Abstract
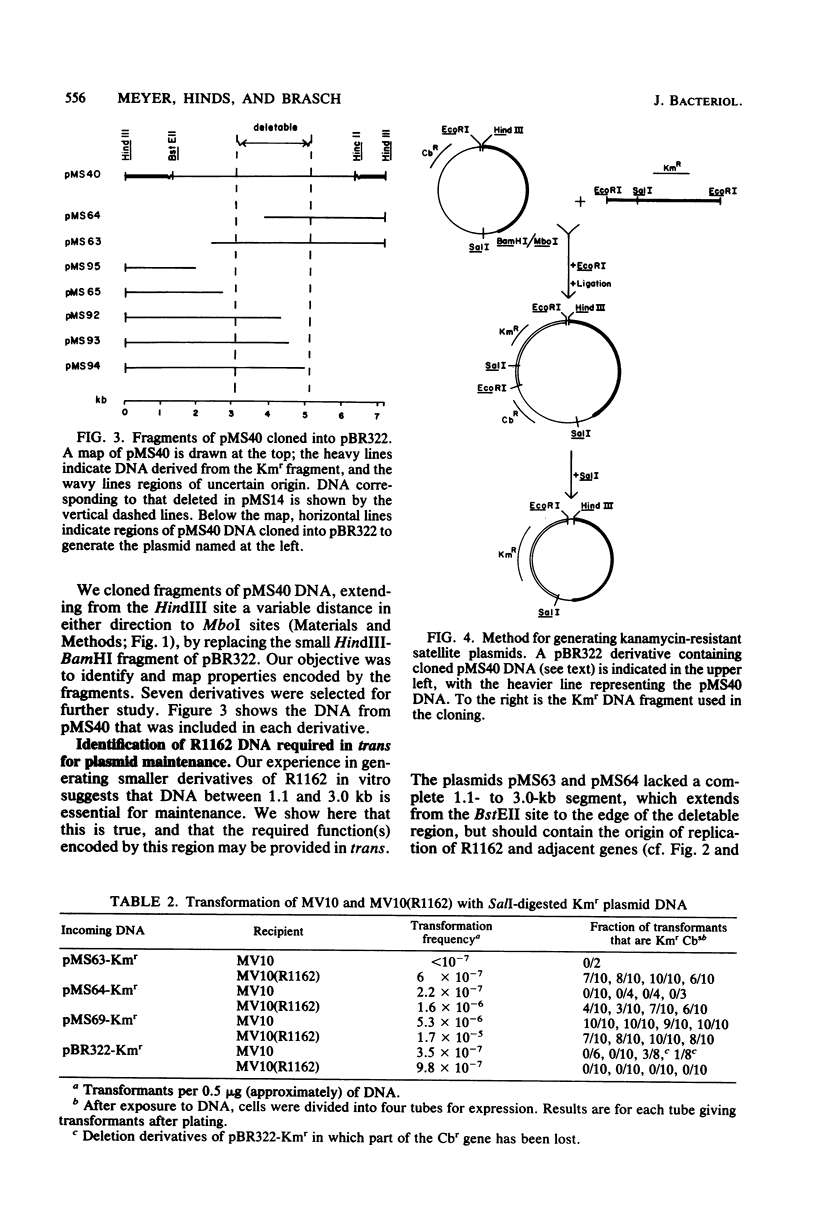
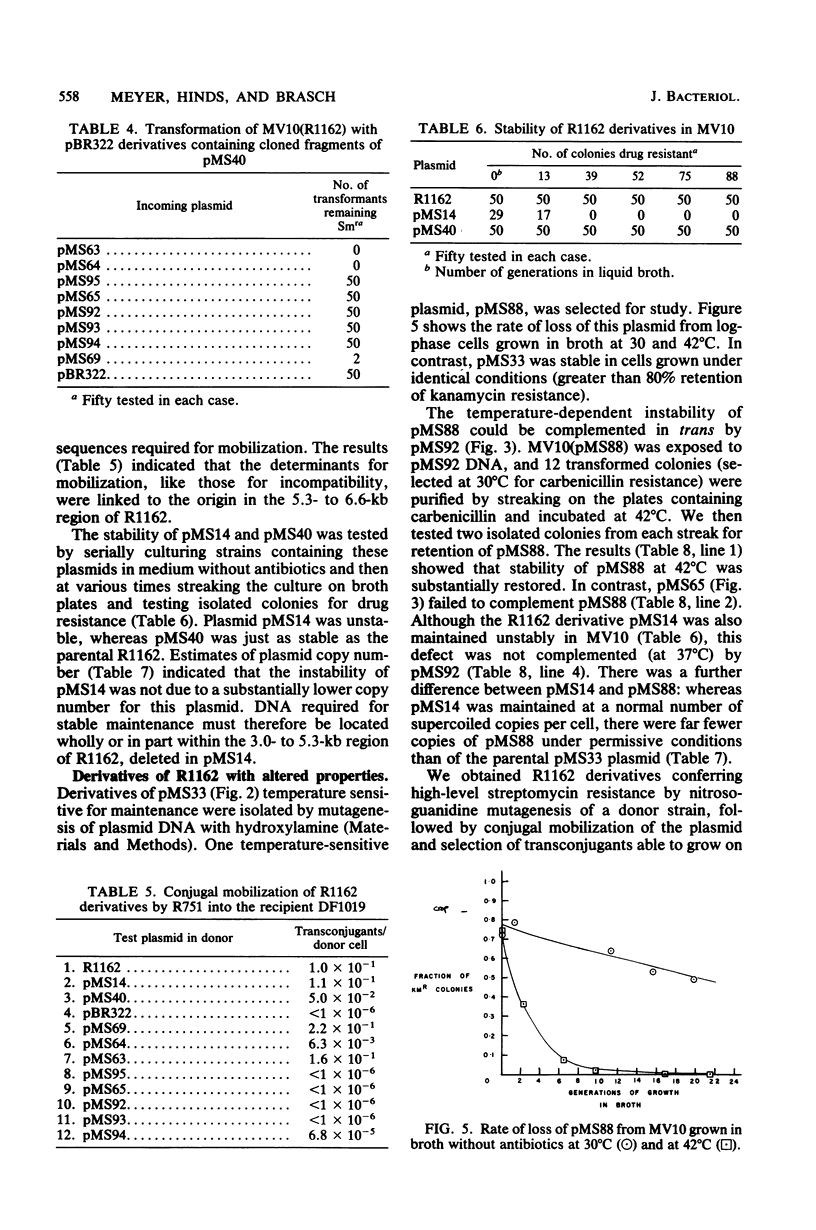
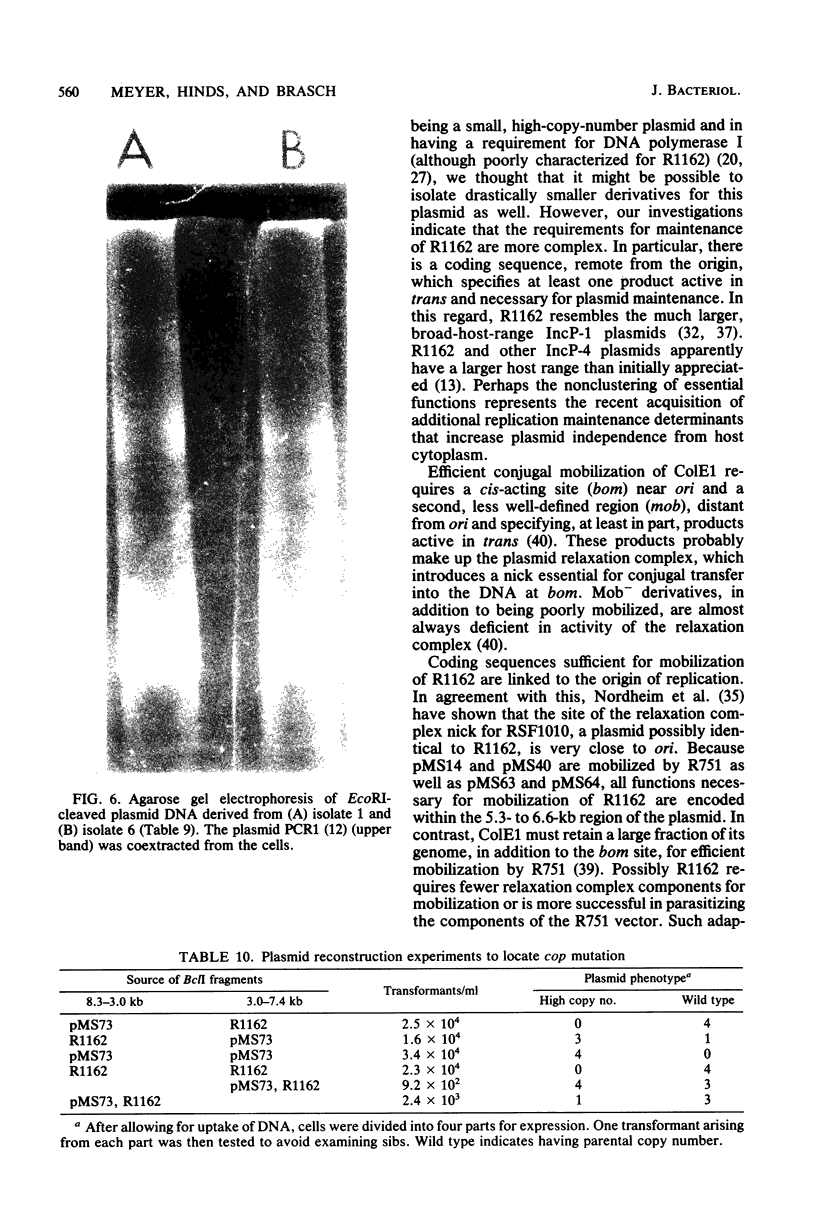
Regions of plasmid DNA encoding characteristic properties of the IncQ (P-4) group plasmid R1162 were identified by mutagenesis and in vitro cloning. Coding sequences sufficient for expression of incompatibility and efficient conjugal mobilization by plasmid R751 were found to be linked to the origin of DNA replication. In contrast, there was a region remote from the origin, and active in trans, that was required for plasmid maintenance. A derivative that was temperature sensitive for stability was isolated. The defect mapped at or near the region required for plasmid maintenance and resulted in far fewer copies of supercoiled plasmid DNA per cell under permissive conditions. A second region required for stability was also identified from the behavior of a deletion derivative of R1162, which did not, however, show an altered number of supercoiled plasmid DNA copies. Finally, a plasmid DNA mutation resulting in a substantially higher copy number was isolated. Plasmid reconstruction experiments suggested that the mutation was linked to the replicative origin.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Hershfield V., Helinski D. R. Gene cloning and containment properties of plasmid Col E1 and its derivatives. Science. 1977 Apr 8;196(4286):172–174. doi: 10.1126/science.322277. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- CAIRNS J. The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol. 1963 Mar;6:208–213. doi: 10.1016/s0022-2836(63)80070-4. [DOI] [PubMed] [Google Scholar]
- Coggins L. W., McCluskey E. Characterization of the ColE1-Km plasmids pCR1 and pCR11 by electron microscope and restriction endonuclease mapping. Plasmid. 1979 Jul;2(3):446–453. doi: 10.1016/0147-619x(79)90028-3. [DOI] [PubMed] [Google Scholar]
