Abstract
RecA-assisted restriction endonuclease (RARE) cleavage is an “Achilles’ heel” approach to restriction mapping whereby a RecA-protein–oligodeoxynucleotide complex protects an individual restriction site from methylation, thus limiting subsequent digestion to a single, predetermined site. We have used RARE cleavage to cut yeast artificial chromosomes (YACs) at specific EcoRI sites located within or adjacent to sequence-tagged sites (STSs). Each cleavage reaction produces two YAC fragments whose sizes are a direct measure of the position of the STS in the YAC. In this fashion, we have positioned 45 STSs within a contig of 19 independent YACs and constructed a detailed RARE-cleavage map that represents 8.4 Mbp of human chromosome 6p21.3–22. By comparing maps of overlapping YACs, we were able to detect seven internal deletions that ranged from ≈75 kbp to ≈1 Mbp in size. Thirteen pairs of EcoRI sites were targeted for double RARE cleavage in uncloned total human DNA. The excised fragments, up to 2 Mbp in size, were resolved by pulsed-field gel electrophoresis and were detected by hybridization. In general, the genomic RARE-cleavage results support the YAC-based map. In one case, the distance in uncloned DNA between the two terminal EcoRI sites of a YAC insert was ≈1 Mbp larger than the YAC itself, indicating a major deletion. The general concept of RARE-cleavage mapping as well as its applications and limitations are discussed.
Clone-based physical genome mapping involves assembling arrays of cloned DNA fragments and constructing a composite map to represent the linear order and position of mapping landmarks along the genome. The use of high capacity cloning vectors has allowed the building of extensive contigs of overlapping clones. Maps have been constructed based on yeast artificial chromosomes (YACs), the clones with the largest inserts (1), which provide almost contiguous coverage along entire human chromosomes (2–8). The preferred mapping landmarks are sequence-tagged sites (STSs; ref. 9), short single-copy DNA sequences whose presence or absence in a given clone can be determined by PCR. Content mapping, the ordering of STSs and clone ends along the chromosome based on the STS content of each clone (10, 11), has become the dominant mapping strategy.
The use of STSs as mapping landmarks provides several practical and conceptual advantages that greatly enhance the utility of genome maps (9). Nonetheless, STS-content mapping merely assigns landmarks to ordered map intervals without actually specifying precise map coordinates. The only distance constraints on an STS-content map arise from the measured clone-insert sizes. Although it is possible, in principle, to calculate bounds on inter-STS spacings from content- and clone-size data alone (11), the prevalence of chimeric clones undermines such distance estimates because the size of the relevant portion of a chimeric YAC is typically unknown. Hence, the precise spacing of landmarks and the extent of clone overlaps remain generally uncertain.
A less obvious implication of the lack of reliable length calibration is that there is no strong test for clonal integrity. The power to detect aberrations such as internal deletions by STS content depends on the average distance between uniquely ordered STSs. Although at least two human chromosomes have been mapped at an ordered-marker density of 100 kbp or better (7, 8), the effective marker resolution of genome-wide maps has been estimated to be close to 1 Mbp (6), leaving even major cloning artifacts often undetected.
One approach to determine precise map coordinates for sequence landmarks is RecA-assisted restriction endonuclease (RARE) cleavage mapping. RARE cleavage is a technique that allows selective cleavage of a DNA molecule at a single, predetermined restriction site (12, 13). The targeted restriction site, usually an EcoRI site, is occupied by a RecA–oligodeoxynucleotide complex that recognizes unique sequence including the restriction site. This site thus is protected from enzymatic methylation and, provided complete methylation of the remaining sites, is the only restriction site that is susceptible to digestion by the cognate restriction endonuclease. By cleaving at an STS and measuring the size of the resulting fragments, one can determine the distance between the STS and either another cleavage site or the ends of the DNA molecule as one would do for a traditional restriction site.
This mapping approach previously has been applied to a short YAC contig (14). That study described an idealized situation in that the clone-inserts were relatively small and none of them was chimeric or grossly rearranged. Here, we report the application of RARE-cleavage mapping to a real-life contig of “MegaYACs” that has served in a positional cloning project (15). Furthermore, we describe RARE-cleavage experiments performed on uncloned total human DNA. By re-measuring physical distances in the underlying genome itself, we were able to confirm, and in some cases reject, the clone-based results.
