Abstract
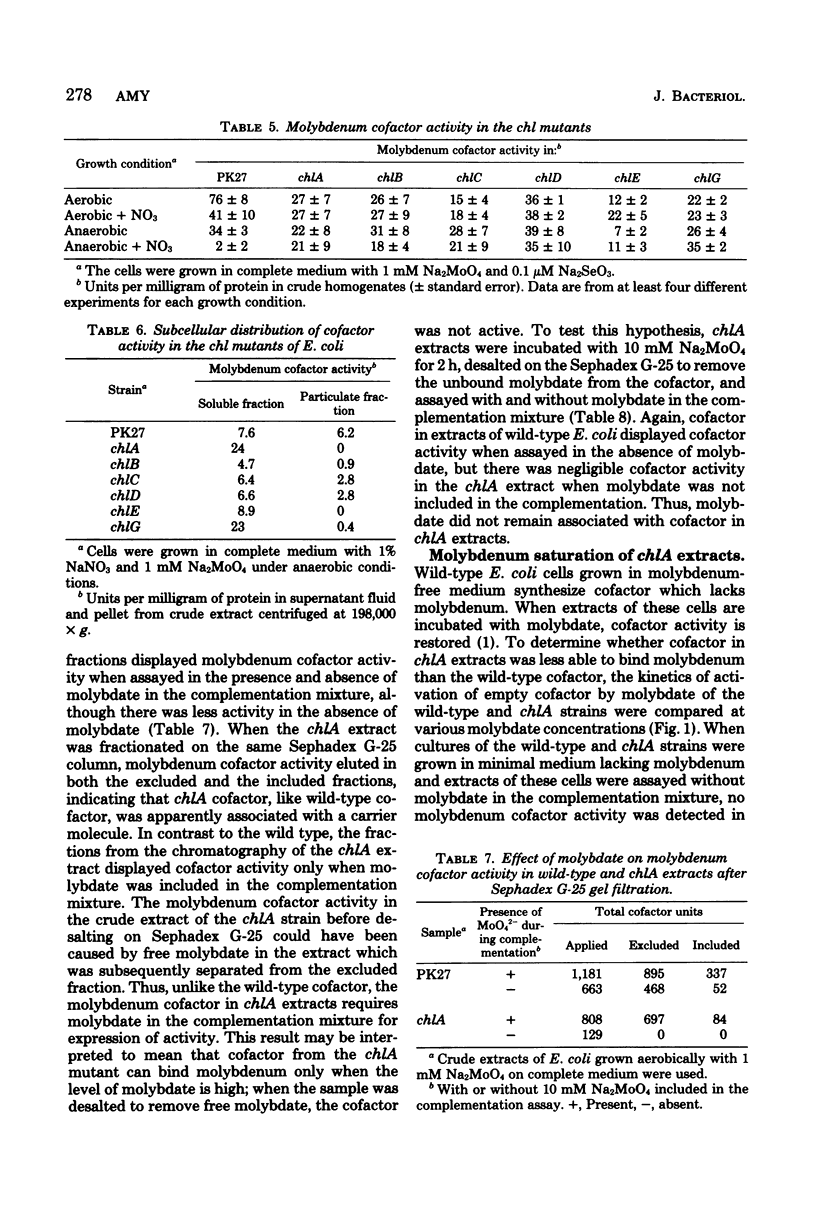
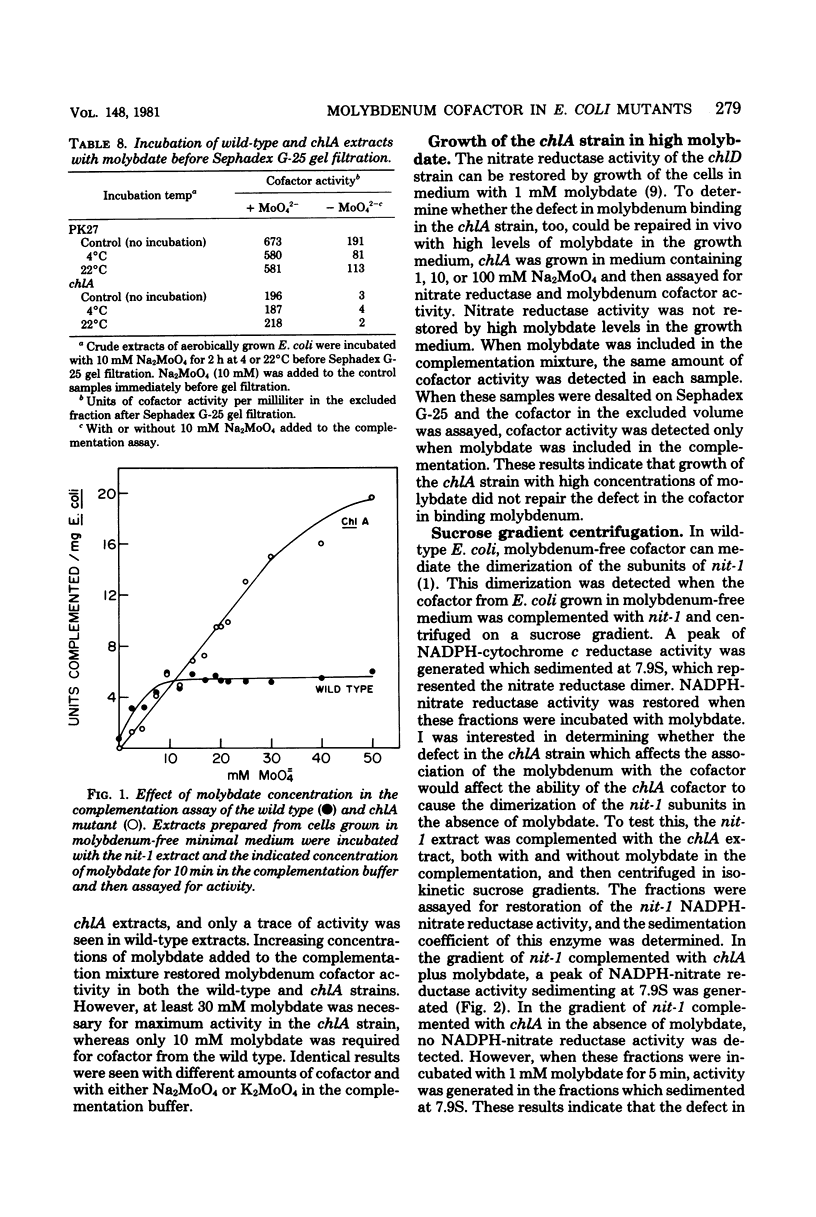
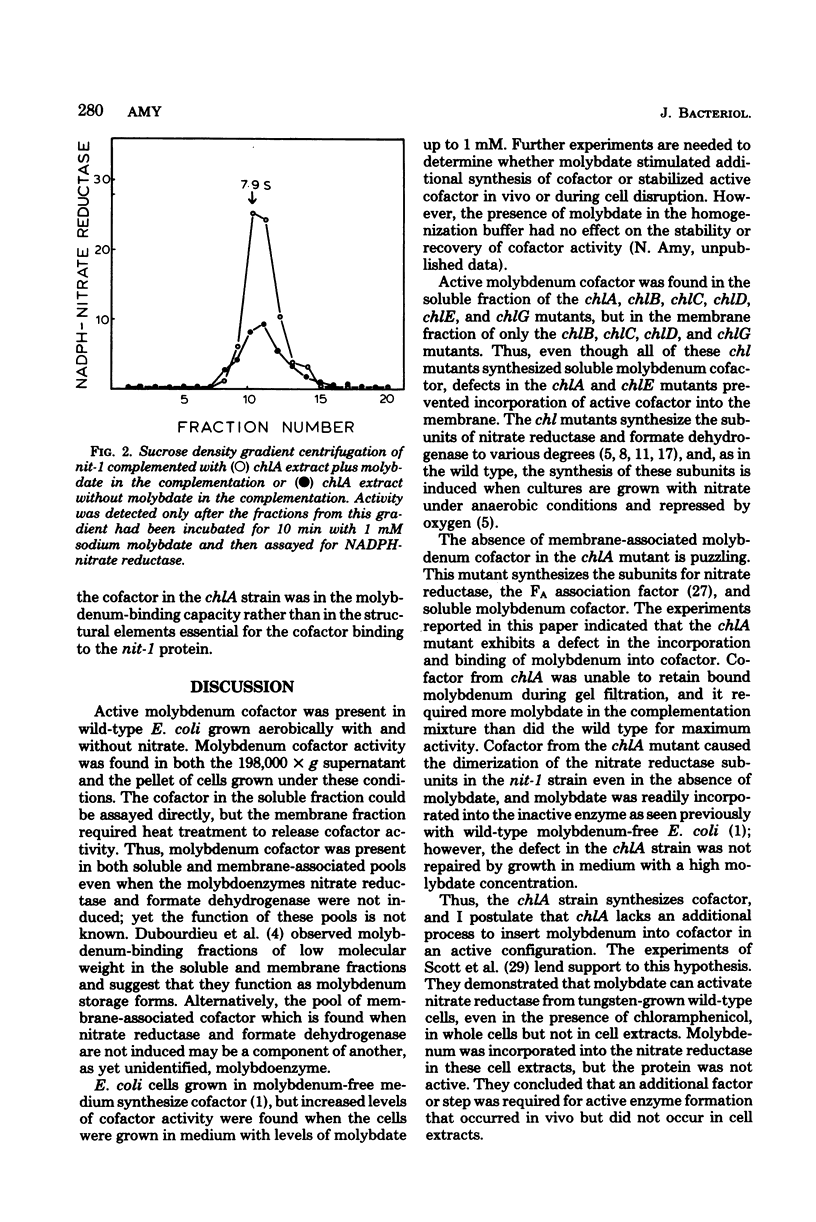
Experiments were performed to determine whether defects in molybdenum cofactor metabolism were responsible for the pleiotropic loss of the molybdoenzymes nitrate reductase and formate dehydrogenase in chl mutants of Escherichia coli. In wild-type E. coli, molybdenum cofactor activity was present in both the soluble and membrane-associated fractions when the cells were grown either aerobically or anaerobically, with and without nitrate. Molybdenum cofactor in the soluble fraction decreased when the membrane-bound nitrate reductase and formate dehydrogenase were induced. In the chl mutants, molybdenum cofactor activity was found in the soluble fraction of chlA, chlB, chlC, chlD, chlE, and chlG, but only chlB, chlC, chlD, and chlG expressed cofactor activity in the membrane fraction. The defect in the chlA mutants which prevented incorporation of the soluble cofactor into the membrane also caused the soluble cofactor to be defective in its ability to bind molybdenum. This cofactor was not active in the absence of molybdate, and it required at least threefold more molybdate than did the wild type in the Neurospora crassa nit-1 complementation assay. However, the cofactor from the chlA strain mediated the dimerization of the nit-1 subunits in the presence and absence of molybdate to yield the 7.9S dimer. Growth of chlA mutants in medium with increased molybdate did not repair the defect in the chlA cofactor nor restore the molybdoenzyme activities. Thus, molybdenum cofactor was synthesized in all the chl mutants, but additional processing steps may be missing in chlA and chlE mutants for proper insertion of cofactor in the membrane.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy N. K., Rajagopalan K. V. Characterization of molybdenum cofactor from Escherichia coli. J Bacteriol. 1979 Oct;140(1):114–124. doi: 10.1128/jb.140.1.114-124.