Abstract
We purified lipoamide dehydrogenase from cells of Pseudomonas putida PpG2 grown on glucose (LPD-glu) and lipoamide dehydrogenase from cells grown on valine (LPD-val), which contained branched-chain keto acid dehydrogenase. LPD-glu had a molecular weight of 56,000 as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and LPD-val had a molecular weight of 49,000. The pH optimum for LPD-glu for reduced nicotinamide adenine dinucleotide oxidation was 7.4, compared with pH 6.5 for LPD-val. When oxidized nicotinamide adenine dinucleotide was included in the assay mixture, the pH optima were 7.1 and 5.7, respectively. There was also a difference in pH optima between the two enzymes for oxidized nicotinamide adenine dinucleotide reduction, but the Michaelis constants and maximum velocities were similar. A purified preparation of branched-chain keto acid dehydrogenase, which was deficient in lipoamide dehydrogenase, was stimulated 10-fold by LPD-val but not by LPD-glu, which suggested that the branched-chain keto acid dehydrogenase of P. putida has a specific requirement for LPD-val. In contrast, a partially purified preparation of 2-ketoglutarate dehydrogenase that was deficient in lipoamide dehydrogenase was stimulated by LPD-glu but not by LPD-val, indicating that this complex has a specific requirement of LPD-glu.
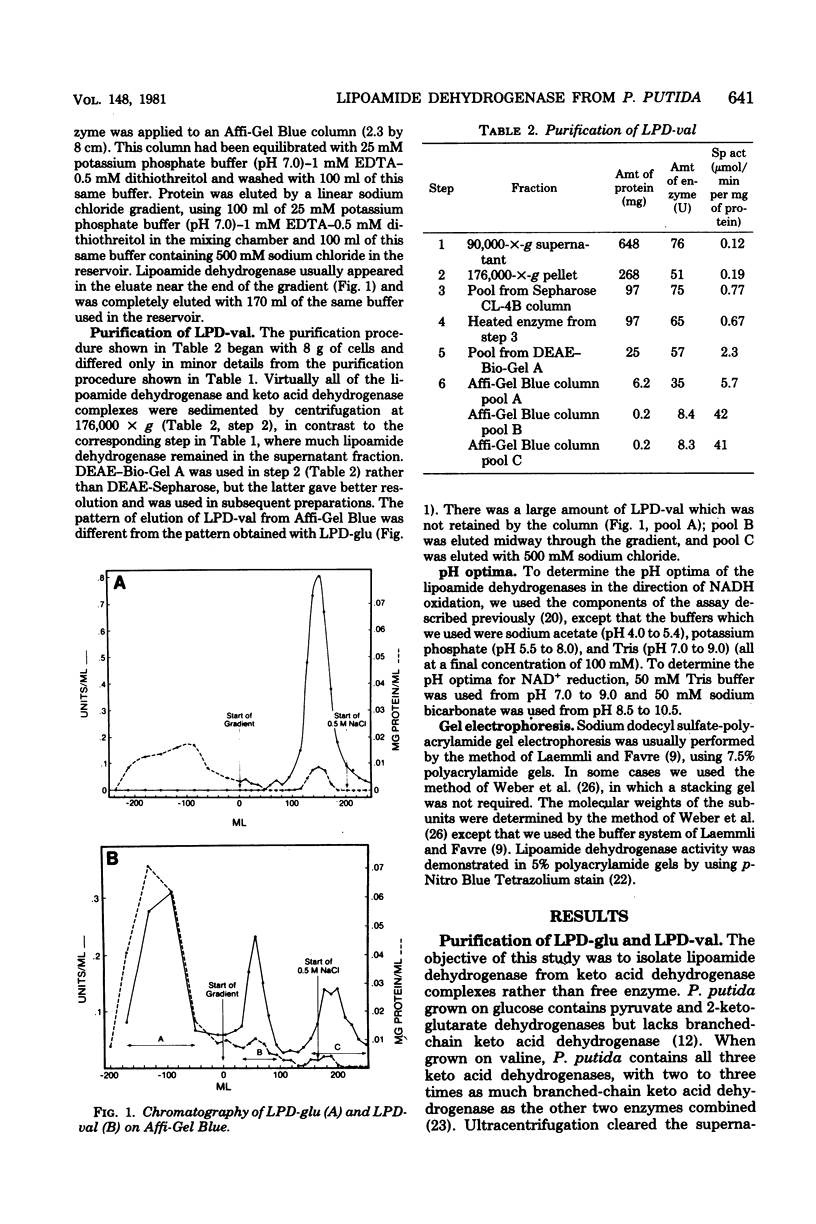
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ATKINSON D. E., HATHAWAY J. A., SMITH E. C. KINETICS OF REGULATORY ENZYMES. KINETIC ORDER OF THE YEAST DIPHOSPHOPYRIDINE NUCLEOTIDE ISOCITRATE DEHYDROGENASE REACTION AND A MODEL FOR THE REACTION. J Biol Chem. 1965 Jun;240:2682–2690. [PubMed] [Google Scholar]
- Alwine J. C., Russell R. M., Murray K. N. Characterization of an Escherichia coli mutant deficient in dihydrolipoyl dehydrogenase activity. J Bacteriol. 1973 Jul;115(1):1–8. doi: 10.1128/jb.115.1.1-8.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera C. R., Namihira G., Hamilton L., Munk P., Eley M. H., Linn T. C., Reed L. J. -Keto acid dehydrogenase complexes. XVI. Studies on the subunit structure of the pyruvate dehydrogenase complexes from bovine kidney and heart. Arch Biochem Biophys. 1972 Feb;148(2):343–358. doi: 10.1016/0003-9861(72)90152-x. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Aspects of the molecular biology of lipoamide dehydrogenase. Adv Neurol. 1978;21:219–244. [PubMed] [Google Scholar]
- Guest J. R., Creaghan I. T. Gene-protein relationships of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: isolation and characterization of lipoamide dehydrogenase mutants. J Gen Microbiol. 1973 Mar;75(1):197–210. doi: 10.1099/00221287-75-1-197. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Gene-protein relationships of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: Chromosomal location of the lipoamide dehydrogenase gene. J Gen Microbiol. 1974 Feb;80(2):523–532. doi: 10.1099/00221287-80-2-523. [DOI] [PubMed] [Google Scholar]
- Jacobson L. A., Bartholomaus R. C., Gunsalus I. C. Repression of malic enzyme by acetate in Pseudomonas. Biochem Biophys Res Commun. 1966 Sep 22;24(6):955–960. doi: 10.1016/0006-291x(66)90343-3. [DOI] [PubMed] [Google Scholar]