MATERIALS AND METHODS
YACs and STSs have been described (15, 16). Total human DNA was prepared from Boleth, a lymphoblastoid cell line from which the Centre d’Etude du Polymorphisme Humain YAC library had been constructed (17). Yeast and human cells were embedded in 0.5% agarose blocks at 1 × 109 and 1.5 × 107 cells per ml, respectively, and high molecular mass DNA was purified as described (18, 19). Oligodeoxynucleotides for RARE cleavage were crude, desalted, commercial preparations, typically 35–45 nt in length. Examples are 5′-AGACTTACAATAAAGTAACCAAAATAGTTTAGAGAATTC and 5′-GATGTCTCATATATCTGTGAATTCTGAATGTGTGGCTCCTTT for cleavage at 950–15 and 950–5, respectively (the targeted EcoRI site is underlined). All other oligonucleotide sequences have been deposited in the Genome Data Base as annotations to their respective STS (see ref. 16 for GDB ID numbers). RARE cleavage was performed on DNA in 50-μl agarose plugs. Detailed protocols and tips for troubleshooting have been published elsewhere (18, 20). Pulsed-field gel (PFG) electrophoresis, DNA transfer, and hybridization were performed as described (14, 21). YAC blots were first probed with the 2.7-kbp BamHI–PvuII fragment of pBR322, which detects the left (centric) YAC-vector arm (1), and subsequently with all of pBR322 to detect both vector arms. Probe Y333 was a mixture of the PCR products of STSs sy935a8-L and sy947f6-R (16). Other hybridization probes including JFp19, a cloned genomic fragment ≈1 kbp in size, RFP7, a ≈1-kbp reverse transcriptase PCR product from the mRNA for the RET finger protein (22), and cDNAs 25 and 30, two ≈2-kbp cDNA clones from the hemochromatosis region (23), were a gift of John Feder (Progenitor, Inc.).
RESULTS
RARE-Cleavage Mapping Strategy.
To construct YAC-based RARE-cleavage maps, we isolated the ends of clone inserts and sequenced across the vector-insert junction. For each clone end we developed an STS as well as a RARE-cleavage assay specific for the EcoRI site at the very end of the cloned insert. The STS was used to identify overlapping clones by PCR. The precise extent of clone overlap was determined by RARE cleavage as illustrated in Fig. 1A. Microsatellite markers and other STSs were developed from subcloned EcoRI fragments, a simple strategy that allows subsequent mapping by RARE cleavage at a flanking EcoRI site (Fig. 1B). To validate the clone-based map, we carried out RARE-cleavage reactions on uncloned genomic DNA. Simultaneous cleavage at two EcoRI sites releases a fragment whose size indicates the distance in the genome between the two cleavage sites (Fig. 1C).
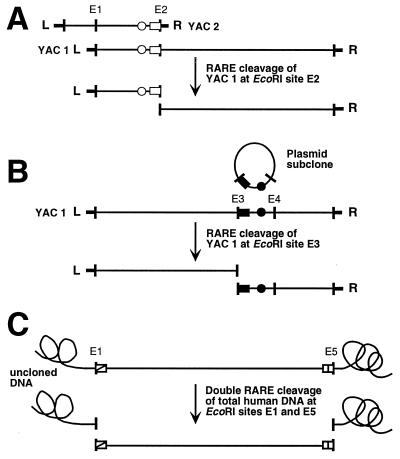
Figure 1.
RARE-cleavage-mapping strategy. (A) Determining the extent of overlap between two YACs. The right end of the insert cloned in YAC 2 is isolated, sequenced, and tagged with an STS (○). An oligodeoxynucleotide (□) containing the end sequence up to and including the EcoRI site at the vector-insert junction (E2) is used to split the overlapping clone YAC 1 at E2. The size of the left RARE-cleavage fragment is a direct measure of the extent of overlap. The clone overlap can also be measured by RARE cleavage of YAC 2 at E1. (B) Generating and placing a microsatellite marker. A plasmid-clone library is prepared from EcoRI-digested YAC-1 DNA and is screened by hybridization with a CA-repeat probe. Once a suitable marker has been found (•), the sequence information necessary for designing a RARE-cleavage oligonucleotide at one of the flanking EcoRI sites (■) can be obtained by simply sequencing the end of the plasmid insert the marker came from. RARE cleavage of YAC 1 at this EcoRI site (E3) produces two fragments whose sizes indicate the position of the microsatellite marker within the YAC. (C) Clone validation. Uncloned genomic DNA is subjected to double RARE cleavage by using two oligonucleotides for the two EcoRI sites defining the ends of the YAC-1 insert at once. The resulting fragment should have the same size as the EcoRI fragment cloned in YAC 1.
Mapping of YACs by RARE Cleavage at EcoRI Sites.
An example for the RARE-cleavage mapping of a YAC is shown in Fig. 2A. In this experiment, we mapped YAC y950h11, which is 1.5 Mbp in size, by RARE cleavage at 15 distinct EcoRI sites, 10 of which were next to microsatellite markers and five of which were defining the ends of overlapping clones. The YAC was split at each site separately. Duplicate reactions were set up that differed in their oligonucleotide concentration (12, 13, 20). The products were resolved on a PFG, and one of the two resulting fragments was detected by a hybridization probe specific for the left YAC-vector arm.
