Abstract
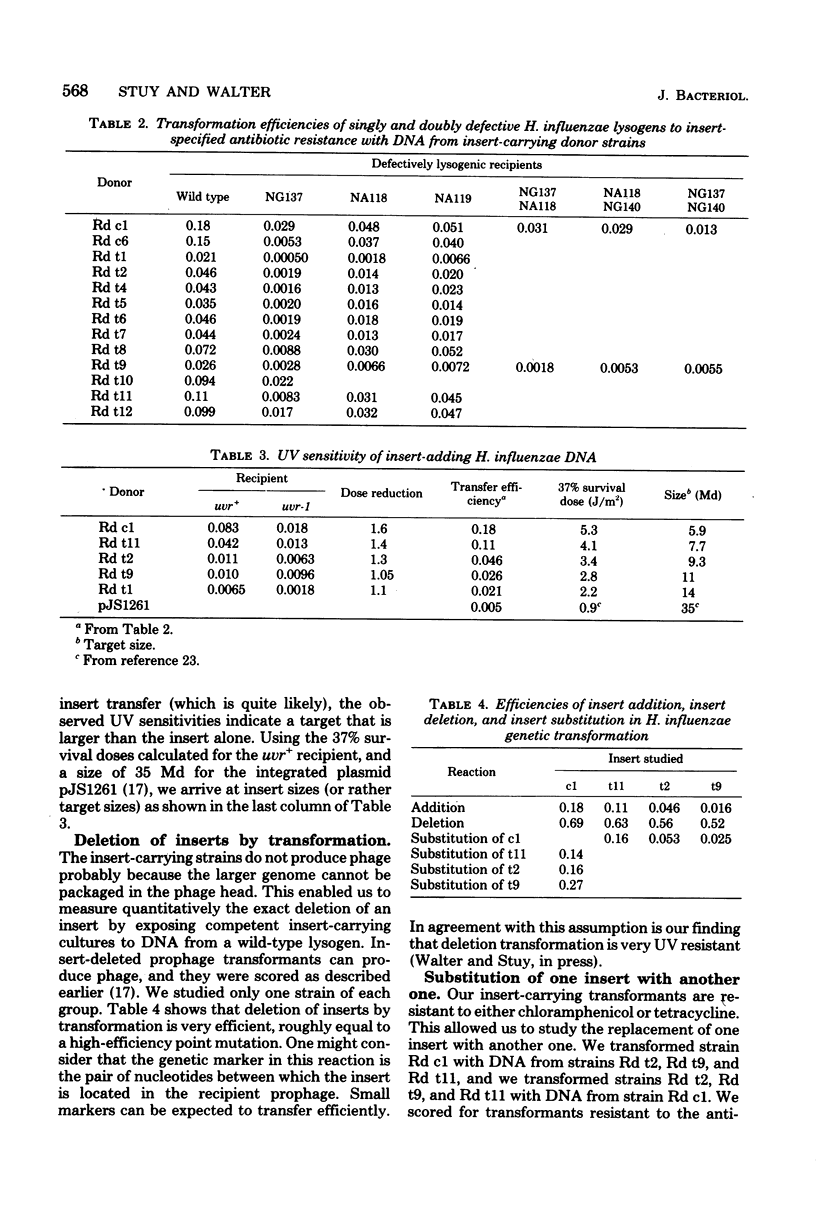
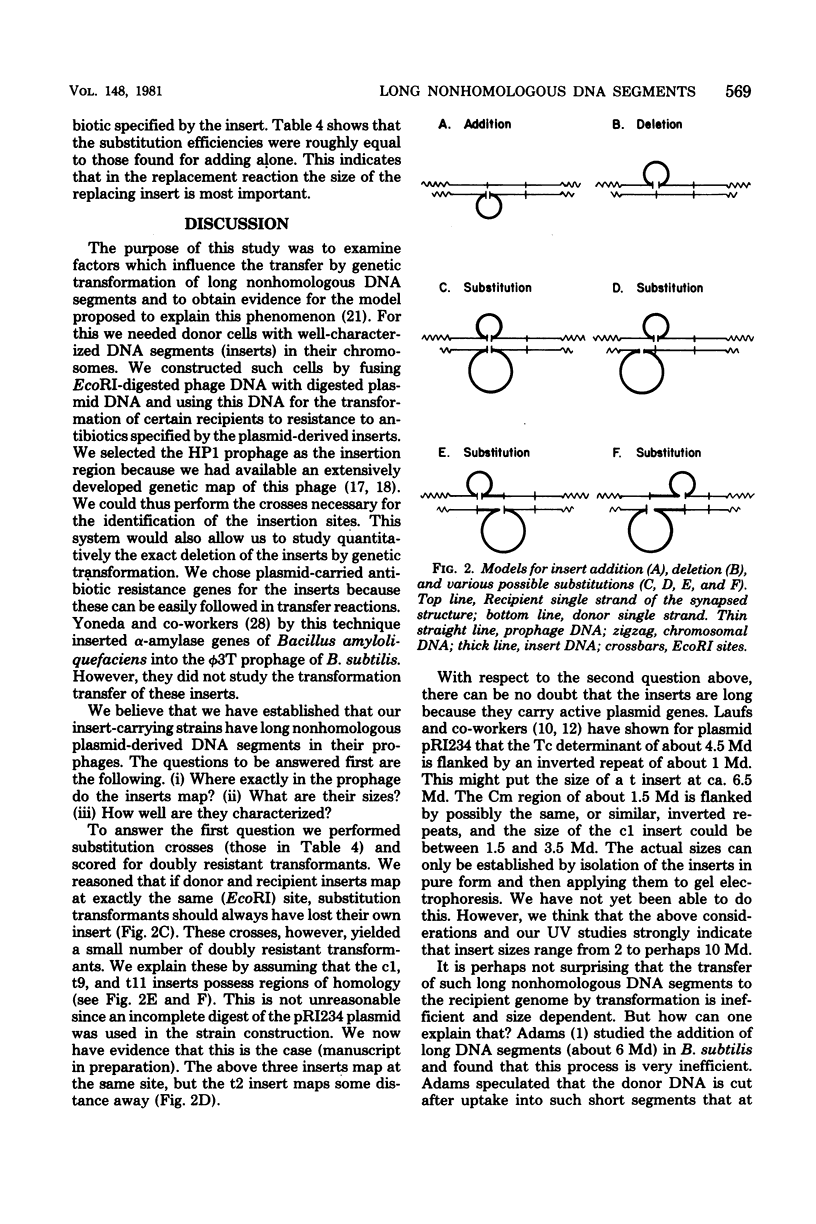
A complete EcoRI digest of Haemophilus influenzae phage HP1 deoxyribonucleic acid (DNA) was mixed with incomplete digests of various H. influenzae R plasmids, sealed with T4 ligase, and transformed into an HP1 lysogen. Most of the chloramphenicol- and tetracycline-resistant transformants did not produce phage although they possessed all the phage genes examined. They also did not transfer antibiotic resistance by conjugation. DNA lysates from them transformed other lysogens to resistance and to loss of phage production at different but quite high frequencies (addition of long DNA segments). They themselves could be transformed efficiently to strains with a wild prophage (deletion of long DNA segments). It was concluded that lysogenic cultures had been constructed with various DNA inserts in their prophages carrying antibiotic resistance genes from the R plasmids. The site of insertion was determined by genetic crosses. DNAs with inserts that transferred with lower efficiency were more sensitive to ultraviolet radiation. This supports the view that insert transfer efficiencies reflect the sizes of the insert.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. Transformation and transduction of a large deletion mutation in Bacillus subtilis. Mol Gen Genet. 1972;118(4):311–320. doi: 10.1007/BF00333566. [DOI] [PubMed] [Google Scholar]
- Barnhart B. J., Cox S. H. Radiation-sensitive and radiation-resistant mutants of Haemophilus influenzae. J Bacteriol. 1968 Jul;96(1):280–282. doi: 10.1128/jb.96.1.280-282.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendler J. W., 3rd Physical size of the donor locus and transmission of Haemophilus influenzae ampicillin resistance genes by deoxyribonucleic acid-mediated transformation. J Bacteriol. 1976 Jan;125(1):197–204. doi: 10.1128/jb.125.1.197-204.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheimer H. P., Wermundsen I. E. Homology in capsular transformation reactions in Pneumococcus. Mol Gen Genet. 1972;116(1):68–83. doi: 10.1007/BF00334261. [DOI] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Mechanism of integrating foreign DNA during transformation of Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3664–3668. doi: 10.1073/pnas.75.