suggest that ERKs participate in negative feedback by limiting the intensity of Stat3 activation, and one would hypothesize that a mutated gp130 molecule that does not activate the MAPK pathway would activate Stat3 more strongly. Indeed, this hypothesis is supported by data showing that mutant gp130 receptors that do not activate ERKs are superactivators of Stat3-regulated target genes (53, 54) and of Stat3 DNA binding (ref. 54 and T.K.S., unpublished data). Because the mutation that blocks ERK activation abolishes the docking site for the tyrosine phosphatase SHP-2, further experiments will be required to resolve the relative roles and possible interactions of ERK activation and other functions mediated by SHP-2. Similar observations on the balance between ERK-dependent proliferation and Stat3-dependent differentiation have been reported in MDCK epithelial cells (24) and in myeloid precursors (27, 29). Greater understanding of the molecular basis for inhibition of IL-6 activation of Stat3 by the MEK-ERK pathway will lead to increased insight into regulation of cell proliferation and differentiation by cytokines and also may lead to novel therapeutic approaches to inflammatory arthritis, in which IL-6 plays an important role in pathogenesis (55).
Acknowledgments
We thank Jim Darnell, Kendall Smith, and Neil Stahl for helpful discussions, and Jackie Bromberg, Curt Horvath, and Akshay Vaishnaw for reviewing the manuscript. We are grateful to F. Giancotti, J. Woodgett, and H. W. Ziegler-Heitbrock for providing plasmids and cells, and J. Darnell for providing STAT antibodies. This work was supported by grants from the American Cancer Society, Arthritis Foundation, and National Institutes of Health.
ABBREVIATIONS
- GM-CSF
granulocyte/macrophage-colony-stimulating factor
- PMA
phorbol 12-myristate 13-acetate
- STAT
signal transducer and activator of transcription
- Jak
Janus kinase
- IL
interleukin
- MAPK
mitogen-activated protein kinase
- ERK
extracellular stimulus-regulated kinase
- EMSA
electrophoretic mobility shift assay
- IP
immunoprecipitation
- MBP
myelin basic protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.O’Shea J J. Immunity. 1997;7:1–11. doi: 10.1016/s1074-7613(00)80505-1. [DOI] [PubMed] [Google Scholar]
- 2.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 3.Azam M, Lee C, Strehlow I, Schindler C. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 4.Kim T K, Maniatis T. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 5.Haspel R L, Salditt-Georgieff M, Darnell J E., Jr EMBO J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 6.Shuai K, Liao J, Song M M. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aragane Y, Kulms D, Luger T A, Schwarz T. Proc Natl Acad Sci USA. 1997;94:11490–11495. doi: 10.1073/pnas.94.21.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung C D, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 9.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 10.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 11.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, et al. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 12.Feldman G M, Chuang E J, Finbloom D S. J Immunol. 1995;154:318–325. [PubMed] [Google Scholar]
- 13.Marth T, Kelsall B L. J Exp Med. 1997;185:1987–1995. doi: 10.1084/jem.185.11.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat G J, Abraham S T, Baker K M. J Biol Chem. 1996;271:22447–22452. doi: 10.1074/jbc.271.37.22447. [DOI] [PubMed] [Google Scholar]
- 15.Bright J J, Kerr L D, Sriram S. J Immunol. 1997;159:175–183. [PubMed] [Google Scholar]
- 16.Pazdrak K, Justement L, Alam R. J Immunol. 1995;155:4454–4458. [PubMed] [Google Scholar]
- 17.Sengupta T K, Schmitt E M, Ivashkiv L B. Proc Natl Acad Sci USA. 1996;93:9499–9504. doi: 10.1073/pnas.93.18.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David M, Petricoin III E, Larner A C. J Biol Chem. 1996;271:4585–4588. doi: 10.1074/jbc.271.9.4585. [DOI] [PubMed] [Google Scholar]
- 19.Petricoin III E, David M, Igarashi K, Benjamin C, Ling L, Goelz S, Finbloom D S, Larner A C. Mol Cell Biol. 1996;16:1419–1424. doi: 10.1128/mcb.16.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulton T G, Nye S H, Robbins D J, Ip N Y, Radziejewska E, Morgenbesser S D, DePinho R A, Panayotatos N, Cobb M H, Yancopoulos G D. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- 21.Woodgett J R, Kyriakis J M, Avruch J, Zon L I, Zanke B, Templeton D J. Philos Trans R Soc London B. 1996;351:135–141. doi: 10.1098/rstb.1996.0009. [DOI] [PubMed] [Google Scholar]