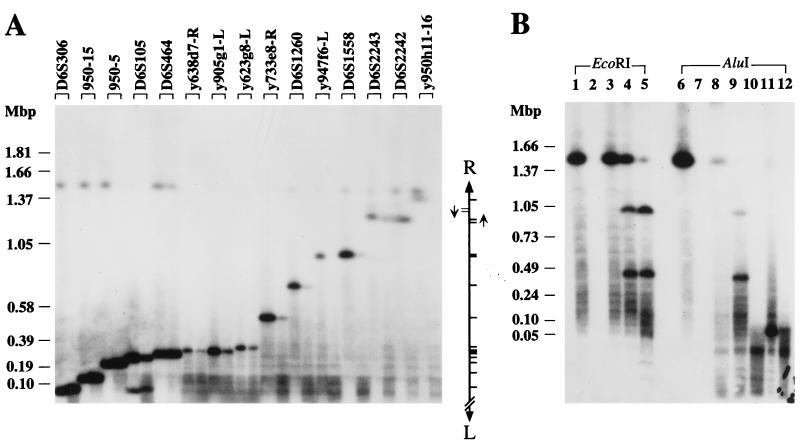
Figure 2.
Mapping of a YAC by RARE cleavage at 15 EcoRI and 2 AluI sites. (A) YAC y950h11 was subjected to RARE cleavage at 15 EcoRI sites. Two reactions were set up for each site containing 40 μg of RecA protein and either 1.65 μg (left lane of each pair) or 0.83 μg (right lane) of oligonucleotide. The mapping landmarks associated with the RARE-cleavage sites are listed along the top. The relative positions of the cleavage sites within the YAC are illustrated on the right. The two arrows denote the inverted duplication that includes D6S2243 and D6S2242. (B) RARE cleavage and control experiments were performed on YAC y950h11 by using either the EcoRI (lanes 1–5) or the AluI restriction/modification system (lanes 6–12). Samples in lanes 1 and 6 were only treated with methylase. Lanes 2 and 7 contain DNA that was completely digested with restriction endonuclease. Samples in lanes 3 and 8 were first methylated and then incubated in the presence of restriction enzyme. Samples in lanes 4 and 5 and 9–12 were complete RARE-cleavage reactions containing 40 μg of RecA protein and 1.32 μg (lanes 4, 9, and 11) or 0.66 μg (lanes 5, 10, and 12) of oligonucleotide. Lanes 4, 5, 9, and 10 contain RARE-cleavage reactions for D6S1558. Samples cleaved at an AluI site near y899 g1-L were loaded in lanes 11 and 12. PFG electrophoresis was performed at 6 V/cm by using a switching time ramped from 60 to 120 s. DNA molecules containing only the left (A) or either YAC-vector arm (B) were detected by hybridization. The positions of selected lambda-concatemer and H. wingei size markers are indicated.
For each RARE-cleavage reaction, a single major band (other than the constant band due to the presence of intact YAC) is visible whose size indicates the distance between the cleavage site—as well as the neighboring STS—and the left end of the YAC. In some lanes (D6S105, D6S2243, and D6S2242), a second, weaker band occurs as well. The most likely explanation for this phenomenon is cross-reaction of the oligonucleotide–RecA complex to a related sequence. D6S2243 and D6S2242 are known to reside in a segment that is duplicated elsewhere in the genome (16). The occurrence and pattern of the slightly larger secondary products after cleavage at these markers also suggest a local duplication within this YAC.
Hybridization with pBR322, which detects both YAC-vector arms, provided a second size measurement for each cleavage site. These data (not shown) confirmed the order and position of landmarks except for D6S2243 and D6S2242, for which the second data set suggested the reversed order. This finding is consistent with a head-to-head orientation of a local duplication encompassing these two markers, i.e., an inverted-repeat structure.
RARE Cleavage at AluI Sites.
In some cases, repetitive sequence immediately adjacent to an EcoRI site or the absence of EcoRI sites in a given STS prevented the development of a specific EcoRI–RARE-cleavage assay. We therefore explored the feasibility of using AluI instead, which has a more frequent 4-bp recognition site. The sequence of the RARE-cleavage oligonucleotide for D6S1558 contains both an AluI and an EcoRI site thereby providing an opportunity for a direct comparison of these two restriction/modification systems.
