Abstract
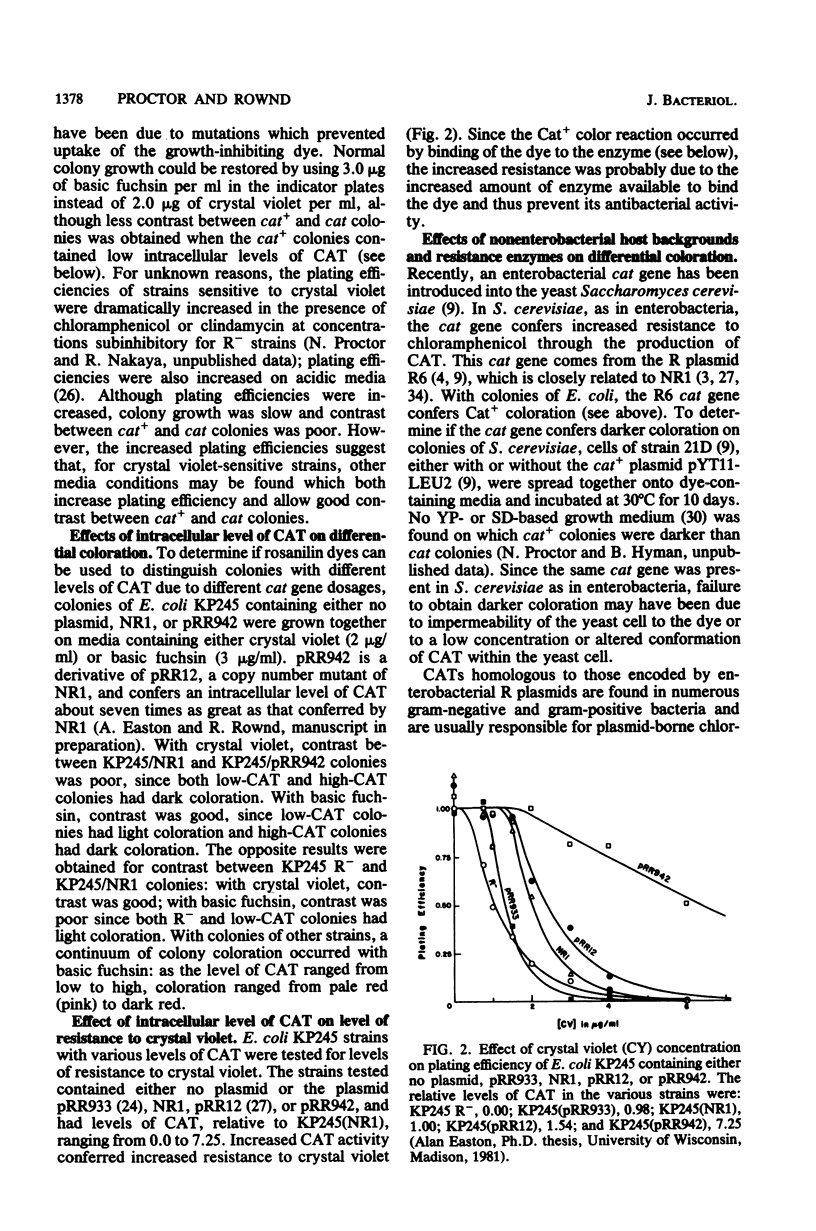
Rosanilin dyes such as crystal violet and basic fuchsin have been used as indicator dyes in solid growth medium for chloramphenicol-resistant enterobacterial colonies containing the enterobacterial resistance enzyme chloramphenicol acetyltransferase (CAT). On certain media containing rosanilins, cells containing CAT formed darker colonies than cells not containing CAT. Contrast was affected by the types and concentrations of complex nutrients, sugars salts, and rosanilin dyes present. When crystal violet was used as the indicator dye, contrast could not be obtained for strains whose growth was partially inhibited by crystal violet. Contrast could not be obtained between yeast colonies with and without the enterobacterial resistance enzyme, between Bacillus subtilis colonies with and without the staphylococcal resistance enzyme, or between enterobacterial colonies with and without the staphylococcal resistance enzyme. The darker coloration of enterobacterial colonies with the enterobacterial enzyme was due to the binding of dye to enzyme. Rosanilin dues have been used to score resistance phenotypes by colony color, to detect chloramphenicol-sensitive sectors in chloramphenicol-resistant colonies, and to screen for occasional chloramphenicol-sensitive cells in a resistant population during cloning by insertional inactivation of the chloramphenicol resistance gene.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Barton C. R., Warren R. L., Jezo P., Easton A. M., Rownd R. H. Sa/I restriction endonuclease maps of FII incompatibility group R plasmids. Plasmid. 1979 Jan;2(1):150–154. doi: 10.1016/0147-619x(79)90013-1. [DOI] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Cohen J. D., Eccleshall T. R., Needleman R. B., Federoff H., Buchferer B. A., Marmur J. Functional expression in yeast of the Escherichia coli plasmid gene coding for chloramphenicol acetyltransferase. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1078–1082. doi: 10.1073/pnas.77.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. DNA cloning in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1433–1436. doi: 10.1073/pnas.75.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Dubnau D. Construction and properties of chimeric plasmids in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1428–1432. doi: 10.1073/pnas.75.3.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T., Shivakumar A. G., Dubnau D. Characterization of chimeric plasmid cloning vehicles in Bacillus subtilis. J Bacteriol. 1980 Jan;141(1):246–253. doi: 10.1128/jb.141.1.246-253.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood J., Smith D. H. Catabolite repression of chloramphenicol acetyl transferase synthesis in E. coli K12. Biochem Biophys Res Commun. 1971 Jan 8;42(1):57–62. doi: 10.1016/0006-291x(71)90361-5. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Moats W. A., Maddox S. E., Jr Effect of pH on the antimicrobial activity of some triphenylmethane dyes. Can J Microbiol. 1978 Jun;24(6):658–661. doi: 10.1139/m78-110. [DOI] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Izaki K., Takahashi H. Some properties of chloramphenicol acetyltransferase, with particular reference to the mechanism of inhibition by basic triphenylmethane dyes. J Biochem. 1974 Nov;76(5):1009–1019. [PubMed] [Google Scholar]
- Tanaka H., Kudo O., Sato K., Izaki K., Takahashi H. Inhibition of chloramphenicol O-acetyltransferase of Escherichia coli by basic triphenylmethane dyes. J Antibiot (Tokyo) 1971 May;24(5):324–325. doi: 10.7164/antibiotics.24.324. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Ogata Y. Genetic stability of various resistance factors in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1970 May;102(2):363–368. doi: 10.1128/jb.102.2.363-368.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winshell E., Shaw W. V. Kinetics of induction and purification of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1969 Jun;98(3):1248–1257. doi: 10.1128/jb.98.3.1248-1257.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidenzaig Y., Shaw W. V. Affinity and hydrophobic chromatography of three variants of chloramphenicol acetyltransferases specified by R factors in Escherichia coli. FEBS Lett. 1976 Mar 1;62(3):266–271. doi: 10.1016/0014-5793(76)80072-5. [DOI] [PubMed] [Google Scholar]



