Abstract
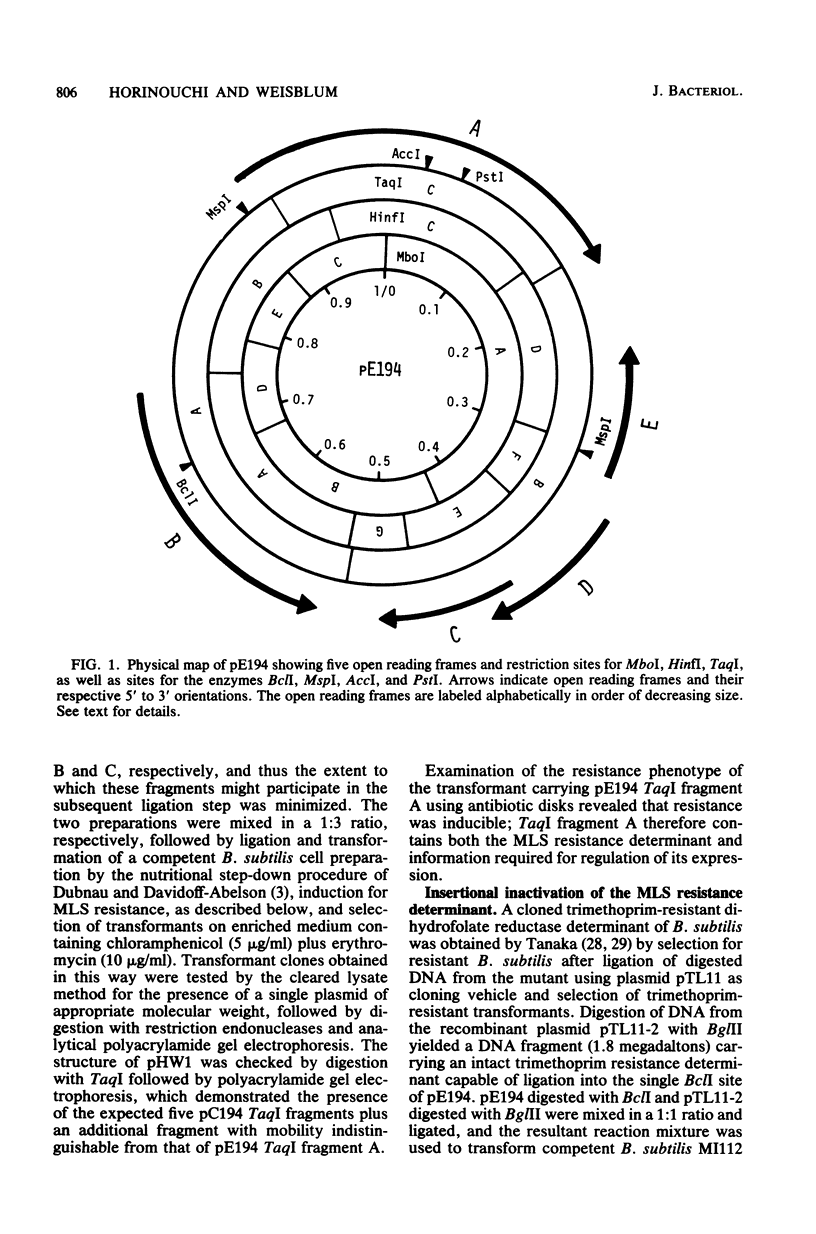
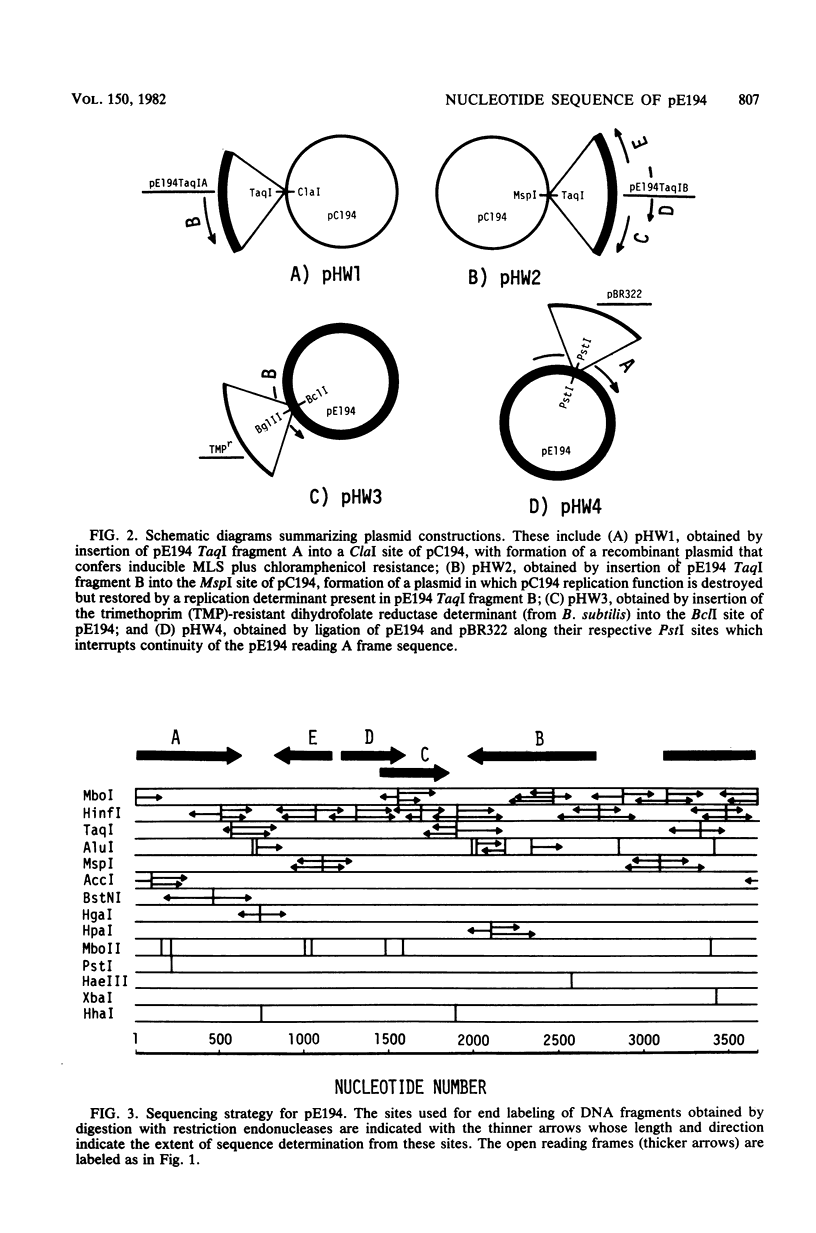
pE194 is a small plasmid (isolated originally in Staphylococcus aureus) which confers erythromycin-inducible resistance to macrolide, lincosamide, and streptogramin type B (MLS) antibiotics. The nucleotide sequence of pE194 contains 3,728 base pairs (bp), corresponding to a molecular mass of 2.4 million daltons. By means of site-specific cleavage with restriction endonucleases and cloning resultant fragments, determinants of the two major biological functions of p E194, i.e., inducible MLS resistance and replication, could be localized and assigned to specific sequences in the plasmid. Restriction endonuclease TaqI cut pE194 at three sites. TaqI fragment A (1,443 bp) contained the determinant for inducible MLS resistance, whereas TaqI fragment B (1,354 bp) contained a determinant necessary for plasmid replication. Regulatory mutations resulting in constitutive expression of MLS resistance mapped in TaqI fragment A, whereas a mutation associated with elevated plasmid copy number was mapped in TaqI fragment B. Also mapping in TaqI fragment B was a plasmid replication determinant comprising two sets of inverted complementary repeat sequences, one of which spanned 124 bp and was adjacent to a second smaller set which was rich in guanine and cytosine residues. pE194 contained six open reading frames which were theoretically capable of coding for proteins with maximum molecular masses as follows (in daltons): A, 48,300; B, 29,200; C, 14,000; D, 13,900; E, 12,600; and F, 2,700. Insertion of plasmid pBR322 into the single PstI site located in frame A of pE194 resulted in a composite plasmid which could replicate in both Bacillus subtilis and Escherichia coli, suggesting that an intact polypeptide A is dispensable for both replication of pE194 and for MLS resistance. Frame B specified inducible MLS resistance, whereas frame F