Abstract
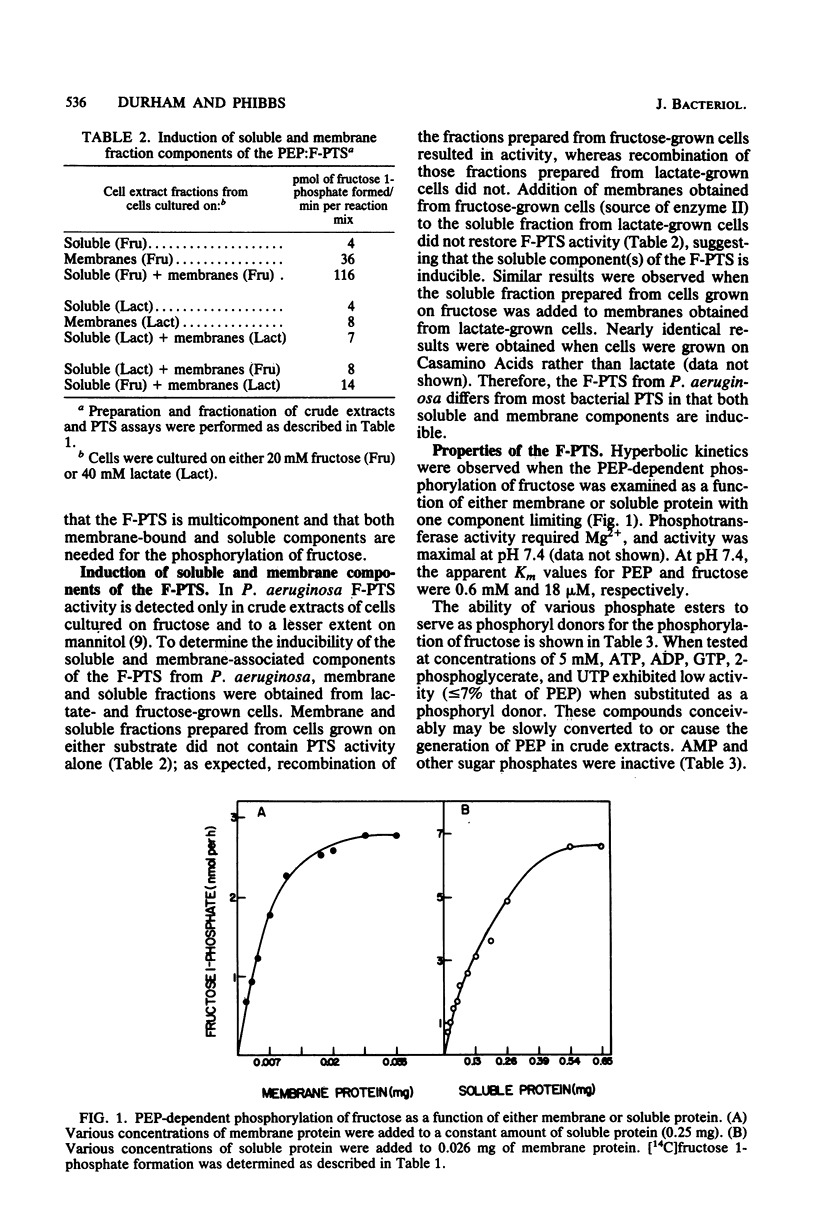
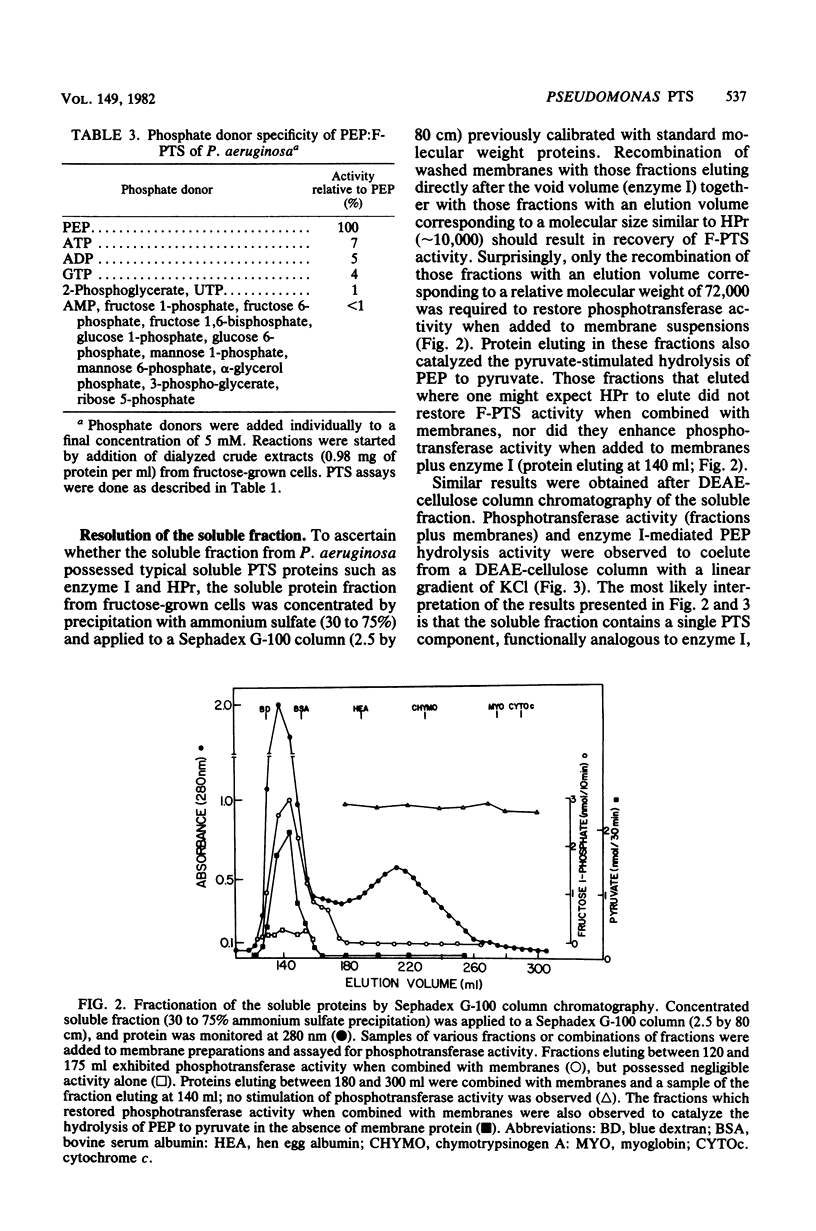
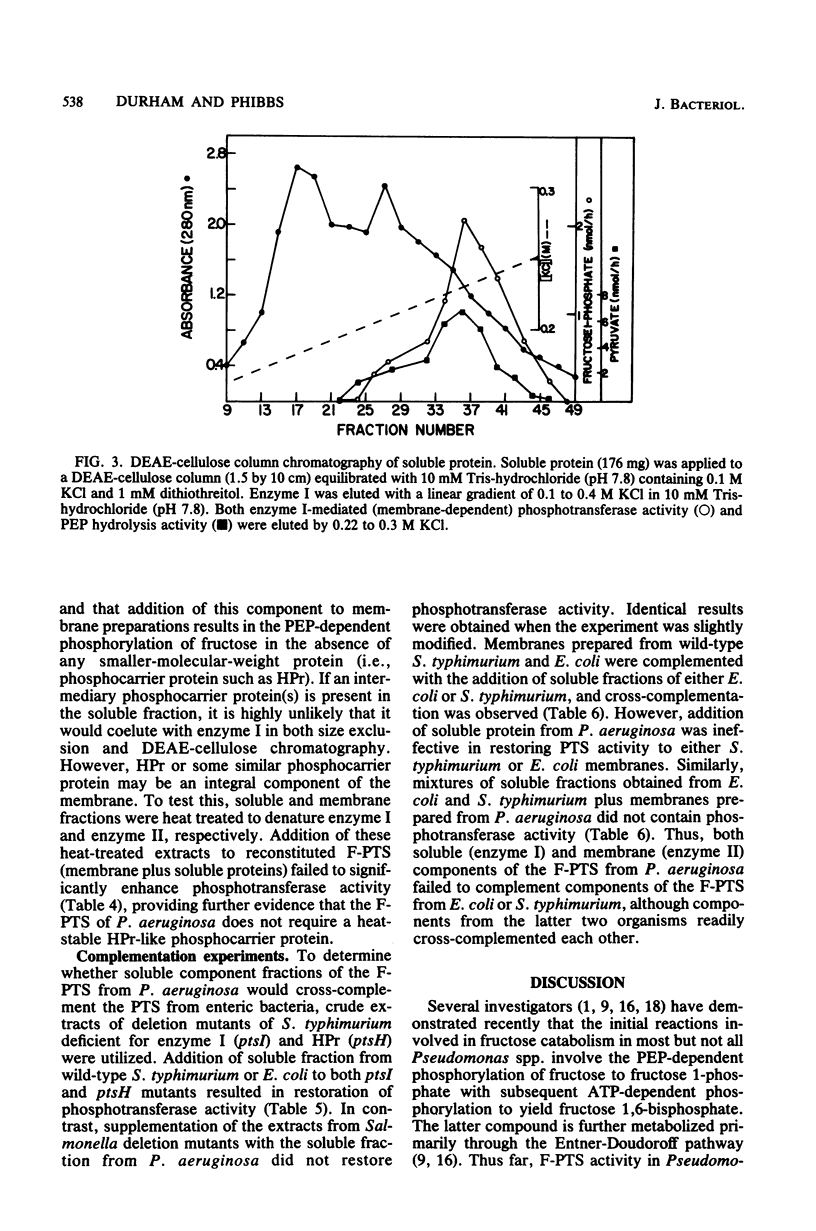
The initial reactions involved in the catabolism of fructose in Pseudomonas aeruginosa include the participation of a phosphoenolpyruvate:fructose 1-phosphotransferase system (F-PTS). Fractionation of crude extracts of fructose-grown cells revealed that both membrane-associated and soluble components were essential for F-PTS activity. Further resolution of the soluble fraction by both size exclusion and ion-exchange chromatography revealed the presence of only one component, functionally analogous to enzyme I. Enzyme I exhibited a relative molecular weight of 72,000, catalyzed the pyruvate-stimulated hydrolysis of phosphoenolpyruvate to pyruvate, and mediated the phosphorylation of fructose when combined with a source of enzyme II (washed membranes). No evidence for the requirement of a phosphate carrier protein, such as HPr, could be demonstrated. Thus, the F-PTS requires a minimum of two components, a soluble enzyme I and a membrane-associated enzyme II complex, and both were shown to be inducible. Reconstituted F-PTS activity was specific for phosphoenolpyruvate as a phosphate donor (Km, approximately -0.6 mM) and fructose as the sugar substrate (Km, approximately 18 microM). Components