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell L. P., De Graaff J., Seibert D., Falkow S. Plasmid-linked ampicillin resistance in haempohilus influenza type b. Infect Immun. 1975 Aug;12(2):404–410. doi: 10.1128/iai.12.2.404-410.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARM W., RUPERT C. S. INFECTION OF TRANSFORMABLE CELLS OF HAEMOPHILUS INFLUENZAE BY BACTERIOPHAGE AND BACTERIOPHAGE DNA. Z Vererbungsl. 1963 Dec 30;94:336–348. doi: 10.1007/BF00897593. [DOI] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Lederberg J. Interspecies transformation in Bacillus: mechanism of heterologous intergenote transformation. J Bacteriol. 1978 Mar;133(3):1246–1253. doi: 10.1128/jb.133.3.1246-1253.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn G., Laufs R., Kaulfers P. M., Kolenda H. Molecular nature of two Haemophilus influenzae R factors containing resistances and the multiple integration of drug resistance transposons. J Bacteriol. 1979 May;138(2):584–597. doi: 10.1128/jb.138.2.584-597.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUY J. H. Transformability of Haemophilus influenzae. J Gen Microbiol. 1962 Nov;29:537–549. doi: 10.1099/00221287-29-3-537. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Brown D. C., Boling M. E., Mattingly A., Gordon M. P. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J Bacteriol. 1968 Feb;95(2):546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Smith M. D., Guild W. R. Organization and transfer of heterologous chloramphenicol and tetracycline resistance genes in pneumococcus. J Bacteriol. 1979 Aug;139(2):432–441. doi: 10.1128/jb.139.2.432-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Chromosomally integrated conjugative plasmids are common in antibiotic-resistant Haemophilus influenzae. J Bacteriol. 1980 Jun;142(3):925–930. doi: 10.1128/jb.142.3.925-930.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Fate of transforming bacteriophage HP1 deoxyribonucleic acid in Haemophilus influenzae lysogens. J Bacteriol. 1975 Jun;122(3):1038–1044. doi: 10.1128/jb.122.3.1038-1044.