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casse F. Mapping of the gene chl-B controlling membran bound nitrate reductase and formic hydrogen-lyase activities in Escherichia coli K 12. Biochem Biophys Res Commun. 1970 May 11;39(3):429–436. doi: 10.1016/0006-291x(70)90596-6. [DOI] [PubMed] [Google Scholar]
- DeMoss J. A. Role of the chlC gene in formation of the formate-nitrate reductase pathway in Escherichia coli. J Bacteriol. 1978 Feb;133(2):626–630. doi: 10.1128/jb.133.2.626-630.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourdieu M., Andrade E., Puig J. Molybdenum and chlorate resistant mutants in Escherichia coli K12. Biochem Biophys Res Commun. 1976 Jun 7;70(3):766–773. doi: 10.1016/0006-291x(76)90658-6. [DOI] [PubMed] [Google Scholar]
- Forget P. Effect of growth conditions on the synthesis of nitrate reductase components in chlorate resistant mutants of Escherichia coli K 12. Biochem Biophys Res Commun. 1979 Jul 27;89(2):659–663. doi: 10.1016/0006-291x(79)90680-6. [DOI] [PubMed] [Google Scholar]
- Garrett R. H., Nason A. Involvement of a B-type cytochrome in the assimilatory nitrate reductase of Neurospora crassa. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1603–1610. doi: 10.1073/pnas.58.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett R. H. The induction of nitrite reductase in Neurospora crassa. Biochim Biophys Acta. 1972 May 16;264(3):481–489. doi: 10.1016/0304-4165(72)90011-6. [DOI] [PubMed] [Google Scholar]
- Giordano G., Grillet L., Pommier J., Terriere C., Haddock B. A., Azoulay E. Precursor forms of the subunits of nitrate reductase in chlA and chlB mutants of Escherichia coli K12. Eur J Biochem. 1980 Apr;105(2):297–306. doi: 10.1111/j.1432-1033.1980.tb04501.x. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Ketchum P. A., Cambier H. Y., Frazier W. A., 3rd, Madansky C. H., Nason A. In vitro assembly of Neurospora assimilatory nitrate reductase from protein subunits of a Neurospora mutant and the xanthine oxidizing or aldehyde oxidase systems of higher animals. Proc Natl Acad Sci U S A. 1970 Jul;66(3):1016–1023. doi: 10.1073/pnas.66.