YAC y950h11 was RARE-cleaved with either EcoRI or AluI. Both left and right YAC fragments were visualized by hybridization (Fig. 2B). After cutting with EcoRI, the two products, ≈0.5 and ≈1 Mbp in size, gave rise to bands of similar intensity, and residual uncleaved YAC DNA was still present in the reaction mixture (Fig. 2B, lanes 4 and 5). This was not the case in the AluI reactions, where incomplete methylation lowered the yield of the larger fragment and where few full-length YAC molecules escaped digestion by the restriction enzyme. Furthermore, lowering the oligonucleotide concentration is much more detrimental in the AluI system (Fig. 2B, compare lanes 9 and 10). Still, at least at the higher oligonucleotide concentration, both products of the AluI digestion are clearly visible on the autoradiograph (Fig. 2B, lane 9).
A similar pattern emerged after RARE cleavage at an AluI site near y899g1-L, a clone end for which we could not use EcoRI because of adjoining repetitive sequence. Cleavage with AluI produced one very small (<100 kbp) and one very large (≈1.4 Mbp) product (Fig. 2B, lanes 11 and 12). Again, the latter is underrepresented in the reaction mixture, and the corresponding band (running just below the full-length YAC) is accordingly faint.
Detection of Clonal Instabilities by Redundant RARE-Cleavage Mapping.
The experiment shown in Fig. 3 is an example of how measuring the size of a RARE-cleavage fragment in two independent clones has led to the detection of an otherwise unrecognized internal deletion. Two YACs, y901a10 and y935a8, overlap such that the right end of y901a10 is within y935a8. Conversely, the left end of y935a8 falls within y901a10 (see Fig. 5 at map position 6–6.5 Mbp). Ideally, after RARE cleavage of each YAC at the EcoRI site defining the overlapping end of the other, the right-end fragment of y901a10 and the left-end fragment of y935a8 should be identical in size (except for the negligible size variation due to the different vector arms attached).
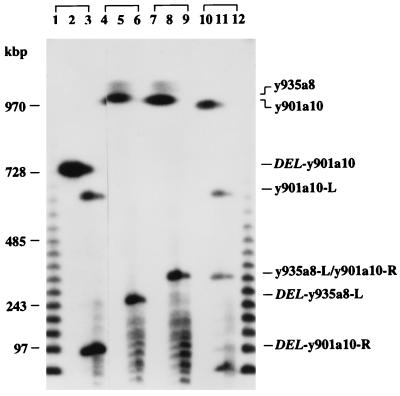
Figure 3.
Comparison of RARE-cleavage fragments from two overlapping YACs. Two yeast colonies containing YAC y901a10 (lanes 1–3 and 10–12) were analyzed by RARE cleavage at the EcoRI site defining the right end of YAC y935a8. Lanes 4–9 contain samples from two different DNA preparations of YAC y935a8 that were cleaved at the EcoRI site at the right end of YAC 901a10. The leftmost lane within each group of three contained undigested DNA. The cleavage reactions contained 40 μg RecA protein and either 1.32 μg (middle) or 0.66 μg (right lanes) of oligonucleotide. The products were separated on a PFG, which was run for 24 h at 6 V/cm, and a switching time ramped from 35 to 70 s. The hybridization probe was a mixture of pBR322 and λ DNA to visualize the λ-concatemer size markers. The positions of YACs and of RARE-cleavage fragments are indicated on the right. y935a8 is 2.3 Mbp in size. Both full length and deleted y935a8 as well as the right-vector-arm containing fragments thereof migrate in the unresolved limiting-mobility band. The right end-fragment of the deleted y901a10 clone (DEL-y901a10-R; lanes 2 and 3) is smaller than the right end-fragment from the intact version (y901a10-R; lanes 11 and 12), which in turn has the same size as the left end-fragment from y935a8 (y935a8-L; lanes 8 and 9). DEL-y935a8-L is a deleted fragment that was observed in one particular y935a8 preparation. The hybridization signal at the bottom of lanes 11 and 12 is due to a spill-over from the λ ladder.
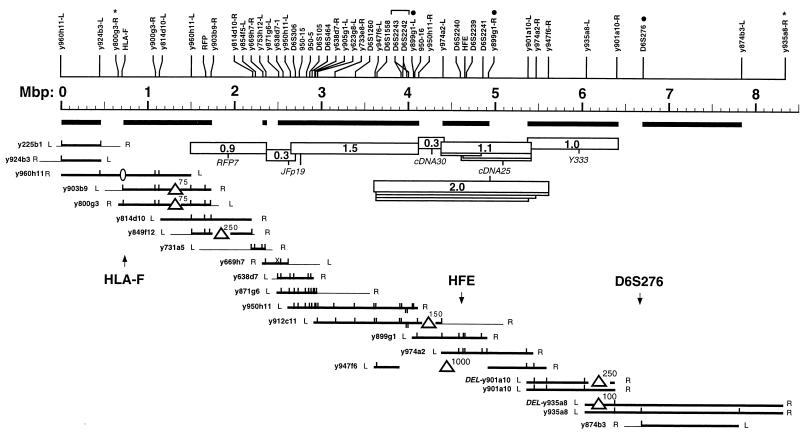
Figure 5.