- Collins J., Yanofsky S., Helinski D. R. Altered DNA-protein relaxation complex in a replication mutant of plasmid ColE1. Mol Gen Genet. 1978 Nov 16;167(1):21–28. doi: 10.1007/BF00270317. [DOI] [PubMed] [Google Scholar]
- Covey C., Richardson D., Carbon J. A method for the deletion of restriction sites in bacterial plasmid deoxyribonucleic acid. Mol Gen Genet. 1976 May 7;145(2):155–158. doi: 10.1007/BF00269587. [DOI] [PubMed] [Google Scholar]
- David M., Tronchet M., Dénarié J. Transformation of Azotobacter vinelandii with plasmids RP4 (IncP-1 group) and RSF1010 (IncQ group). J Bacteriol. 1981 Jun;146(3):1154–1157. doi: 10.1128/jb.146.3.1154-1157.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D., Meyer R., Miller D. S., Helinski D. R. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1(1):107–119. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bonewald R. The use of plasmid R1162 and derivatives for gene cloning in the methanol-utilizing Pseudomonas AM1. Mol Gen Genet. 1980;178(2):375–380. doi: 10.1007/BF00270487. [DOI] [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Goebel W., Helinski D. R. Nicking activity of an endonuclease. I. Transfer ribonucleic acid complex of Escherichia coli. Biochemistry. 1970 Nov 24;9(24):4793–4801. doi: 10.1021/bi00826a025. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Kelley W. S. Effects of different alleles of the E. coli K12 pol A gene on the replication of non-transferring plasmids. Mol Gen Genet. 1976 Feb 2;143(3):311–318. doi: 10.1007/BF00269409. [DOI] [PubMed] [Google Scholar]