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A., Normansell D. E. Purification and properties of nitrate reductase from Escherichia coli K12. J Biol Chem. 1974 Aug 25;249(16):5321–5327. [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A. Restoration of reduced nicotinamide adenine dinucleotide phosphate-nitrate reductase activity of a Neurospora mutant by extracts of various chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1972 Oct;112(1):388–391. doi: 10.1128/jb.112.1.388-391.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1975 Mar;121(3):1117–1121. doi: 10.1128/jb.121.3.1117-1121.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. Invitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate: nitrate reductase from a Neurospora mutant and a component of molybdenum-enzymes. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3242–3246. doi: 10.1073/pnas.68.12.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATEMAN J. A., COVE D. J., REVER B. M., ROBERTS D. B. A COMMON CO-FACTOR FOR NITRATE REDUCTASE AND XANTHINE DEHYDROGENASE WHICH ALSO REGULATES THE SYNTHESIS OF NITRATE REDUCTASE. Nature. 1964 Jan 4;201:58–60. doi: 10.1038/201058a0. [DOI] [PubMed] [Google Scholar]
- Pienkos P. T., Shah V. K., Brill W. J. Molybdenum cofactors from molybdoenzymes and in vitro reconstitution of nitrogenase and nitrate reductase. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5468–5471. doi: 10.1073/pnas.74.12.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J., Azoulay E. Etude génétique et biochimique des mutants résistant au Clo minus 3 (gènes chl A, chl B, chl C) C R Acad Sci Hebd Seances Acad Sci D. 1967 Apr 10;264(15):1916–1918. [PubMed] [Google Scholar]
- Puig J., Azoulay E., Gendre J., Richard E. Etude génétique des mutants de la région chl A chez l'Escherichia coli k12. C R Acad Sci Hebd Seances Acad Sci D. 1969 Jan 6;268(1):183–184. [PubMed] [Google Scholar]