- 22.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 23.David M, Petricoin E, 3rd, Benjamin C, Pine R, Weber M J, Larner A C. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 24.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P M. Nature (London) 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 25.Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank D A, Rozovsky I, Stahl N, Yancopoulos G D, Greenberg M E. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 26.Czerniecki B J, Carter C, Rivoltini L, Koski G K, Kim H I, Weng D E, Roros J G, Hijazi Y M, Xu S, Rosenberg S A, Cohen P A. J Immunol. 1997;159:3823–3837. [PubMed] [Google Scholar]
- 27.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 28.Ihara S, Nakajima K, Fukada T, Hibi M, Nagata S, Hirano T, Fukui Y. EMBO J. 1997;16:5345–5352. doi: 10.1093/emboj/16.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 30.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung J, Uchida E, Grammer T C, Blenis J. Mol Cell Biol. 1997;17:6508–6516. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beadling C, Ng J, Babbage J W, Cantrell D A. EMBO J. 1996;15:1902–1913. [PMC free article] [PubMed] [Google Scholar]
- 33.Boulton T G, Zhong Z, Wen Z, Darnell J E, Jr, Stahl N, Yancopoulos G D. Proc Natl Acad Sci USA. 1995;92:6915–6919. doi: 10.1073/pnas.92.15.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eilers A, Georgellis D, Klose B, Schindler C, Ziemiecki A, Harpur A G, Wilks A F, Decker T. Mol Cell Biol. 1995;15:3579–3586. doi: 10.1128/mcb.15.7.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceresa B P, Horvath C M, Pessin J E. Endocrinology. 1997;138:4131–4137. doi: 10.1210/endo.138.10.5266. [DOI] [PubMed] [Google Scholar]
- 36.Ng J, Cantrell D. J Biol Chem. 1997;272:24542–24549. doi: 10.1074/jbc.272.39.24542. [DOI] [PubMed] [Google Scholar]
- 37.Zhu X, Wen Z, Xu L Z, Darnell J E., Jr Mol Cell Biol. 1997;17:6618–6623. doi: 10.1128/mcb.17.11.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Blenis J, Li H C, Schindler C, Chen-Kiang S. Science. 1995;267:1990–1994. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 39.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 40.Ihle J N. BioEssays. 1996;18:95–98. doi: 10.1002/bies.950180204. [DOI] [PubMed] [Google Scholar]
- 41.Welham M J, Duronio V, Sanghera J S, Pelech S L, Schrader J W. J Immunol. 1992;149:1683–1693. [PubMed] [Google Scholar]
- 42.Ziegler-Heitbrock H W, Thiel E, Futterer A, Herzog V, Wirtz A, Riethmuller G. Int J Cancer. 1988;41:456–461. doi: 10.1002/ijc.2910410324. [DOI] [PubMed] [Google Scholar]
- 43.Sengupta T K, Chen A, Zhong Z, Darnell J E, Jr, Ivashkiv L B. J Exp Med. 1995;181:1015–1025. doi: 10.1084/jem.181.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Z, Wen Z, Darnell J E., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 45.Mainiero F, Murgia C, Wary K K, Curatola A M, Pepe A, Blumemberg M, Westwick J K, Der C J, Giancotti F G. EMBO J. 1997;16:2365–2375. doi: 10.1093/emboj/16.9.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanke B W, Rubie E A, Winnett E, Chan J, Randall S, Parsons M, Boudreau K, McInnis M, Yan M, Templeton D J, et al. J Biol Chem. 1996;271:29876–29881. doi: 10.1074/jbc.271.47.29876. [DOI] [PubMed] [Google Scholar]
- 47.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 48.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taga T, Kishimoto T. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 50.Stahl N, Farruggella T J, Boulton T G, Zhong Z, Darnell J E, Jr, Yancopoulos G D. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 51.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 52.Ogata A, Chauhan D, Teoh G, Treon S P, Urashima M, Schlossman R L, Anderson K C. J Immunol. 1997;159:2212–2221. [PubMed] [Google Scholar]
- 53.Symes A, Stahl N, Reeves S A, Farruggella T, Servidei T, Gearan T, Yancopoulos G, Fink J S. Curr Biol. 1997;7:697–700. doi: 10.1016/s0960-9822(06)00298-3. [DOI] [PubMed] [Google Scholar]
- 54.Kim H, Hawley T S, Hawley R G, Baumann H. Mol Cell Biol. 1998;18:1525–1533. doi: 10.1128/mcb.18.3.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivashkiv L B. Adv Immunol. 1996;63:337–376. doi: 10.1016/s0065-2776(08)60859-7. [DOI] [PubMed] [Google Scholar]