Abstract
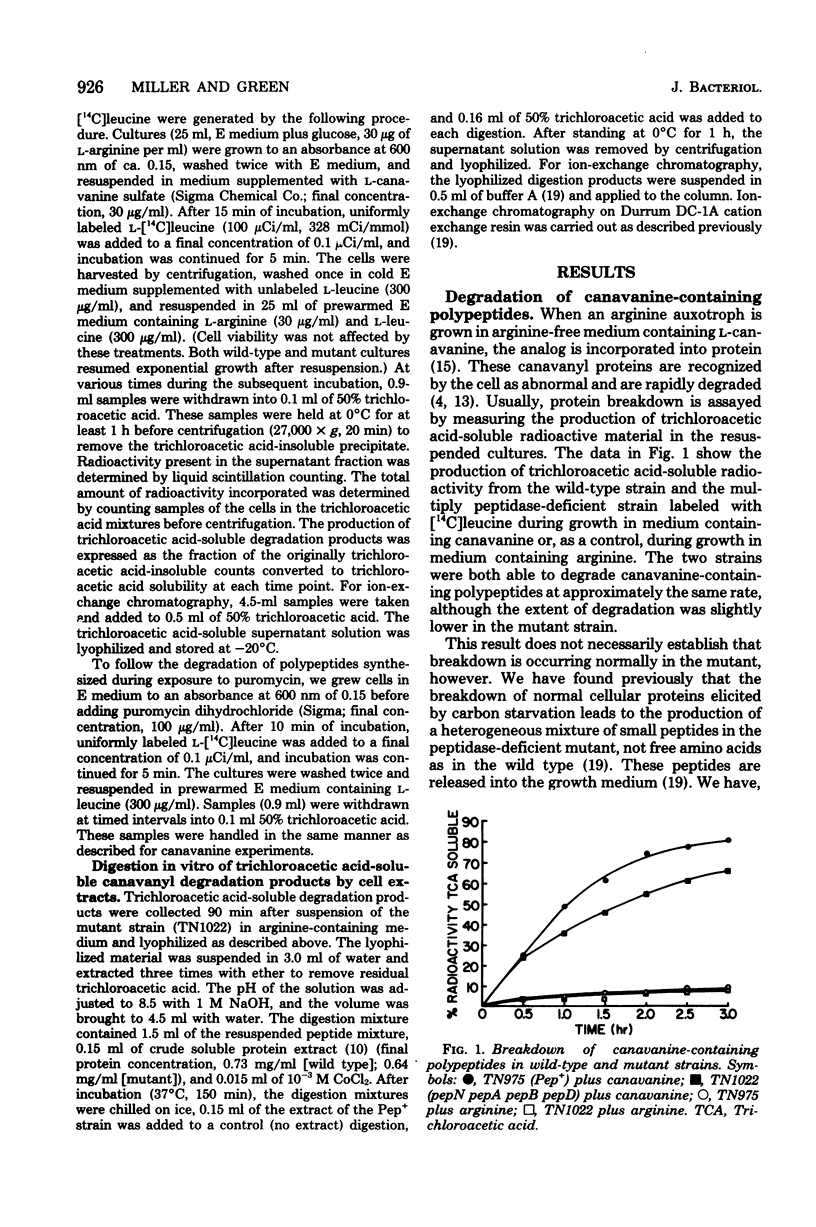
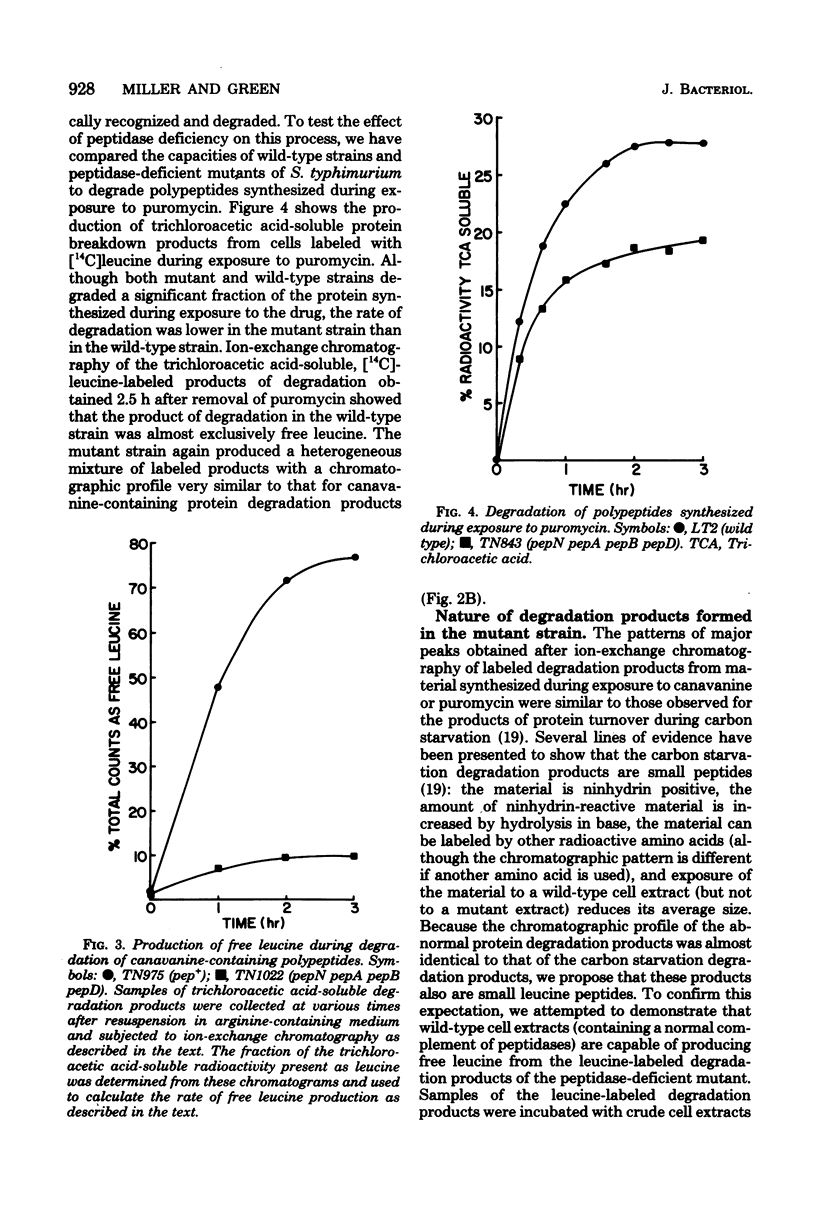
The degradation of abnormal proteins produced as a result of incorporation of the arginine analog L-canavanine or generated by exposure to puromycin was studied in wild-type and multiply peptidase-deficient strains of Salmonella typhimurium. Both types of abnormal protein were rapidly degraded during growth of Pep+ strains of this organism. Peptidase--deficient mutants (lacking peptidases N, A, B, and D) could also degrade these abnormal proteins, although the rate of production of trichloroacetic acid-soluble degradation products was slower in the mutant strain than in a strain carrying a normal complement of peptidases. Analysis of these trichloroacetic acid-soluble degradation products of ion-exchange chromatography showed that free amino acid was the major breakdown product produced by the wild-type strain. The acid-soluble degradation product produced by the mutant strain, however, was a complex mixture that contained a variety of small peptides as well as free amino acids. These results indicate that the same group of peptidases shown previously to function in the degradation of exogenously supplied peptides and in protein turnover during carbon starvation also lie on the pathway by which abnormal proteins are degraded.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ciechanover A., Elias S., Heller H., Ferber S., Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem. 1980 Aug 25;255(16):7525–7528. [PubMed] [Google Scholar]
- Etlinger J. D., Goldberg A. L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A. 1977 Jan;74(1):54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin). Proc Natl Acad Sci U S A. 1972 Feb;69(2):422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L., St John A. C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. In vivo degradation of nonsense fragments in E. coli. Nature. 1970 Dec 19;228(5277):1151–1154. doi: 10.1038/2281151a0. [DOI] [PubMed] [Google Scholar]
- Kemshead J. T., Hipkiss A. R. Degradation of abnormal proteins in Escherichia coli: relative susceptibility of canavanyl proteins and puromycin peptides to proteolysis in vitro. Eur J Biochem. 1974 Jun 15;45(2):535–540. doi: 10.1111/j.1432-1033.1974.tb03578.x. [DOI] [PubMed] [Google Scholar]