of the Pseudomonas F-PTS did not restore activity to extracts of deletion mutants of Salmonella typhimurium deficient in individual proteins of the PTS or to fractionated membrane and soluble components of the F-PTS of Escherichia coli. Similarly, membrane and soluble components of E. coli and S. typhimurium would not cross-complement the F-PTS components from P. aeruginosa.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumann P., Baumann L. Catabolism of D-fructose and D-ribose by Pseudomonas doudoroffii. I. Physiological studies and mutant analysis. Arch Microbiol. 1975 Nov 7;105(3):225–240. doi: 10.1007/BF00447141. [DOI] [PubMed] [Google Scholar]
- Dills S. S., Apperson A., Schmidt M. R., Saier M. H., Jr Carbohydrate transport in bacteria. Microbiol Rev. 1980 Sep;44(3):385–418. doi: 10.1128/mr.44.3.385-418.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüdig H., Hengstenberg W. The bacterial phosphoenolpyruvate dependent phosphotransferase system (PTS): solubilisation and kinetic parameters of the glucose-specific membrane bound enzyme II component of Streptococcus faecalis. FEBS Lett. 1980 May 19;114(1):103–106. doi: 10.1016/0014-5793(80)80869-6. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Kundig W., Roseman S. Sugar transport. II. Characterization of constitutive membrane-bound enzymes II of the Escherichia coli phosphotransferase system. J Biol Chem. 1971 Mar 10;246(5):1407–1418. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Perret J., Gay P. Kinetic study of a phosphoryl exchange reaction between fructose and fructose 1-phosphate catalyzed by the membrane-bound enzyme II of the phosphoenolpyruvate-fructose 1-phosphotransferase system of Bacillus subtilis. Eur J Biochem. 1979 Dec;102(1):237–246. doi: 10.1111/j.1432-1033.1979.tb06285.x. [DOI] [PubMed] [Google Scholar]
- Phibbs P. V., Jr, McCowen S. M., Feary T. W., Blevins W. T. Mannitol and fructose catabolic pathways of Pseudomonas aeruginosa carbohydrate-negative mutants and pleiotropic effects of certain enzyme deficiencies. J Bacteriol. 1978 Feb;133(2):717–728. doi: 10.1128/jb.133.2.717-728.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W., Roseman S. The bacterial phosphoenolpyruvate: sugar phosphotransferase system. Biochim Biophys Acta. 1976 Dec 14;457(3-4):213–257. doi: 10.1016/0304-4157(76)90001-0. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr Bacterial phosphoenolpyruvate: sugar phosphotransferase systems: structural, functional, and evolutionary interrelationships. Bacteriol Rev. 1977 Dec;41(4):856–871. doi: 10.1128/br.41.4.856-871.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr, Feucht B. U., Roseman S. Phosphoenolpyruvate-dependent fructose phosphorylation in photosynthetic bacteria. J Biol Chem. 1971 Dec 25;246(24):7819–7821. [PubMed] [Google Scholar]