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H., Hoffmann J. F., Duket L. H. Chromosomal recombination in Haemophilus influenzae. Genetics. 1972 Aug;71(4):507–520. doi: 10.1093/genetics/71.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H., Hoffmann J. F. Influence of transformability on the formation of superinfection double lysogens in Haemophilus influenzae. J Virol. 1971 Jan;7(1):127–136. doi: 10.1128/jvi.7.1.127-136.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Mechanism of Haemophilus influenzae transfection by single and double prophage deoxyribonucleic acid. J Bacteriol. 1980 Dec;144(3):1003–1008. doi: 10.1128/jb.144.3.1003-1008.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Mechanism of additive genetic transformation in Haemophilus influenzae. J Bacteriol. 1980 Dec;144(3):999–1002. doi: 10.1128/jb.144.3.999-1002.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Origin and direction of Haemophilus bacteriophage HP1 DNA replication. J Virol. 1974 Mar;13(3):757–759. doi: 10.1128/jvi.13.3.757-759.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Plasmid transfer in Haemophilus influenzae. J Bacteriol. 1979 Aug;139(2):520–529. doi: 10.1128/jb.139.2.520-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Prophage mapping by transformation. Virology. 1969 Aug;38(4):567–572. doi: 10.1016/0042-6822(69)90177-9. [DOI] [PubMed] [Google Scholar]
- Stuy J. H. Tetracyclin-stimulated expression of ampicillin resistance in Haemophilus influenzae. J Bacteriol. 1980 Nov;144(2):823–825. doi: 10.1128/jb.144.2.823-825.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Intergenotic transformation of the Bacillus subtilis genospecies. J Bacteriol. 1972 Sep;111(3):705–716. doi: 10.1128/jb.111.3.705-716.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y., Graham S., Young F. E. Cloning of a foreign gene coding for alpha-amylase in Bacillus subtilis. Biochem Biophys Res Commun. 1979 Dec 28;91(4):1556–1564. doi: 10.1016/0006-291x(79)91242-7. [DOI] [PubMed] [Google Scholar]
- van Klingeren B., van Embden J. D., Dessens-Kroon M. Plasmid-mediated chloramphenicol resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1977 Mar;11(3):383–387. doi: 10.1128/aac.11.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]