- LUSTY C. J. LIPOYL DEHYDROGENASE FROM BEEF LIVER MITOCHONDRIA. J Biol Chem. 1963 Oct;238:3443–3452. [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- MASSEY V., GIBSON Q. H., VEEGER C. Intermediates in the catalytic action of lipoyl dehydrogenase (diaphorase). Biochem J. 1960 Nov;77:341–351. doi: 10.1042/bj0770341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASSEY V. The identity of diaphorase and lipoyl dehydrogenase. Biochim Biophys Acta. 1960 Jan 15;37:314–322. doi: 10.1016/0006-3002(60)90239-0. [DOI] [PubMed] [Google Scholar]
- MASSEY V., VEEGER C. Studies on the reaction mechanism of lipoyl dehydrogenase. Biochim Biophys Acta. 1961 Mar 18;48:33–47. doi: 10.1016/0006-3002(61)90512-1. [DOI] [PubMed] [Google Scholar]
- Marshall V. D., Sokatch J. R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972 Jun;110(3):1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit F. H., Reed L. J. Alpha-keto acid dehydrogenase complexes. 8. Comparison of dihydrolipoyl dehydrogenases from pyruvate and alpha-ketoglutarate dehydrogenase complexes of Escherichia coli. Proc Natl Acad Sci U S A. 1967 Sep;58(3):1126–1130. doi: 10.1073/pnas.58.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit F. H., Yeaman S. J., Reed L. J. Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REED L. J., KOIKE M., LEVITCH M. E., LEACH F. R. Studies on the nature and reactions of protein-bound lipoic acid. J Biol Chem. 1958 May;232(1):143–158. [PubMed] [Google Scholar]
- Sakurai Y., Fekuyoshi Y., Hamada M., Hayakawa T., Koike M. Mammalian alpha-keto acid dehydrogenase complexes. VI. Nature of the multiple forms of pig heart lipoamide dehydrogenase. J Biol Chem. 1970 Sep 10;245(17):4453–4462. [PubMed] [Google Scholar]
- Scouten W. H., McManus I. R. Microbial lipoamide dehydrogenase. Purification and some characteristics of the enzyme derived from selected microorganisms. Biochim Biophys Acta. 1971 Feb 10;227(2):248–263. doi: 10.1016/0005-2744(71)90058-1. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Roberts C. M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser J., Strating M. Separation of lipoamide dehydrogenase isoenzymes by affinity chromatography. Biochim Biophys Acta. 1975 Mar 28;384(1):69–80. doi: 10.1016/0005-2744(75)90096-0. [DOI] [PubMed] [Google Scholar]
- Weber K., Pringle J. R., Osborn M. Measurement of molecular weights by electrophoresis on SDS-acrylamide gel. Methods Enzymol. 1972;26:3–27. doi: 10.1016/s0076-6879(72)26003-7. [DOI] [PubMed] [Google Scholar]
- Wilson J. E. A comparative study of the multiple forms of pig heart lipoyl dehydrogenase. Arch Biochem Biophys. 1971 May;144(1):216–223. doi: 10.1016/0003-9861(71)90471-1. [DOI] [PubMed] [Google Scholar]
- v Muiswinkel-Voetberg H., Veeger C. Conformational studies on lipoamide dehydrogenase from pig heart. 4. The binding of NAD + to non-equivalent sites. Eur J Biochem. 1973 Mar 1;33(2):285–291. doi: 10.1111/j.1432-1033.1973.tb02682.x. [DOI] [PubMed] [Google Scholar]