RARE-cleavage map representing 8.4 Mbp of human chromosome 6p21.3–22. YACs are represented as horizontal lines whose lengths are proportional to their size. Thin lines indicate noncontiguous end pieces of chimeric clones or YAC ends whose chromosomal origin is unknown. L and R refer to the left (centric) and right (acentric) YAC-vector arm, respectively. Vertical ticks mark the positions of RARE-cleavage sites within each YAC. Maps of individual YACs were aligned graphically. A consensus map indicating the coordinates of 47 cleavage sites and the inferred positions of two clone ends for which no RARE-cleavage assay has been developed (∗) is shown at the top. The solid dots mark RARE-cleavage assays that require AluI. The bracket at ≈4 Mbp denotes the inverted repeat structure mentioned in the text. Bold horizontal lines are map segments whose apparent length is consistent, within experimental error, between at least two independent clones. Triangles indicate length inconsistencies that were interpreted as internal deletions with the approximate extent of the deletion indicated in kilobase pairs. The absence of HLA-F in YAC y960h11 (ellipse) is known from its STS content; the extent of this deletion is not known. Other missing ticks simply mean that these assays have not been performed on a particular YAC, except for the “X” at 2.5 Mbp in y669h7 where cleavage attempts have failed consistently. Open bars represent fragments generated by double RARE cleavage of uncloned total human DNA along with their respective hybridization probes with the approximate fragment sizes indicated in megabase pairs. Only the size of the largest fragment is given for sets of nested fragments (drawn as “stacks”).
The result was that the fragment clipped off y901a10 was much smaller (≈100 kbp; Fig. 3, lanes 2 and 3) than that from y935a8 (≈350 kbp; Fig. 3, lanes 8 and 9), suggesting that y901a10 had suffered an internal deletion. Indeed, after analyzing an additional six yeast colonies, we were able to find another version of YAC y901a10, ≈250 kbp larger than the first isolate. This longer YAC gave rise to a fragment of the expected size (Fig. 3, lanes 11 and 12). At least one preparation of y935a8 DNA, too, produced a smaller fragment (Fig. 3, lanes 5 and 6), indicating a stretch of human DNA that is unstable in the yeast host regardless of the clonal context. Of note, the deletion in YAC y901a10 was not obvious in the published STS-content map (16), which in fact lists the shorter, deleted version of y901a10.
Validation of YAC-Based Maps by RARE Cleavage of Uncloned Human DNA.
Although redundant RARE-cleavage analysis of overlapping YAC clones can detect many cloning artifacts and is therefore a powerful method for map validation, the ultimate test for clone integrity is to compare distances that were determined in cloned material with the corresponding measurements in uncloned DNA.
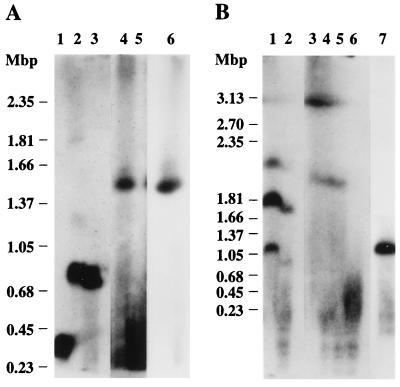
To test whether YAC y950h11 was a faithful replica of human DNA, we performed RARE cleavage of total human DNA at the two EcoRI sites that define the ends of the y950h11 clone insert. This double digest should release a fragment whose size indicates the true distance between the two YAC-insert ends. To avoid length inconsistencies caused by polymorphisms rather than cloning artifacts, the human DNA used for validation purposes was from the same human cell line from which the YAC library had been constructed. The reaction mixture was separated on a PFG along with YAC y950h11 itself, and the RARE-cleavage fragment was detected with an internal hybridization probe. The genomic RARE-cleavage fragment (Fig. 4A, lane 4) has essentially the same electrophoretic mobility as the YAC itself (Fig. 4A, lane 6). This finding excludes any gross clonal deletions and suggests that y950h11 propagates an authentic genome fragment, 1.5 Mbp in size.
Figure 4.