- Grinter N. J., Barth P. T. Characterization of SmSu plasmids by restriction endonuclease cleavage and compatibility testing. J Bacteriol. 1976 Oct;128(1):394–400. doi: 10.1128/jb.128.1.394-400.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick J. L., Allard R. W. Microgeographical Variation in Allozyme Frequencies in Avena barbata. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2100–2104. doi: 10.1073/pnas.69.8.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Sekiguchi M. Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine. J Bacteriol. 1976 Sep;127(3):1561–1563. doi: 10.1128/jb.127.3.1561-1563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobanputra R. S., Datta N. Trimethoprim R factors in enterobacteria from clinical specimens. J Med Microbiol. 1974 May;7(2):169–177. doi: 10.1099/00222615-7-2-169. [DOI] [PubMed] [Google Scholar]
- Kahn M., Helinski D. R. Construction of a novel plasmid-phage hybrid: use of the hybrid to demonstrate ColE1 DNA replication in vivo in the absence of a ColE1-specified protein. Proc Natl Acad Sci U S A. 1978 May;75(5):2200–2204. doi: 10.1073/pnas.75.5.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Activity of the replication terminus of plasmid R6K in hybrid replicons in Escherichia coli. J Mol Biol. 1978 Sep 25;124(3):425–441. doi: 10.1016/0022-2836(78)90180-8. [DOI] [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Construction of plasmid R6K derivatives in vitro: characterization of the R6K replication region. Plasmid. 1978 Sep;1(4):571–580. doi: 10.1016/0147-619x(78)90014-8. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. J. Expression of incompatibility by derivatives of the broad host-range inc P-1 plasmid RK2. Mol Gen Genet. 1979;177(1):155–161. doi: 10.1007/BF00267265. [DOI] [PubMed] [Google Scholar]