specified the putative peptide associated with the proposed B determinant translational attenuator. The extent to which frames C, D, and E, all contained in TaqI fragment B, were translated into polypeptide products is not known; however, a base change in frame E was found in a comparison between the high-copy-number mutant, cop-6, and the wild-type strains.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Dubnau D. Construction and properties of chimeric plasmids in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1428–1432. doi: 10.1073/pnas.75.3.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T., Shivakumar A. G., Dubnau D. Characterization of chimeric plasmid cloning vehicles in Bacillus subtilis. J Bacteriol. 1980 Jan;141(1):246–253. doi: 10.1128/jb.141.1.246-253.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Uozumi T., Hoshino T., Ozaki A., Nakajima S., Beppu T., Arima K. Molecular cloning and in vitro transcription of Bacillus subtilis plasmid in Escherichia coli. Mol Gen Genet. 1977 Nov 29;157(2):175–182. doi: 10.1007/BF00267395. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. The control region for erythromycin resistance: free energy changes related to induction and mutation to constitutive expression. Mol Gen Genet. 1981;182(2):341–348. doi: 10.1007/BF00269681. [DOI] [PubMed] [Google Scholar]
- Iordanescu S., Surdeanu M., Della Latta P., Novick R. Incompatibility and molecular relationships between small Staphylococcal plasmids carrying the same resistance marker. Plasmid. 1978 Sep;1(4):468–479. doi: 10.1016/0147-619x(78)90005-7. [DOI] [PubMed] [Google Scholar]
- Iordănescu S., Surdeanu M. New incompatibility groups of Staphylococcus aureus plasmids. Plasmid. 1980 Nov;4(3):256–260. doi: 10.1016/0147-619x(80)90064-5. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Queen C. L., Wegman M. N. Computer analysis of nucleic acid regulatory sequences. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4401–4405. doi: 10.1073/pnas.74.10.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Dahlberg J. E., Weisblum B. Structure of an inducibly methylatable nucleotide sequence in 23S ribosomal ribonucleic acid from erythromycin-resistant Staphylococcus aureus. Biochemistry. 1973 Jan 30;12(3):457–460. doi: 10.1021/bi00727a015. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975 Mar;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Schaller H., Gray C., Herrmann K. Nucleotide sequence of an RNA polymerase binding site from the DNA of bacteriophage fd. Proc Natl Acad Sci U S A. 1975 Feb;72(2):737–741. doi: 10.1073/pnas.72.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Gryczan T. J., Kozlov Y. I., Dubnau D. Organization of the pE194 genome. Mol Gen Genet. 1980;179(2):241–252. doi: 10.1007/BF00425450. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Grandi G., Kozlov Y., Dubnau D. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3903–3907. doi: 10.1073/pnas.77.7.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Szalay A. A., Grohmann K., Sinsheimer R. L. Separation of the complementary strands of DNA fragments on polyacrylamide gels. Nucleic Acids Res. 1977;4(5):1569–1578. doi: 10.1093/nar/4.5.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawano N. Cloning vehicles for the homologous Bacillus subtilis host-vector system. Gene. 1980 Jul;10(2):131–136. doi: 10.1016/0378-1119(80)90130-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T. recE4-Independent recombination between homologous deoxyribonucleic acid segments of Bacillus subtilis plasmids. J Bacteriol. 1979 Sep;139(3):775–782. doi: 10.1128/jb.139.3.775-782.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Siddhikol C., Lai C. J., Demohn V. Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J Bacteriol. 1971 Jun;106(3):835–847. doi: 10.1128/jb.106.3.835-847.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]