Test of YAC integrity by RARE cleavage of uncloned human DNA. Total human DNA was cut by RARE cleavage at two pairs of EcoRI sites that define the ends of two YACs. (A) Lanes 1–3 contain total human DNA that has been digested with SfiI (lane 1) or NotI (lane 2) or that has been first methylated with EcoRI methylase and then treated with both NotI and EcoRI restriction endonucleases (lane 3). The RARE-cleavage reactions (lanes 4 and 5) contained 80 μg of RecA protein and 1.32 μg or 0.66 μg of each of the two oligonucleotides specific for the insert ends of y950h11. Lane 6 contains y950h11 DNA. The PFG was run for 28 h at 6 V/cm with a reorientation angle of 120° and 75- to 150-s switching times. (B) Lane 1 contains NotI-digested total human DNA. The sample in lane 2 was first methylated with EcoRI methylase and subsequently exposed to both EcoRI and NotI restriction endonucleases. The RARE-cleavage reactions (lanes 3–6) contained 80 μg of RecA protein and 2.64, 1.98, 1.32, or 0.66 μg of each of the two oligonucleotides directed at the terminal EcoRI sites of y947f6. Lane 7 contains y947f6 DNA. The gel was run for 60 h at 3 V/cm, 106° with 300–600 s switching time. Each gel was blotted to a nylon membrane that was cut between the human and yeast-DNA containing lanes. Fragments produced by digestion of human DNA were detected by hybridization with the JFp19 (A) or the cDNA 25 probe (B). Autoradiography was for 2 d. The complete digests serve as positive controls for the sensitivity of hybridization and degree of methylation (20). The multiple bands in the NotI digest in B may be due to partial digestion, partial methylation, or cross-hybridization. The 3.13-Mbp size marker in B runs at the limiting mobility where the high local DNA concentration may cause unspecific hybridization. The yield of RARE-cleavage product decreases at lower oligonucleotide concentrations, and no product band is visible at the lowest concentration (lane 5 in A, lane 6 in B). This effect is due to unspecific binding to DNA of excess RecA protein leading to incomplete methylation and unspecific cleavage. The YAC-containing filter strips were probed with pBR322 and were exposed for less than 5 h to match the intensity of the bands in lanes containing human DNA.
We performed the same analysis on YAC y947f6, which carries a 1-Mbp insert. y947f6, however, did not pass this test; the corresponding genomic fragment was about twice as long (Fig. 4B, compare lanes 3–5 with lane 7). This clone evidently has lost ≈1 Mbp of DNA. A distance in the genome of 2 Mbp between the two YAC termini is consistent with the RARE-cleavage map based on other YAC clones from this region, which predicts a spacing of 2.0 Mbp (see Fig. 5).
We also tested the 1-Mbp version of YAC y901a10 (see above) and another clone, y974a2, which holds a cloned EcoRI fragment of ≈1.1 Mbp in size. In both cases, the distance in uncloned DNA between the two terminal EcoRI sites was found to be the same as in the YAC itself (data not shown). In a similar fashion, we measured the distance between another 10 pairs of EcoRI sites (data not shown). All 13 pairs of EcoRI sites tested on total human DNA using five different hybridization probes gave rise to specific bands that were clearly visible after 1-day autoradiography.
RARE-Cleavage Map.
A length-calibrated YAC-contig and STS map is shown in Fig. 5. This map comprises 19 independent YACs and 47 RARE-cleavage sites. Together, the YAC clones represent 8.4 Mbp from the short arm of human chromosome 6 and cover the region between the class I region at the distal end of the major histocompatibility complex in 6p21.3 (see HLA-F at map position 0.7 Mbp) and D6S276 (at 6.7 Mbp) in 6p22.
Map intervals that account for a total of 5.8 Mbp displayed no obvious clone-to-clone variation. Five intervals were notably different when compared among at least two independent clones. In these cases, the larger distance was assumed to be the correct one, and the presumed deletions, ranging from ≈75 kbp to ≈1 Mbp in size, are indicated. The genomic RARE-cleavage map, which is based on the analysis of uncloned total human DNA, spans 4.9 Mbp (between 1.5 and 6.4 on the Mbp scale) and is in excellent agreement with the YAC-based map.
This map was constructed for the positional cloning of a gene involved in hereditary hemochromatosis (15). The hemochromatosis gene, now termed HFE, is located at map position 4.6, precisely within the one Mbp of human DNA missing in YAC y947f6. This YAC was the only clone listed in the public YAC map as covering this region, and the gap in clone coverage probably has hampered the hunt for the hemochromatosis gene.
DISCUSSION
We have used RARE cleavage at individual restriction sites within or adjacent to STSs as a means to construct and validate YAC-based physical maps. This approach combines some of the virtues of two classes of mapping landmarks, STSs and restriction sites. The resulting RARE-cleavage map still is formatted with STSs as landmarks. Unlike in an STS-content map, however, the STS positions are fixed, and spacings between the STSs are defined.
This distance calibration not only increases the information content but is also beneficial for map validation as length consistency is a much stronger test for clonal integrity than consistent STS content alone. Furthermore, by using the very same landmarks in uncloned DNA, the clone-based map can be compared with a RARE-cleavage map of the underlying genomic DNA, thereby providing additional validation.