- Meyer R. J., Shapiro J. A. Genetic organization of the broad-host-range IncP-1 plasmid R751. J Bacteriol. 1980 Sep;143(3):1362–1373. doi: 10.1128/jb.143.3.1362-1373.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahari K. Deletion plasmids from transformants of Pseudomonas aeruginosa trp cells with the RSF1010-trp hybrid plasmid and high levels of enzyme activity from the gene on the plasmid. J Bacteriol. 1978 Oct;136(1):312–317. doi: 10.1128/jb.136.1.312-317.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N., Yamagishi H., Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene. Gene. 1978 Feb;3(1):39–51. doi: 10.1016/0378-1119(78)90006-9. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Hashimoto-Gotoh T., Timmis K. N. Location of two relaxation nick sites in R6K and single sites in pSC101 and RSF1010 close to origins of vegetative replication: implication for conjugal transfer of plasmid deoxyribonucleic acid. J Bacteriol. 1980 Dec;144(3):923–932. doi: 10.1128/jb.144.3.923-932.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. Nucleotide sequence of the region required for maintenance of colicin E1 plasmid. Mol Gen Genet. 1979 Oct 3;176(2):161–170. doi: 10.1007/BF00273210. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J Bacteriol. 1980 Jan;141(1):213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. J., Saul M. W., Sherratt D. J. ColE1 plasmid mobility: essential and conditional functions. Mol Gen Genet. 1979 Feb 16;170(1):103–107. doi: 10.1007/BF00268585. [DOI] [PubMed] [Google Scholar]
- Warren G. J., Twigg A. J., Sherratt D. J. ColE1 plasmid mobility and relaxation complex. Nature. 1978 Jul 20;274(5668):259–261. doi: 10.1038/274259a0. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]
- de Graaff J., Crosa J. H., Heffron F., Falkow S. Replication of the nonconjugative plasmid RSF1010 in Escherichia coli K-12. J Bacteriol. 1978 Jun;134(3):1117–1122. doi: 10.1128/jb.134.3.1117-1122.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