- Kirsh M., Dembinski D. R., Hartman P. E., Miller C. G. Salmonella typhimurium peptidase active on carnosine. J Bacteriol. 1978 May;134(2):361–374. doi: 10.1128/jb.134.2.361-374.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh G. L., Miller C. G. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):364–371. doi: 10.1128/jb.120.1.364-371.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G., Mackinnon K. Peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G. Peptidases and proteases of Escherichia coli and Salmonella typhimurium. Annu Rev Microbiol. 1975;29:485–504. doi: 10.1146/annurev.mi.29.100175.002413. [DOI] [PubMed] [Google Scholar]
- Murakami K., Voellmy R., Goldberg A. L. Protein degradation is stimulated by ATP in extracts of Escherichia coli. J Biol Chem. 1979 Sep 10;254(17):8194–8200. [PubMed] [Google Scholar]
- Pine M. J. Response of intracellular proteolysis to alteration of bacterial protein and the implications in metabolic regulation. J Bacteriol. 1967 May;93(5):1527–1533. doi: 10.1128/jb.93.5.1527-1533.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine M. J. Turnover of intracellular proteins. Annu Rev Microbiol. 1972;26:103–126. doi: 10.1146/annurev.mi.26.100172.000535. [DOI] [PubMed] [Google Scholar]
- Schachtele C. F., Rogers P. Canavanine death in Escherichia coli. J Mol Biol. 1965 Dec;14(2):474–489. doi: 10.1016/s0022-2836(65)80197-8. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vogt V. M. Purification and properties of an aminopeptidase from Escherichia coli. J Biol Chem. 1970 Sep 25;245(18):4760–4769. [PubMed] [Google Scholar]
- Yaron A., Mlynar D., Berger A. A dipeptidocarboxypeptidase from E. coli. Biochem Biophys Res Commun. 1972 May 26;47(4):897–902. doi: 10.1016/0006-291x(72)90577-3. [DOI] [PubMed] [Google Scholar]
- Yen C., Green L., Miller C. G. Degradation of intracellular protein in Salmonella typhimurium peptidase mutants. J Mol Biol. 1980 Oct 15;143(1):21–33. doi: 10.1016/0022-2836(80)90122-9. [DOI] [PubMed] [Google Scholar]
- Yen C., Green L., Miller C. G. Peptide accumulation during growth of peptidase deficient mutants. J Mol Biol. 1980 Oct 15;143(1):35–48. doi: 10.1016/0022-2836(80)90123-0. [DOI] [PubMed] [Google Scholar]
- Zipser D., Bhavsar P. Missense mutations in the lacZ gene that result in degradation of beta-galactosidase structural protein. J Bacteriol. 1976 Sep;127(3):1538–1542. doi: 10.1128/jb.127.3.1538-1542.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]