- Puig J., Azoulay E., Pichinoty F., Gendre J. Genetic mapping of the chl C gene of the nitrate reductase A system in Escherichia coli K12. Biochem Biophys Res Commun. 1969 Jun 6;35(5):659–662. doi: 10.1016/0006-291x(69)90455-0. [DOI] [PubMed] [Google Scholar]
- Riviere C., Giordano G., Pommier J., Azoulay E. Membrane reconstitution in chl-r mutants of Escherichia coli K 12. VIII. Purification and properties of the FA factor, the product of the chl B gene. Biochim Biophys Acta. 1975 May 6;389(2):219–235. doi: 10.1016/0005-2736(75)90317-x. [DOI] [PubMed] [Google Scholar]
- Scott R. H., DeMoss J. A. Formation of the formate-nitrate electron transport pathway from inactive components in Escherichia coli. J Bacteriol. 1976 Apr;126(1):478–486. doi: 10.1128/jb.126.1.478-486.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. H., Sperl G. T., DeMoss J. A. In vitro incorporation of molybdate into demolybdoproteins in Escherichia coli. J Bacteriol. 1979 Feb;137(2):719–726. doi: 10.1128/jb.137.2.719-726.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V. K., Brill W. J. Isolation of an iron-molybdenum cofactor from nitrogenase. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3249–3253. doi: 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer A. H. Biochemistry and genetics of nitrate reductase in bacteria. Adv Microb Physiol. 1976;14(11):315–375. doi: 10.1016/s0065-2911(08)60230-1. [DOI] [PubMed] [Google Scholar]
- Tomsett A. B., Garrett R. H. The isolation and characterization of mutants defective in nitrate assimilation in Neurospora crassa. Genetics. 1980 Jul;95(3):649–660. doi: 10.1093/genetics/95.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W. A., Guest J. R. Transduction of nitrate reductase loci of Escherichia coli by phages P-1 and lambda. Mol Gen Genet. 1968;103(2):127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]