The map precision and hence the power to detect clonal abnormalities is only limited by one’s ability to accurately size and compare DNA fragments. In practice, size measurements on PFGs are often compromised by gel-smiling (or frowning) effects as well as lane-to-lane differences in DNA concentration that affect the electrophoretic mobility. For example, the high DNA concentration of complex DNA samples such as total human DNA may lead to a systematic overestimation of fragment sizes when the mobility is compared with that of DNA size markers or yeast chromosomes of lower concentration. Overall, we estimate the experimental error in raw sizing data to be ≈10% of a given fragment length.
Furthermore, it should be noted that we are using an indirect end-label strategy for YAC mapping in which distances between cleavage sites are calculated by subtraction of two measured fragment sizes. The inferred spacings can be inaccurate, particularly for closely spaced sites, thereby limiting the power to recognize subtle clone-to-clone variations. The smallest deviation that was indicated in Fig. 5 as an inconsistency was ≈75 kbp in a 1-Mbp YAC. Discrepancies of <50 kbp were not considered significant. More subtle cloning artifacts may be revealed by comparing fragments side-by-side on the same gel. However, such a direct comparison requires either an arrangement similar to that in the experiment shown in Fig. 3 or RARE cleavage at two restriction sites at once.
These limitations in detecting subtle length deviations notwithstanding, the ability to compare distances determined in cloned DNA directly to the corresponding measurements in uncloned material is perhaps the most significant advantage of RARE cleavage over conventional restriction digestion. Restriction mapping is affected much more by factors such as differences in methylation at restriction sites in cloned vs. uncloned DNA, CpG islands, lack of suitable restriction sites, as well as the presence of noncontiguous DNA in chimeric clones, all of which may complicate the analysis and often preclude a meaningful comparison of conventional restriction maps of cloned and uncloned DNA.
RARE-cleaved yeast DNA usually gives rise to discrete bands when separated on a PFG. In contrast, more complex DNA such as total human DNA typically produces a background smear of unspecific cleavage products that is comparable in intensity to a complete NotI digest (see for example figure 16 in ref. 20). Given this high background and the relatively low yield of specific product, we regard RARE cleavage of uncloned complex DNA more as an analytical tool than a preparative technique.
Currently, by far the best restriction/modification system for RARE cleavage is EcoRI, for which both methylase and endonuclease are commercially available at sufficient purity and modest price. One caveat is that the vast majority of existing STSs do not harbor an EcoRI site. In our experience, it is a relatively minor effort to generate STSs in a way that ensures the presence of a neighboring EcoRI site for RARE cleavage, and making new RARE-cleavable STSs is generally more practical than resorting to other restriction/modification systems such as M.HhaII/HinfI (13), HhaI (14) or AluI. Unavailability or high costs of commercial methylases, difficulties achieving near-complete methylation of agarose-embedded DNA, and traces of nuclease activity in some enzyme preparations are the main obstacles to implementing these alternative systems for routine mapping.
Despite its conceptual appeal, RARE cleavage has not been widely used for physical mapping purposes. This may be in part due to the complexity of the procedure, which requires three consecutive enzymatic reactions. Another reason may be the relative inconvenience of using agarose beads or rods, the DNA format prescribed in early RARE-cleavage protocols (12, 13, 24).
In our hands, RARE-cleavage mapping of a YAC is a routine experiment. We use a protocol that has been optimized for DNA in standard agarose plugs (18). Our procedure does not require specialized hybridization probes nor does it require more than one specific cut per molecule. In contrast to conventional restriction digests, RARE cleavage is almost never complete. Consequently, single-cleavage reactions are much more robust than experiments that require simultaneous cleavage at two different sites on the same DNA molecule. We find that even marginal RARE-cleavage assays or suboptimal reaction conditions generally yield clearly interpretable results after single RARE cleavage of DNA samples of moderate complexity such as the DNA of YAC-containing yeast strains.
RARE-cleavage mapping of uncloned total human DNA is less forgiving. Unless the cleavage site is near the end of a chromosome, genomic RARE cleavage requires two cuts (which lowers the yield significantly), a dedicated hybridization probe, and sufficient detection sensitivity to visualize the excised fragment in the complex sample. Nonetheless, we and others (12, 19, 24, 25) have demonstrated the feasibility of such experiments with a high success rate even for fragments that exceed 1 Mbp in size.
Acknowledgments
We thank S. Iadonato for the original in-plug RARE-cleavage protocol, J. Feder for hybridization probes, W. Thomas for CA repeat markers, D. Ruddy for sequencing of clone ends, E. Morikang for culturing the Boleth lymphoblastoid cell line, and R. Wolff and M. Ellis for discussion and critical reading of the manuscript.