- Saier M. H., Jr, Newman M. J., Rephaeli A. W. Properties of a phosphoenolpyruvate: mannitol phosphotransferase system in Spirochaeta aurantia. J Biol Chem. 1977 Dec 25;252(24):8890–8898. [PubMed] [Google Scholar]
- Saier M. H., Jr, Schmidt M. R., Lin P. Phosphoryl exchange reaction catalyzed by enzyme I of the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Kinetic characterization. J Biol Chem. 1980 Sep 25;255(18):8579–8584. [PubMed] [Google Scholar]
- Sawyer M. H., Baumann P., Baumann L., Berman S. M., Cánovas J. L., Berman R. H. Pathways of D-fructose catabolism in species of Pseudomonas. Arch Microbiol. 1977 Feb 4;112(1):49–55. doi: 10.1007/BF00446653. [DOI] [PubMed] [Google Scholar]
- Simoni R. D., Smith M. F., Roseman S. Resolution of a staphylococcal phosphotransferase system into four protein components and its relation to sugar transport. Biochem Biophys Res Commun. 1968 Jun 10;31(5):804–811. doi: 10.1016/0006-291x(68)90634-7. [DOI] [PubMed] [Google Scholar]
- Van Dijken J. P., Quayle J. R. Fructose metabolism in four Pseudomonas species. Arch Microbiol. 1977 Sep 28;114(3):281–286. doi: 10.1007/BF00446874. [DOI] [PubMed] [Google Scholar]