ABBREVIATIONS
- YAC
yeast artificial chromosome
- STS
sequence-tagged site
- RARE
recA-assisted restriction endonuclease
- PFG
pulsed-field gel
Footnotes
Formerly Mercator Genetics, Inc.
References
- 1.Burke D T, Carle G F, Olson M V. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 2.Chumakov I, Rigault P, Guillou S, Ougen P, Billaut A, Guasconi G, Gervy P, LeGall I, Soularue P, Grinas L, et al. Nature (London) 1992;359:380–387. doi: 10.1038/359380a0. [DOI] [PubMed] [Google Scholar]
- 3.Chumakov, I. M., Rigault, P., Le Gall, I., Bellanné-Chantelot, C., Billault, A., Guillou, S., Soularue, P., Guasconi, G., Poullier, E., Gross, I., et al. (1995) Nature (London) 377, Suppl., 175–183. [DOI] [PubMed]
- 4.Foote S, Vollrath D, Hilton A, Page D C. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- 5.Cohen D, Chumakov I, Weissenbach J. Nature (London) 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- 6.Hudson T J, Stein L D, Gerety S S, Ma J, Castle A B, Silva J, Slonim D K, Baptista R, Kruglyak L, Xu S-H, et al. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- 7.Nagaraja R, MacMillan S, Kere J, Jones C, Griffin S, Schmatz M, Terrell J, Shomaker M, Jermak C, Hott C, et al. Genome Res. 1997;7:210–222. doi: 10.1101/gr.7.3.210. [DOI] [PubMed] [Google Scholar]
- 8.Bouffard G G, Idol J R, Braden V V, Iyer L M, Cunningham A F, Weintraub L A, Touchman J W, Mohr-Tidwell R M, Peluso D C, Fulton R S, et al. Genome Res. 1997;7:673–692. doi: 10.1101/gr.7.7.673. [DOI] [PubMed] [Google Scholar]
- 9.Olson M, Hood L, Cantor C, Botstein D. Science. 1989;245:1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- 10.Green E D, Olson M V. Science. 1990;250:94–98. doi: 10.1126/science.2218515. [DOI] [PubMed] [Google Scholar]
- 11.Green E D, Green P. PCR Methods Appl. 1991;1:77–90. doi: 10.1101/gr.1.2.77. [DOI] [PubMed] [Google Scholar]
- 12.Ferrin L J, Camerini-Otero R D. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 13.Koob M, Burkiewicz A, Kur J, Szybalski W. Nucleic Acids Res. 1992;20:5831–5836. doi: 10.1093/nar/20.21.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnirke A, Iadonato S P, Kwok P-Y, Olson M V. Genomics. 1994;24:199–210. doi: 10.1006/geno.1994.1607. [DOI] [PubMed] [Google Scholar]
- 15.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Jr, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 16.Lauer P, Meyer N C, Prass C E, Starnes S M, Wolff R K, Gnirke A. Genome Res. 1997;7:457–470. doi: 10.1101/gr.7.5.457. [DOI] [PubMed] [Google Scholar]
- 17.Albertsen H M, Abderrahim H, Cann H M, Dausset J, Le Paslier D, Cohen D. Proc Natl Acad Sci USA. 1990;87:4256–4260. doi: 10.1073/pnas.87.11.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iadonato S P, Gnirke A. Methods Mol Biol. 1996;54:75–85. doi: 10.1385/0-89603-313-9:75. [DOI] [PubMed] [Google Scholar]
- 19.Gnirke A, Huxley C, Peterson K, Olson M V. Genomics. 1993;15:659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- 20.Riethman H, Birren B, Gnirke A. In: Genome Analysis: A Laboratory Manual. Birren B, Green E D, Klapholz S, Myers R M, Roskams J, editors. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1997. pp. 83–248. [Google Scholar]
- 21.Gnirke A, Huxley C. Somat Cell Mol Genet. 1991;17:573–580. doi: 10.1007/BF01233622. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi M, Inaguma Y, Hiai H, Hirose F. Mol Cell Biol. 1988;8:1853–1856. doi: 10.1128/mcb.8.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruddy D A, Lee V K, Kronmal G S, Mintier G A, Quintana L, Domingo R, Jr, Meyer N C, Irrinki A, McClelland E E, Fullan A, et al. Genome Res. 1997;7:441–456. doi: 10.1101/gr.7.5.441. [DOI] [PubMed] [Google Scholar]
- 24.Ferrin L J, Camerini-Otero R D. Nat Genet. 1994;6:379–383. doi: 10.1038/ng0494-379. [DOI] [PubMed] [Google Scholar]
- 25.Macina R A, Barr F G, Galili N, Riethman H C. Genomics. 1995;26:1–8. doi: 10.1016/0888-7543(95)80076-x. [DOI] [PubMed] [Google Scholar]